Abstract
Trichothecene mycotoxins synthesized by Fusarium species are potent inhibitors of eukaryotic translation. They are encountered in both the environment and in food, posing a threat to human and animal health. They have diverse roles in the cell that are not limited to the inhibition of protein synthesis. To understand the trichothecene mechanism of action, we screened the yeast knockout library to identify genes whose deletion confers resistance to trichothecin (Tcin). The largest group of resistant strains affected mitochondrial function, suggesting a role for fully active mitochondria in trichothecene toxicity. Tcin inhibited mitochondrial translation in the wild-type strain to a greater extent than in the most resistant strains, implicating mitochondrial translation as a previously unrecognized site of action. The Tcin-resistant strains were cross-resistant to anisomycin and chloramphenicol, suggesting that Tcin targets the peptidyltransferase center of mitochondrial ribosomes. Tcin-induced cell death was partially rescued by mutants that regulate mitochondrial fusion and maintenance of the tubular morphology of mitochondria. Treatment of yeast cells with Tcin led to the fragmentation of the tubular mitochondrial network, supporting a role for Tcin in disruption of mitochondrial membrane morphology. These results provide genome-wide insight into the mode of action of trichothecene mycotoxins and uncover a critical role for mitochondrial translation and membrane maintenance in their toxicity.
Keywords: Fusarium head blight, ribosome, translation, deoxynivalenol, mycotoxin
The trichothecenes are toxic sesquiterpenoids produced by various Fusarium species. Fusarium head blight (FBH), also known as scab, caused by Fusarium graminearum, is a devastating disease that results in yield reductions and in the contamination of cereals with trichothecene mycotoxins, the most prominent being deoxynivalenol (DON) (1, 2). The trichothecenes are potent cytotoxins of eukaryotic cells and are a public health concern because contamination of human foods and animal feed with these toxins is a worldwide problem (1, 2). Toxic effects of trichothecenes include growth retardation, hemorrhagic lesions, reproductive disorders, immune dysfunction, vomiting, and reduced weight gain (3, 4). Trichothecenes all share a 12,13-epoxytrichothecene core structure and have been classified based on their substitution pattern of specific side groups. Trichothecenes that have a hydroxyl group, an ester group, or no side chain at all at C8 have been classified as Type A (T-2 toxin and diacetoxyscirpenol), while trichothecenes that have a ketone functional group at C8 are classified as Type B (DON, trichodermin, and trichothecin). Type C are characterized by a second epoxide function (crotocin), while type D trichothecenes contain a macrocyclic ring (verrucarin and satratoxins) (1, 2, 4). A mutation in the RPL3 gene, encoding ribosomal protein L3, referred to as tcm1 conferred resistance to trichodermin (5, 6). Other trichothecenes were shown to target L3 at the peptidyltransferase center in Saccharomyces cerevisiae and inhibit peptidyltransferase activity of eukaryotic ribosomes (7–9). Trichodermin-resistant yeast mutants were also resistant to anisomycin (9) and the trichodermin binding site on the 60S ribosomal subunit was closely related to the binding site of anisomycin (9, 10).
Trichothecenes and translation inhibitors like anisomycin that target the peptidyltransferase center are known to induce the ribotoxic stress response that leads to rapid activation of mitogen activated protein kinases (MAPKs), induction of proinflammatory responses, and cell death (11, 12). In the macrophage, DON induced mobilization of MAPKs to the ribosome (13) and promoted 28S rRNA cleavage at the peptidyltransferase center (14), suggesting that ribosome interactions of trichothecenes play an important role in their cytotoxicity.
Differences have been reported in the mode of action of trichothecenes, including mechanisms of translation inhibition (3, 10) and different signaling pathways that lead to cytotoxicity and apoptosis (15, 16). These observations suggest that toxicity of trichothecenes might not be a simple function of translational arrest and that they may have multiple mechanisms of toxicity. Despite the early studies that implicated DNA synthesis (2), respiration (17), mitochondrial protein synthesis (17), and membrane structure and integrity (18) in the cytotoxicity of trichothecenes, the molecular mechanisms of their toxicity are not well understood, and the proteins targeted by trichothecenes other than L3 have not been identified. To develop a better understanding of the trichothecene mechanism of action, we used a chemical genomics approach in S. cerevisiae to identify genes that when deleted confer resistance to trichothecin (Tcin). Our results provide genome-wide insight into the mechanism of toxicity of a trichothecene mycotoxin and demonstrate that its toxicity is mediated by the mitochondria.
Results
High-Throughput Screen of the Yeast Deletion Library Reveals a Role for Mitochondrial Translation in Trichothecin Sensitivity.
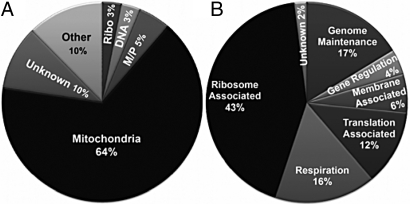
To identify the genes involved in mediating sensitivity to trichothecenes, we carried out a systematic screen of the 4,720 viable S. cerevisiae diploid gene deletion collection for Tcin resistance. The screen was carried out using 4 μM Tcin, which severely inhibited the growth of the parental strain, BY4743 (>90%) with an IC50 of 2.5 μM in liquid YPD media (Fig. 1). We identified 138 deletion strains as resistant to 4 μM Tcin from three independent experiments (Table S1). The largest group of deletions (89/138 or 64%) that showed resistance to 4 μM Tcin encoded proteins associated with the mitochondria or affected mitochondrial function (Fig. 2A). Strains associated with mitochondrial ribosomes constituted the largest group (43%) within the mitochondria category (Fig. 2B). Approximately 25% (35/138) of the Tcin-resistant strains associated with the mitochondria had abnormal mitochondrial morphology based on the Saccharomyces genome database (SGD) (http://www.yeastgenome.org/).
