Abstract
It is well established that all of the cis-acting sequences required for fully regulated human α-globin expression are contained within a region of ≈120 kb of conserved synteny. Here, we show that activation of this cluster in erythroid cells dramatically affects expression of apparently unrelated and noncontiguous genes in the 500 kb surrounding this domain, including a gene (NME4) located 300 kb from the α-globin cluster. Changes in NME4 expression are mediated by physical cis-interactions between this gene and the α-globin regulatory elements. Polymorphic structural variation within the globin cluster, altering the number of α-globin genes, affects the pattern of NME4 expression by altering the competition for the shared α-globin regulatory elements. These findings challenge the concept that the genome is organized into discrete, insulated regulatory domains. In addition, this work has important implications for our understanding of genome evolution, the interpretation of genome-wide expression, expression-quantitative trait loci, and copy number variant analyses.
Keywords: chromosome looping, copy number variants, globin gene expression, allele-specific expression, 4C
Recent global analyses of mammalian genomes have revised our view of the relationship between genome organization and the regulation of gene expression. It was previously thought that the genome might be arranged as a series of independently regulated chromosomal domains flanked by boundary elements (1). In contrast, it is now clear that cis-acting regulatory elements (locus control regions, enhancers, silencers, enhancer blockers, and chromatin barrier elements), controlling tissue- or developmental stage-specific genes, may be dispersed over tens to thousands of kilobases (2, 3). Furthermore, we now know that in gene-rich regions such elements are commonly interspersed with widely expressed genes (2). These observations raise important general questions, such as: How does the activation of specialized regulatory elements and their cognate genes influence the expression of other apparently unrelated genes in a shared chromosomal environment? How do common structural variants which alter genome architecture affect gene expression? What, if any, are the consequences of such apparently adventitious effects on gene expression?
To investigate these issues in detail we have examined the pattern of gene expression across a large segment of the human genome and studied how polymorphic variation in this region may influence long-range patterns of gene expression. In particular, we analyzed a well-characterized, gene-dense, telomeric region of the genome (16p13.3) containing the human α-like globin genes (ζ, α2, and α1), which are activated and transcribed at very high levels only in erythroid cells (4, 5). We have previously shown that critical, remote regulatory elements controlling α-globin expression, MCS-R1 to -R4 (representing previously identified DNaseI hypersensitive sites HS-48, HS-40, HS-33, and HS-10, respectively), three of which lie within the introns of a widely expressed gene (C16orf35) lying 50 to 70 kb upstream the α-like genes (4). In addition, we have also shown that a 120-kb region of conserved synteny containing the human α-like globin genes, together with their major upstream regulatory element (MCS-R2, also called HS-40), is sufficient to obtain optimal tissue- and developmental stage-specific expression in a mouse model (6). However, here we have asked whether globin gene activation within this region has more far reaching consequences, by affecting the expression of apparently unrelated genes in the surrounding chromosomal neighborhood.
To investigate this hypothesis, we examined the expression of 14 genes in an extensive region (500 kb) surrounding the α-globin cluster in nonerythroid cells (when the α-globin genes are silent) and in erythroid cells (when the α-globin genes and their regulatory elements are fully active). When the α-globin genes are switched on, expression of the functionally unrelated gene (C16orf35), containing the α-globin regulatory elements, is increased by ≈30-fold. In addition, we have shown that another apparently unrelated gene (NME4), located 300 kb from the α-globin cluster (in which we have identified a potential erythroid cis-acting element) physically interacts with, and is regulated by, MCS-R2, such that its expression is also increased 10-fold in erythroid cells. All other genes lying between MCS-R2 and NME4 are unaffected. When the α-globin genes are deleted from this chromosomal region, expression of NME4 (300 kb away) is further increased by 8-fold, as a result of increased competition for the shared regulatory element (MCS-R2). Because α-globin deletions have been selected to reach high frequencies in many populations (as they cause α-thalassemia, which protects against falciparum malaria), the levels of NME4 will be expected to vary in such populations in parallel with changes in the number of cis-linked α-globin genes.
This study therefore demonstrates a common mechanism by which patterns and levels of gene expression across a large chromosomal region may radically change in an unexpected way. Common structural polymorphisms in the α-globin genes, which have been selected during evolution, have a dramatic effect on expression of an unrelated gene (NME4) lying 300 kb away in what appears to be a shared chromosomal environment. These findings have important, general implications for the evolution of the genome, and for understanding how common expression quantitative trait loci and copy number variants (CNVs) may influence gene expression across long segments of the human genome.
Results
Analysis of the Expression of Genes in the Telomeric Region of Chromosome 16.
The expression of 14 genes contained in the terminal 500-kb region of chromosome 16 (16p13.3) (Fig. 1A) was examined in human embryonic stem cells (hES), where the α genes are largely silent, and in erythroid cells, in which they are fully activated. Most genes showed no increase in expression in erythroid cells (Fig. 1B). However, two genes within this 500-kb region (in addition to the α-globin genes) are up-regulated to levels similar to that of the erythroid-specific control gene, EPOR. C16orf35 (up-regulated by a factor of 27) is a highly conserved gene of unknown function containing the erythroid MCS-R elements, which become activated during erythropoiesis (4). A second gene specifically up-regulated in erythroid cells, NME4 [a nucleoside diphosphate kinase (7); expression increased by a factor of 12] lies ≈300-kb downstream of the α cluster, far beyond the region of conserved synteny. Erythroid-specific up-regulation of both C16orf35 and NME4 was also confirmed by comparison with another nonerythroid cell type (EBV-transformed B lymphocytes) (Fig. S1).
Fig. 1.