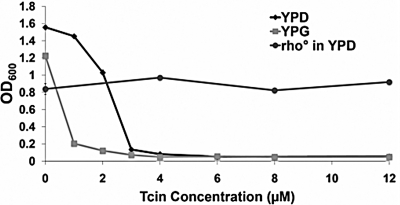
Fig. 1.
Growth comparison of BY4743 in YPD or YPG media. The parental strain BY4743 was grown in liquid media supplemented with 2% dextrose (YPD) for 20 h or with 3% glycerol (YPG) for 48 h at 30 °C in the presence of different concentrations of Tcin. The rho0 version of BY4743 was grown for 48 h at 30 °C. The experiments were performed in triplicate and repeated twice. Growth is represented by mean OD600 values (± SE).
Fig. 2.
Functional classification of yeast deletion mutants that conferred resistance to 4 μM Tcin. (A) Yeast deletion mutants exhibiting resistance to 4 μM Tcin. The figure shows the GO terms (www.yeastgenome.org) most associated with the genes identified. Genes associated with heat shock protein, signal transduction, proteasome/ubiquitination, transcription and RNA metabolism constitute the “Other” category. M/P and Ribo refer to membrane/phospholipid and cytosolic ribosomes. (B) Classification of the mitochondria-associated deletion mutants.
The relatively large proportion of resistant strains that had defects associated with the mitochondria were surprising in light of the reported role of trichothecenes in inhibition of cytosolic, but not mitochondrial protein synthesis (2, 4) and their lack of inhibition of bacterial growth and translation (19, 20). To determine whether mitochondria were critical for Tcin sensitivity, the parental strain, BY4743, was grown in liquid media containing glycerol (YPG), which forces the cells to depend on mitochondria for energy. The IC50 of BY4743 for Tcin was significantly reduced from 2.5 to 0.75 μM in YPG media (Fig. 1) reflecting a distinct mitochondrial role in the sensitivity to Tcin. To confirm this, we tested the sensitivity of a rho0 strain derived from BY4743 by ethidium bromide treatment. Unlike the parental strain, the rho0 strain was resistant to Tcin, confirming the role of mitochondria in Tcin sensitivity (Fig. 1).
To determine whether any of the strains that showed resistance to 4 μM Tcin had mitochondrial defects, the 138 resistant strains were grown on YPDG plates containing 3% glycerol and 0.2% dextrose. A large fraction (62%) of the strains that showed resistance to 4 μM Tcin was confirmed to be petite, consistent with their classification in the SGD (Table S1). Only four (ndi1Δ, ups1Δ, ydc1Δ, and cir2Δ) deletion strains resistant to 4 μM Tcin and associated with the mitochondria displayed the wild-type phenotype when grown on the YPDG media. When the Tcin-resistant petite mutants were grouped and analyzed using FUNSpec (http://funspec.med.utoronto.ca/), 32% (27/85) encoded mitochondrial ribosomal proteins. In addition, six mitochondrial aminoacyl-tRNA-synthetases (SLM5, AIM10, MSR1, MSK1, MSD1, and MSF1), one mitochondrial translation initiation (IFM1), one elongation (MEF2), one termination (RRF1) factor and two translational control proteins (CBP6 and AEP1) were included in the resistant group, providing evidence that mitochondrial translation may be a target of Tcin.
The genome-wide screen was repeated with 8, 12, and 24 μM Tcin to identify the most resistant strains. Twenty-seven out of the 138 strains resistant to 4 μM Tcin grew better than the wild-type on 8 μM Tcin, and 24 of these strains grew well on 12 μM Tcin. Only six of these strains (spo13Δ, ups1Δ, tma7Δ, tof2Δ, ict1Δ, and ynl011cΔ) were able to grow on 24 μM Tcin. Table S2 shows the scores for the 15 strains that had the highest level of resistance based on their growth ratios on 4–24 μM Tcin. These results were confirmed by examining growth of these strains on different concentrations of Tcin in liquid (Fig. S1) or solid YPD media (Fig. S2). The differences in the viability of these strains were apparent at the higher concentrations of Tcin (Fig. S2), indicating a dose-dependent response.
Tcin Affects Mitochondria at Low Doses.
Because a large fraction of the resistant strains were associated with the mitochondria, we characterized the effects of Tcin on yeast growth by growing the parental strain and the top 15 deletion mutants (all rho+) in liquid media containing glycerol (YPG). Growth of BY4743 was inhibited by 2 μM Tcin on YPG media, but not YPD media, indicating that the wild-type strain was more sensitive to Tcin when it depended on mitochondria for energy (Fig. S3). When the concentration of Tcin was doubled to 4 μM, growth of BY4743 was inhibited on both the YPG and the YPD media (Fig. S3). The most resistant strains grew well on YPG media, even at 12 μM Tcin (Fig. 3A), suggesting that the resistance in these strains was due to the mitochondria.
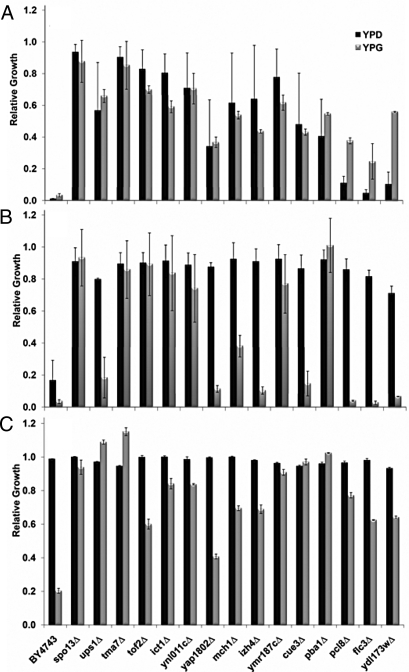
Fig. 3.
Growth of the most resistant 15 deletion strains in the presence of different antibiotics in YPD or YPG media. The Tcin-resistant strains were grown in liquid YPD and YPG media containing (A) 12 μM Tcin, (B) 10 μg/mL anisomycin, or (C) 1 mg/mL chloramphenicol. Relative growth was calculated as the ratio between growth of treated cells to that of untreated cells with a ratio of 1.0, indicating no effect on growth. The experiments were performed in triplicate and repeated twice.
Tcin-Resistant Strains Are Cross-Resistant to Anisomycin, but Not to Other Translation Inhibitors.
To test the specificity of the resistance, we screened the 15 most resistant deletion strains against other translation inhibitors. BY4743 and the resistant mutants failed to grow on 0.5 or 1 μg/mL cycloheximide (Fig. S4A) and were sensitive to hygromycin B (100 or 200 μg/mL) on YPD media, while 10 μg/mL hygromycin B inhibited their growth on YPG media (Fig. S4B). These results indicated that the observed resistance in the deletion strains did not extend to protein inhibitors in general. The Tcin-resistant mutants were, however, resistant to 10 μg/mL anisomycin on YPD media (Fig. 3B). We examined growth on YPG media to determine if anisomycin resistance was due to a possible effect on mitochondrial translation. As shown in Fig. S5, 2.5 μg/mL anisomycin inhibited growth of BY4743 by 52% on YPG media relative to 12% inhibition on YPD, suggesting that anisomycin targets mitochondrial translation at low doses. The Tcin-resistant strains were resistant to 5 μg/mL anisomycin on YPG media when growth of BY4743 was inhibited by 94% (Fig. S5). Several Tcin-resistant strains (ups1Δ, yap1802Δ, izh4Δ, cue3Δ, pcl8Δ, flc3Δ, and ydl173wΔ) were more sensitive to 10 μg/mL anisomycin on YPG media than on YPD media (Fig. 3B), suggesting that anisomycin and Tcin have overlapping, but not identical effects on the mitochondria.
Tcin-Resistant Strains Are Cross-Resistant to Chloramphenicol.
To further investigate the role of the mitochondria in resistance to Tcin, we examined the sensitivity of the Tcin-resistant strains to chloramphenicol, a known inhibitor of mitochondrial translation in eukaryotes, which binds to the A site and occupies the same position as the aminoacyl-tRNA (aa-tRNA), preventing peptide bond formation (21). One mg/mL chloramphenicol reduced the growth of BY4743 and the top 15 resistant mutants by less than 10% in YPD media (Fig. 3C), but was sufficient to inhibit BY4743 growth by 80% in YPG media, consistent with inhibition of mitochondrial translation. In contrast, growth of the top 15 mutants was either not inhibited at all or was markedly better than the parental strain on 1 mg/ml chloramphenicol (Fig. 3C). These results provided evidence that mitochondrial translation was critical for the toxicity of Tcin.
Tcin Inhibits Mitochondrial Translation.
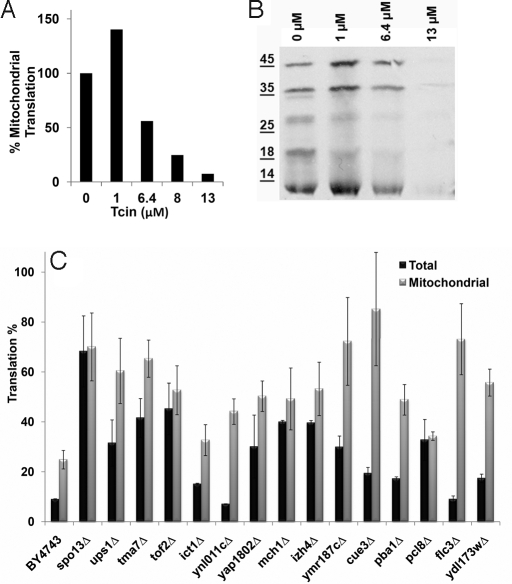
The wild-type cells were exposed to different concentrations of Tcin for 1 h and the mitochondrial translation rate was determined by measuring [35S]-Met incorporation into mitochondrial protein (22). At 6.4 μM Tcin, the mitochondrial translation efficiency was reduced by >50% and at 13 μM Tcin, nearly complete inhibition of mitochondrial protein synthesis was observed (Fig. 4A). A general decrease in mitochondrial protein synthesis, which was not specific to any one particular protein, was observed by SDS/PAGE analysis followed by autoradiography (Fig. 4B). These results demonstrated that mitochondrial translation in the wild-type strain is sensitive to Tcin.
Fig. 4.
Effect of Tcin on mitochondrial translation. (A) Mitochondrial translation rate of the BY4743 cells was determined by measuring the rate of [35S]-methionine incorporation in vivo at increasing concentrations of Tcin. All treatments were performed for 1 h and the rate of translation in the absence of Tcin was normalized to 100%. (B) Autoradiogram of labeled mitochondrial translation products separated on a 15% SDS-polyacrylamide gel. (C) The ratio of translation between Tcin-treated and untreated samples for both total and mitochondrial translation is represented as percent of the untreated control. Each graph represents the mean of four independent experiments with error bars representing the standard deviation.
Because the top 15 mutants were resistant to 8 μM or higher concentrations of Tcin in YPD media (Fig. S3), we examined the total and the mitochondrial translation efficiencies of the top 15 resistant strains in the presence of a final concentration of 8 μM Tcin/OD600 cells. To ensure accurate estimation of the translation rate, the [35S]-Met incorporation was normalized against the protein concentrations. Tcin significantly inhibited total translation in the wild-type (91%) and in all of the resistant strains (55–93%) except spo13Δ, which showed minimal inhibition of total translation (32%) (Fig. S6). The ynl011cΔ and ict1Δ, in which total translation was 93 and 85% affected respectively, were resistant to the highest concentration of Tcin tested (24 μM), (Table S2) indicating that Tcin resistance can be separated from the inhibition of total translation.
Analysis of mitochondrial translation by [35S]-Met incorporation indicated that 8 μM Tcin/OD600 significantly inhibited mitochondrial translation (75%) in BY4743. In contrast, mitochondrial translation was inhibited only 15–51% in the most resistant 15 strains except in ict1Δ and pcl8Δ, which had 66–67% inhibition (Fig. S6B). Mitochondrial translation rates of mutants resistant to 24 μM Tcin (spo13Δ, ups1Δ, tma7Δ, tof2Δ, and ict1Δ) were similar to the parental strain in the absence of Tcin (Fig. S6B). Mutants that had reduced mitochondrial translation efficiencies (ymr187cΔ, cue3Δ, and flc3Δ) were also not any more vulnerable to Tcin (Fig. S6B). Therefore, the resistance observed in these strains was not a result of a general defect in mitochondrial translation due to the individual gene deletion.
Translation percentages were calculated by comparing the translation rate of toxin-treated samples to their untreated controls and are presented as percentage of the untreated control in Fig. 4C. Comparison of the total and the mitochondrial translation in each mutant indicated that mitochondrial translation was inhibited less by Tcin than total translation in the 15 resistant strains (Fig. 4C). Total translation and mitochondrial translation were minimally affected in spo13Δ, tma7Δ, and tof2Δ. Total translation was affected more than the mitochondrial translation in ups1Δ, ict1Δ, ynl011cΔ, yap1802Δ, ymr187cΔ, cue3Δ, pba1Δ, flc3Δ, and ydl173wΔ, while in mch1Δ, izh4Δ, and pcl8Δ, both cytosolic and mitochondrial translation were affected, although less than the wild-type. These results indicated that inhibition of mitochondrial translation was critical for the toxicity of Tcin. The reduction in inhibition of mitochondrial protein synthesis did not correlate with the level of resistance in several strains, suggesting that Tcin had additional effects. A general defect in drug uptake or metabolism was unlikely, because the mutants did not show any resistance to cycloheximide or hygromycin B.
Tcin Affects Mitochondrial Morphology.
Five strains that showed resistance to Tcin (pcp1Δ, mdm12Δ, mdm10Δ, fzo1Δ, and mmm1Δ) displayed severe mitochondrial morphology defects as defined by the absence of cells with wild-type mitochondrial morphology, consistent with their classification in SGD. Several Tcin-resistant deletion strains were defective in mitochondrial fusion (fzo1Δ and pcp1Δ) or interfered with mitochondrial morphology, leading to a fusion/fission imbalance and resulting in the development of fragmented mitochondria (ups1Δ) (23). Deletion of the proteins required for the maintenance of proper tubular morphology and fusion of the mitochondria led to resistance to 4 μM Tcin, implicating disruption of mitochondrial membranes in the toxicity of Tcin.
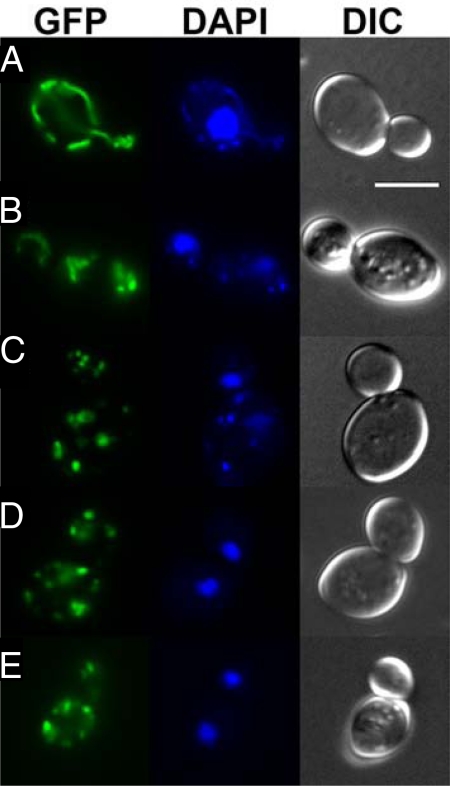
We investigated the effect of Tcin on mitochondrial morphology by transforming BY4743 with pVT100U-mtGFP encoding a constitutively expressed GFP targeted to the mitochondrial matrix (24). Mitochondria in BY4743-mtGPF cells showed the characteristic morphology of a uniformly tubular network and normal nuclear morphology (Fig. 5A). Exposure to 8 μM Tcin for 4 h led to the fragmentation of the mitochondrial network (Fig. 5B). Tcin treated cells contained highly fragmented mitochondria that were very short tubules or spheres. Increasing the concentration of Tcin to 80 μM led to further mitochondrial fragmentation (Fig. 5C). These cells had fragmented mitochondria similar to those observed in fzo1Δ mutant in the fusion pathway (Fig. 5D). Treatment of fzo1Δ with 8 μM Tcin did not lead to further fragmentation (Fig. 5E). Similar results were also observed with ups1Δ. These results demonstrated that Tcin causes fragmentation of the tubular mitochondrial network. The mutants in the fusion machinery, but not the mutants in the fission machinery, were resistant to Tcin (Table S1), suggesting that the observed fragmentation is mediated by the mitochondrial fission machinery.
Fig. 5.
Effect of Tcin on mitochondrial morphology. Mitochondrial morphology of (A) BY4743 control, (B) BY4743 treated with 8 μM Tcin for 4 h, (C) BY4743 treated with 80 μM Tcin for 4 h, (D) fzo1Δ control, and (E) fzo1Δ treated with 8 μM Tcin for 4 h were analyzed using epifluorescence microscopy. Cells were transformed with pVT100U-mtGFP, containing GFP targeted to the mitochondrial matrix. Nuclear and mitochondrial DNA was stained with DAPI. Differential interference contrast (DIC) images of each cell are presented. (Scale bar, 5 μm.)
Discussion
In this study, we performed a genome-wide screen of the homozygous diploid yeast deletion library to gain insight into the mode of action of trichothecenes and identified genes involved in mitochondrial translation, membrane organization, respiration, cellular transport, cell cycle, signal transduction, RNA, DNA, and lipid metabolism (Table S1). Because S. cerevisiae is more sensitive to Tcin (IC50 ≈2.5 μM) than DON (IC50 is >2 mM), we used Tcin as a representative type B trichothecene. The cytosolic ribosomes are reported to be the primary target of trichothecene action (2, 4). However, our results indicated that S. cerevisiae was more sensitive to Tcin when grown in media with glycerol than glucose as the sole carbon source. Since utilization of a nonfermentable carbon source, such as glycerol requires mitochondrial function, this observation provided evidence for the involvement of mitochondria in the cytotoxicity of Tcin. We confirmed this by demonstrating that the rho0 strains derived from BY4743, unlike rho+ were resistant up to 12 μM Tcin (Fig. 1). Loss of mitochondrial genome (rho0) is known to activate the pleiotropic drug resistance pathway via the overexpression of PDR5, the gene encoding the ATP-binding cassette transporter (25). Up-regulation of PDR5 in rho0 mutants depends on the presence of PDR3 (25). In our screen, deletion of PDR3 did not alter Tcin sensitivity of the parental rho+ strain, nor did deletion of PDR1 or PDR5. Both the rho0 and rho+ strains remained sensitive to 0.5–10 μg/mL cycloheximide. Taken together, these results imply that the acquired resistance due to loss of the mitochondrial genome is not simply a consequence of the activation of the pleiotropic drug resistance pathway, but rather it is specific to Tcin and its effect on the mitochondria.
The largest group of the resistant strains associated with mitochondria encoded ribosomal proteins and translation factors, suggesting that Tcin affected mitochondrial translation. Although trichothecenes do not inhibit bacterial translation (19, 20), Tcin inhibited mitochondrial translation in the wild-type yeast. Mitochondrial translation was only minimally affected by Tcin in a majority of the most resistant 15 strains in comparison with the isogenic wild-type strain (Fig. 4C). The Tcin-resistant mutants were cross-resistant to anisomycin, but not to cycloheximide or hygromycin B. Chloramphenicol, an inhibitor of mitochondrial translation, did not inhibit growth of the Tcin-resistant mutants at a concentration sufficient to inhibit growth of the wild-type cells. Chloramphenicol and anisomycin bind at the active site crevice of the ribosome and block peptidyltransferase activity by preventing binding of aminoacyl tRNA (aa-tRNA) to the A site (26), while hygromycin and cycloheximide inhibit translocation (27, 28). The inhibitory effect of anisomycin on mitochondria has not been previously reported. At low doses, anisomycin inhibited growth of the wild-type strain when glycerol was used as the carbon source, indicating that it inhibited mitochondrial translation. Taken together, these results suggest that Tcin and anisomycin target the peptidyltransferase center of mitochondrial ribosomes and inhibit an A site associated function. Consistent with this, deletion of MRPL9, encoding mitochondrial ribosomal protein L3 led to Tcin resistance (Table S1), suggesting that Tcin may inhibit mitochondrial peptidyltransferase activity by targeting the mitochondrial L3. The deletion mutant of the cytosolic ribosomal protein L3 was not included in the library, because it is an essential gene. The Tcin resistant mutants identified from this screen were either resistant to both Tcin and anisomycin or resistant solely to Tcin when glycerol was used as the carbon source, indicating that Tcin and anisomycin have common, as well as unique actions on the mitochondria.
Deletion of TMA7, associated with the 40S ribosomal subunit, conferred a high level of resistance to anisomycin, consistent with previous findings (29). The tma7Δ was highly resistant to Tcin. However, we did not observe a decrease in the total protein synthesis in tma7Δ in the absence of Tcin (29). Total translation and mitochondrial translation were inhibited less by Tcin in tma7Δ than in the wild-type strain. Similarly, spo13Δ and tof2Δ also showed a high level of resistance to Tcin and anisomycin and minimal inhibition of total and mitochondrial translation. The spo13Δ and tof2Δ were identified as highly resistant to the antimicrobial dermaseptins (30). Dermaseptin-induced nuclear DNA fragmentation, reactive oxygen species (ROS) production, and caspase independent cell death were minimized in these strains (30). Tof2 is required for ribosomal DNA (rDNA) silencing and mitotic rDNA condensation, while Spo13 is a meiosis specific protein. Both are involved in cell cycle progression and their deletion increases levels of Rad52 foci, a marker for homologous recombination, which functions to maintain the integrity of genome through repair of DNA double-strand breaks (31). Additional genes involved in recombination and repair of double stranded DNA breaks (RAD57, IRC19, and MHR1), and cell cycle progression (MBP1, RAD57, and HFM1) were identified in our screen, suggesting that DNA damage, a process known to trigger apoptosis, may be a consequence of the effects of Tcin and anisomycin on mitochondrial ribosomes. The absence of SPO13 and TOF2 might prevent the Tcin and anisomycin-induced cell death by enhancing the DNA damage repair response.
Several mutants (ups1Δ, yap1802Δ, izh4Δ, cue3Δ, pcl8Δ, flc3Δ, and ydl173wΔ) were uniquely resistant to Tcin, but not to anisomycin when mitochondria were used as the source of energy (Fig. 3B). These mutants were involved in maintaining the integrity of the mitochondrial membrane (ups1Δ), clathrin cage assembly (yap1802Δ), sterol metabolism (izh4Δ), monoubiquitination (cue3Δ), generation of energy (pcl8Δ) or FAD transport (flc3Δ). Additional genes identified (CEM1, FAB1, ICT1, UPS1, LCB5, SUR1, and YDC1) encoded proteins involved in phospholipid metabolism (Table S1). Trichothecenes were shown to interfere with sphingolipid metabolism by inducing accumulation of glucosylceramide in neural cells (32). YDC1, LCB5, and SUR1 identified in our screen encode proteins involved in the sphingolipid metabolism (33). Yeast mutants lacking YDC1, an alkaline dihydroceramidase had increased chronological life span, while overexpression of YDC1 led to fragmentation of mitochondria, reduced life span, and increased apoptotic cell death (34). These results implicate sphingolipid metabolism in the toxicity of Tcin and suggest that membrane sphingolipids are either directly required for Tcin mediated growth inhibition or are indirectly affected.
In contrast to the wild-type cells, which contain tubular mitochondria, in Tcin-treated cells the tubular structure of the mitochondria was largely broken. Deletion of genes involved in mitochondrial fusion (FZO1, PCP1, and AFG3) and maintenance of the tubular morphology of mitochondria (MMM1, MDM10, and MDM12) conferred resistance to Tcin, suggesting a possible effect on the mitochondrial membrane dynamics. Fzo1 is an outer mitochondrial membrane GTPase essential for mitochondrial fusion. In fzo1Δ yeast cells, mitochondria are highly fragmented due to a deficiency in mitochondrial fusion, a defect that in turn leads to loss of mitochondrial DNA and therefore respiratory activity (35). Pcp1 and Ups1 are associated with the inner membrane and their deletion results in loss of fusion activity (35). Ups1 regulates cardiolipin metabolism, a mitochondria specific phospholipid required for the integrity of several protein complexes in the inner membrane and is essential for respiration (36). The Mmm1-Mdm10-Mdm12-Mdm34 complex has recently been shown to connect the endoplasmic reticulum (ER) and mitochondria (37). Mutations in this complex reduce cardiolipin levels and impair phospholipid biosynthesis (37). These observations imply a possible inhibitory effect of Tcin on the membranes that connect the ER and mitochondria, which would lead to impairment of the interorganellar phospholipid exchange. The mechanistic details by which trichothecene mycotoxins kill target cells are not yet fully delineated. Here, we identified genes in a number of cellular pathways previously unknown to play a role in trichothecene toxicity and demonstrated that mitochondrial membrane maintenance and translation are critical for sensitivity to a trichothecene mycotoxin. Orthologs of these genes may be involved in the toxicity of trichothecene mycotoxins in plants. Our work draws attention to the similarities between the toxic effects of trichothecenes on mammalian cells and toxin induced cell death in the yeast model.
Materials and Methods
Tcin Isolation.
Tcin was extracted from liquid yeast extract-peptone-dextrose shake cultures of Trichothecium roseum (no. A27955 or NRRL3704), purified on silica gel columns, and eluted with hexane:ethyl acetate (3:1).
Yeast Deletion Library Screen.
The yeast genome homozygous diploid gene deletion set (Open Biosystems) was screened for resistance to 4 μM Tcin. The strains from the deletion library were replica plated into 96-deep-well plates containing YPD (2% glucose) liquid media with or without Tcin. Experiments were performed in triplicate. Cells were grown for 20 h in the HiGro shaker (GeneMachines) at 250 rpm at 30 °C. The OD600 for treated and untreated samples was obtained and the relative growth determined as the growth ratio of toxin treatment to the control. A ratio of 1 indicated that the treatment had no effect on the strain. The coefficient of variation (CV) calculated by taking the standard deviation of the ratio scores and dividing by the mean, was used as a measure of variability. The resistance level of a particular strain was based jointly on the mean ratio score and the CV. The threshold for 4 μM Tcin resistance was defined as strains which had a ratio of 0.25 or higher (at least 25% growth relative to the nontoxin treatment of that same strain) and a CV of <0.5.
Antibiotic Assays.
Overnight cultures were transferred into 96-well plates containing Tcin or the different antibiotics in liquid YPG (3% glycerol) or YPD. Antibiotics were used at the following concentrations: Tcin (1, 2, 4, 8, 12, 16, and 24 μM), cycloheximide (0.5, 1, and 10 μg/mL), anisomycin (2.5, 5, 7.5, and 10 μg/mL), hygromycin B (10, 50, 100, and 200 μg/mL) and chloramphenicol at 1 mg/mL. Cells were grown for 20 h in YPD and for 48 h in YPG media. The relative growth was measured as described above. All experiments were performed in triplicate and repeated twice.
Strain Verification.
Yeast strain verification was carried out by PCR using knockout cassette specific primers for each of the 15 Tcin-resistant deletion mutants (yeastdeletion.stanford.edu) (Fig. S7A).
rho0 Strains.
rho0 versions of BY4743 were generated as described in ref. 38. The respiratory deficiency of the rho0 strains was confirmed by the complete lack of growth on YPG media. The absence of mitochondrial DNA was confirmed by staining paraformaldehyde-fixed cells with DAPI and by Southern blot analysis (Fig. S7B) using a mitochondrial COX1 probe (39).
Total Translation Assay.
Cultures of BY4743 and the top 15 resistant mutants were grown in SD-Met containing MSG as the nitrogen source and 2% glucose. Each culture, grown to an OD600 of 0.5–0.6 (4.5–5.4 × 107 cells/mL), was then split into two halves. One-half was treated with 8 μM Tcin/OD600 cells and the other half with an equivalent amount of ethanol (0.03%) for 1 h, shaking at 30 °C. Cold methionine (50 μM) and 1 μL [35S]-Met (Perkin-Elmer, NEG-009A, >1,000 Ci/mmol) was added. The reaction was stopped after 60 min, and [35S]-Met incorporation was measured as described in ref. 40. The rate of incorporation of [35S]-Met was linear up to 60 min.
Mitochondrial Translation Assay.
Mitochondrial translation was measured using a modified assay developed by Fox et al. (22). Fresh YPR (2% raffinose) media was added to an overnight yeast culture, grown in YPR, and incubated for 2–3 h. An equivalent of 10 OD600 cells were pelleted, washed twice and resuspended in SD-Met media supplemented with 2% raffinose and treated with 8 μM Tcin/OD600 or an equivalent amount of ethanol (control) for 1 h at 30 °C. Cycloheximide was immediately added to each sample to stop cytosolic translation followed by addition of 25μCi [35S]-Met. Mitochondria were isolated, and [35S]-Met incorporation was determined as described in ref. 22.
Microscopy.
Mitochondrial morphology was examined by epifluorescence microscopy (Olympus BX41) using cells transformed with pVT100U-mtGFP, which contains green fluorescent protein (GFP) targeted to the mitochondria with the presequence from the subunit 9 of the F0-ATPase of Neurospora crassa (24).
Supplementary Material
Acknowledgments.
We thank Dr. Andreas Ivessa for help with verification of the rho0 strain, Dr. Thomas Fox for the mitochondrial translation protocol, Dr. Benedikt Westermann for pVT100U-mtGFP, and Drs. Jennifer Nielsen Kahn, Xiao-Ping Li, and Zhennan Xu for critical reading of the manuscript. This is a cooperative project No. 59-0790-6-069 supported by the United States Department of Agriculture in cooperation with the United States Wheat and Barley Scab Initiative. N.M. was supported by a National Institutes of Health Research Supplement (AI072425) to promote Diversity in a Health Related Research Program.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0909777106/DCSupplemental.
References
- 1.Desjardins AE, Hohn TM, McCormick SP. Trichothecene biosynthesis in Fusarium species: Chemistry, genetics, and significance. Microbiol Rev. 1993;57:595–604. doi: 10.1128/mr.57.3.595-604.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rocha O, Ansari K, Doohan FM. Effects of trichothecene mycotoxins on eukaryotic cells: A review. Food Addit Contam. 2005;22:369–378. doi: 10.1080/02652030500058403. [DOI] [PubMed] [Google Scholar]
- 3.Ueno Y. The toxicology of mycotoxins. Crit Rev Toxicol. 1985;14:99–132. doi: 10.3109/10408448509089851. [DOI] [PubMed] [Google Scholar]
- 4.Pestka JJ. Mechanisms of deoxynivalenol-induced gene expression and apoptosis. Food Addit Contam. 2008;22:1128–1140. doi: 10.1080/02652030802056626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fried HM, Warner JR. Cloning of yeast gene for trichodermin resistance and ribosomal protein L3. Proc Natl Acad Sci USA. 1981;78:238–242. doi: 10.1073/pnas.78.1.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wickner RB, Ridley SP, Fried HM, Ball SG. Ribosomal protein L3 is involved in replication or maintenance of the killer double-stranded RNA genome of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1982;79:4706–4708. doi: 10.1073/pnas.79.15.4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carrasco L, Barbacid M, Vazquez D. The trichodermin group of antibiotics, inhibitors of peptide bond formation by eukaryotic ribosomes. Biochim Biophys Acta. 1973;312:368–376. doi: 10.1016/0005-2787(73)90381-x. [DOI] [PubMed] [Google Scholar]
- 8.Fernandez-Lobato M, Cannon M, Mitlin JA, Mount RC, Jimenez A. Characterization of Saccharomyces cerevisiae strains displaying high-level or low-level resistance to trichothecene antibiotics. Biochem J. 1990;267:709–713. doi: 10.1042/bj2670709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jimenez A, Sanchez L, Vazquez D. Simultaneous ribosomal resistance to trichodermin and anisomycin in Saccharomyces cerevisiae mutants. Biochim Biophys Acta. 1975;383:427–434. doi: 10.1016/0005-2787(75)90312-3. [DOI] [PubMed] [Google Scholar]
- 10.Cannon M, Jimenez A, Vazquez D. Competition between trichodermin and several other sesquiterpene antibiotics for binding to their receptor site(s) on eukaryotic ribosomes. Biochem J. 1976;160:137–145. doi: 10.1042/bj1600137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shifrin VI, Anderson P. Trichothecene mycotoxins trigger a ribotoxic stress response that activates c-Jun N-terminal kinase and p38 mitogen-activated protein kinase and induces apoptosis. J Biol Chem. 1999;274:13985–13992. doi: 10.1074/jbc.274.20.13985. [DOI] [PubMed] [Google Scholar]
- 12.Iordanov MS, et al. Ribotoxic stress response: Activation of the stress-activated protein kinase JNK1 by inhibitors of the peptidyl transferase reaction and by sequence-specific RNA damage to the alpha-sarcin/ricin loop in the 28S rRNA. Mol Cell Biol. 1997;17:3373–3381. doi: 10.1128/mcb.17.6.3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bae HK, Pestka JJ. Deoxynivalenol induces p38 interaction with the ribosome in monocytes and macrophages. Toxicol Sci. 2008;105:59–66. doi: 10.1093/toxsci/kfn102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li M, Pestka JJ. Comparative induction of 28S ribosomal RNA cleavage by ricin and the trichothecenes deoxynivalenol and T-2 toxin in the macrophage. Toxicol Sci. 2008;105:67–78. doi: 10.1093/toxsci/kfn111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bae HK, Shinozuka J, Islam Z, Pestka JJ. Satratoxin G interaction with 40S and 60S ribosomal subunits precedes apoptosis in the macrophage. Toxicol Appl Pharmacol. 2009;237:137–145. doi: 10.1016/j.taap.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bouaziz C, et al. Fusarial toxin-induced toxicity in cultured cells and in isolated mitochondria involves PTPC-dependent activation of the mitochondrial pathway of apoptosis. Toxicol Sci. 2009;110:363–375. doi: 10.1093/toxsci/kfp117. [DOI] [PubMed] [Google Scholar]
- 17.Pace JG, Watts MR, Canterbury WJ. T-2 mycotoxin inhibits mitochondrial protein synthesis. Toxicon. 1988;26:77–85. doi: 10.1016/0041-0101(88)90139-0. [DOI] [PubMed] [Google Scholar]
- 18.Schappert KT, Khachatourians GG. Influence of the membrane on T-2 toxin toxicity in Saccharomyces spp. Appl Environ Microbiol. 1984;47:681–684. doi: 10.1128/aem.47.4.681-684.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ueno Y, Nakajima M, Sakai K, Ishii K, Sato N. Comparative toxicology of trichothecene mycotoxins: Inhibition of protein synthesis in animal cells. J Biochem. 1973;74:285–296. [PubMed] [Google Scholar]
- 20.Burmeister HR, Hesseltine CW. Biological assays for two mycotoxins produced by Fusarium tricinctum. Appl Microbiol. 1970;20:437–440. doi: 10.1128/am.20.3.437-440.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schlunzen F, et al. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature. 2001;413:814–821. doi: 10.1038/35101544. [DOI] [PubMed] [Google Scholar]
- 22.Fox TD, et al. Analysis and manipulation of yeast mitochondrial genes. Methods Enzymol. 1991;194:149–165. doi: 10.1016/0076-6879(91)94013-3. [DOI] [PubMed] [Google Scholar]
- 23.Sesaki H, et al. Ups1p, a conserved intermembrane space protein, regulates mitochondrial shape and alternative topogenesis of Mgm1p. J Cell Biol. 2006;173:651–658. doi: 10.1083/jcb.200603092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Westermann B, Neupert W. Mitochondria-targeted green fluorescent proteins: Convenient tools for the study of organelle biogenesis in Saccharomyces cerevisiae. Yeast. 2000;16:1421–1427. doi: 10.1002/1097-0061(200011)16:15<1421::AID-YEA624>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 25.Hallstrom TC, Moye-Rowley WS. Multiple signals from dysfunctional mitochondria activate the pleiotropic drug resistance pathway in Saccharomyces cerevisiae. J Biol Chem. 2000;275:37347–37356. doi: 10.1074/jbc.M007338200. [DOI] [PubMed] [Google Scholar]
- 26.Hansen JL, Moore PB, Steitz TA. Structures of five antibiotics bound at the peptidyl transferase center of the large ribosomal subunit. J Mol Biol. 2003;330:1061–1075. doi: 10.1016/s0022-2836(03)00668-5. [DOI] [PubMed] [Google Scholar]
- 27.Gonzalez A, Jimenez A, Vazquez D, Davies JE, Schindler D. Studies on the mode of action of hygromycin B, an inhibitor of translocation in eukaryotes. Biochim Biophys Acta. 1978;521:459–469. doi: 10.1016/0005-2787(78)90287-3. [DOI] [PubMed] [Google Scholar]
- 28.Rao SS, Grollman AP. Cycloheximide resistance in yeast: A property of the 60s ribosomal subunit. Biochem Biophys Res Commun. 1967;29:696–704. doi: 10.1016/0006-291x(67)90273-2. [DOI] [PubMed] [Google Scholar]
- 29.Fleischer TC, Weaver CM, McAfee KJ, Jennings JL, Link AJ. Systematic identification and functional screens of uncharacterized proteins associated with eukaryotic ribosomal complexes. Genes Dev. 2006;20:1294–1307. doi: 10.1101/gad.1422006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morton CO, Dos Santos SC, Coote P. An amphibian-derived, cationic, alpha-helical antimicrobial peptide kills yeast by caspase-independent but AIF-dependent programmed cell death. Mol Microbiol. 2007;65:494–507. doi: 10.1111/j.1365-2958.2007.05801.x. [DOI] [PubMed] [Google Scholar]
- 31.Alvaro D, Lisby M, Rothstein R. Genome-wide analysis of Rad52 foci reveals diverse mechanisms impacting recombination. PLoS Genet. 2007;3:2439–2449. doi: 10.1371/journal.pgen.0030228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kralj A, Gurgui M, Konig GM, van Echten-Deckert G. Trichothecenes induce accumulation of glucosylceramide in neural cells by interfering with lactosylceramide synthase activity. Toxicol Appl Pharmacol. 2007;225:113–122. doi: 10.1016/j.taap.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 33.Obeid LM, Okamoto Y, Mao C. Yeast sphingolipids: Metabolism and biology. Biochim Biophys Acta. 2002;1585:163–171. doi: 10.1016/s1388-1981(02)00337-2. [DOI] [PubMed] [Google Scholar]
- 34.Aerts AM, et al. Ydc1p ceramidase triggers organelle fragmentation, apoptosis and accelerated ageing in yeast. Cell Mol Life Sci. 2008;65:1933–1942. doi: 10.1007/s00018-008-8129-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cerveny KL, Tamura Y, Zhang Z, Jensen RE, Sesaki H. Regulation of mitochondrial fusion and division. Trends Cell Biol. 2007;17:563–569. doi: 10.1016/j.tcb.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 36.Tamura Y, Endo T, Iijima M, Sesaki H. Ups1p and Ups2p antagonistically regulate cardiolipin metabolism in mitochondria. J Cell Biol. 2009;185:1029–1045. doi: 10.1083/jcb.200812018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kornmann B, et al. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science. 2009;325:477–481. doi: 10.1126/science.1175088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rasmussen AK, Chatterjee A, Rasmussen LJ, Singh KK. Mitochondria-mediated nuclear mutator phenotype in Saccharomyces cerevisiae. Nucleic Acids Res. 2003;31:3909–3917. doi: 10.1093/nar/gkg446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheng X, Qin Y, Ivessa AS. Loss of mitochondrial DNA under genotoxic stress conditions in the absence of the yeast DNA helicase Pif1p occurs independently of the DNA helicase Rrm3p. Mol Genet Genomics. 2009;281:635–645. doi: 10.1007/s00438-009-0438-6. [DOI] [PubMed] [Google Scholar]
- 40.Parikh BA, Coetzer C, Tumer NE. Pokeweed antiviral protein regulates the stability of its own mRNA by a mechanism that requires depurination but can be separated from depurination of the alpha-sarcin/ricin loop of rRNA. J Biol Chem. 2002;277:41428–41437. doi: 10.1074/jbc.M205463200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.