Overview and expression analysis of the terminal 500 kb of human chromosome 16p, containing the α-globin cluster. (A) Representation of the genes contained within this chromosomal region. Conserved synteny with the mouse region and the MCS-R region, which is conserved and required for full expression of the α-globin genes are shown. The minimal regions deleted, in all cases of α-thalassemia affecting the MCS-R elements (ΔMCS-R) one (-α) or two (--) α genes, are shown. ChIP for the activating chromatin mark H4ac, the erythroid-specific binding factors GATA1 and SCL, and RNA polymerase II were carried out in erythroid cells and hybridized to a tiled microarray covering this region (ChIP-chip). Tracks are representative of a minimum of two biological replicates. (B) Expression of genes contained within this region, and an erythroid control gene EPOR, in hES and erythroid cells. Expression was normalized to 18S. Values represent an average of three biological replicates ± 1 standard deviation. The y axis is a log scale. (C) Schematic representation of the genomic structure of NME4 and eNME4. (Black boxes) Exons; (gray box) alternative erythroid-specific exon; (full line) introns; (dashed lines) splicing of mature transcript. Amplicons used for expression analysis are shown; further information can be found in Tables S1–S3. The highly polymorphic SNP used for allele-specific expression (rs14293) is shown in red.
Although widely expressed (8), NME4 acquires an increase in activating chromatin modifications (H4ac, H3ac, H3K4me2, H3K4me3) in erythroid cells (Fig. S2). In addition, in erythroid cells, this gene is bound by GATA1 and SCL, which are components of the pentameric erythroid-specific transcription factor complex (consisting of SCL, GATA1, LDB1, E2A, and LMO2) (5). High levels of RNA polymerase II binding were also observed (see Fig. 1A and Fig. S2). Further characterization of NME4 identified a GATA1 binding site within intron 3 (Fig. S3A), which colocalizes with the observed binding of GATA1 and SCL (see Fig. 1A and Fig. S2B). This erythroid-specific transcription factor binding site lies within an internal promoter, which directs expression of a truncated, erythroid-specific transcript which we refer to as eNME4 (Fig. 1C and Fig. S3B). Both the eNME4 and the full-length NME4 transcript are up-regulated in erythroid cells. All other genes tested in this 500-kb chromosomal region, including five genes lying between the α-globin cluster and NME4, are expressed at similar levels in both erythroid and nonerythroid (hES and EBV) cells (see Fig. 1B and Fig. S1).
Expression of a Gene Located 300 kb from the α-Cluster Is Controlled by the α-Globin Regulatory Elements.
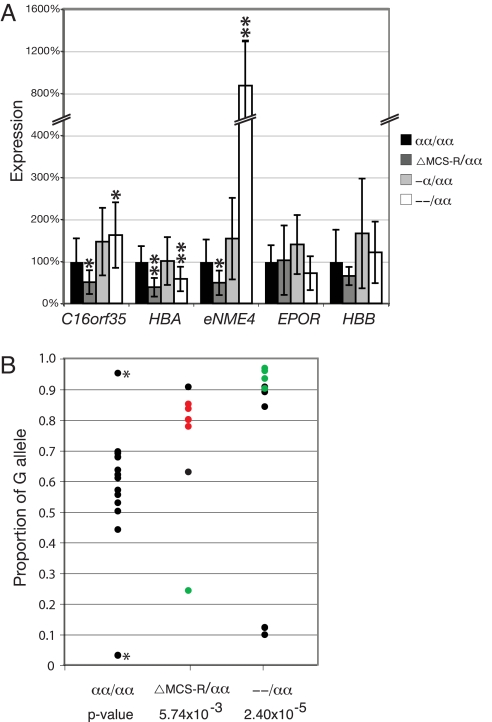
To determine whether expression of eNME4 in erythroid cells is regulated by the α-globin MCS-R elements, we analyzed expression of eNME4 mRNA transcripts in red blood cells obtained from rare individuals, each with a different deletion of the MCS-Rs on one allele (ΔMCS-R/αα) (9) (see Fig. 1A). We found that while the expression of control erythroid-specific genes [EPOR and β-globin (HBB)] was not significantly affected by these deletions, eNME4 was significantly reduced (n = 7, P = 0.01) to ≈50% of its normal level of expression (Fig. 2A). This suggested that enhanced expression of the NME4 erythroid-specific transcript, like α-globin, depends on the α-globin MCS-R elements located ≈300 kb upstream of this gene.
Fig. 2.
Allele-specific effect on the expression of eNME4 by various deletions of the α-globin cluster. (A) Expression of the three erythroid-specific genes in the terminal 500 kb of chromosome 16p, and two erythroid-specific control genes, EPOR and HBB. For each gene, the expression in control samples is set to 100%, and each group of deletions is calculated relative to controls. Student's t test P values are calculated for each gene for each group of samples compared to controls; *, P < 0.05; **, P < 0.005. Controls, n = 15; ΔMCS-R/αα, n = 7; -α/αα, n = 14; --/αα, n = 22. (B) The proportion of the G allele of NME4 contributing to total transcription, as determined by pyrosequencing (see Tables S4 and S5 for details). All samples are heterozygous (A/G) for SNP rs14293 (controls, n = 12; ΔMCS-R/αα, n = 7; --/αα, n = 11) except for an A/A and a G/G control (indicated by *). Samples for which phase of the α-globin locus deletion and the SNP allele could be determined are shown in color; deletion in phase with the A allele are shown in red; deletion in phase with the G allele are shown in green; samples where phase could not be determined are shown in black. The P value is calculated by an f test for differences in variation.
Given the relatively large distance between NME4 and the α-globin regulatory elements, it seemed possible that the effect on expression might be mediated in cis or in trans [as suggested for other long-range interactions (reviewed in ref. 10)]. To establish the effect of the MCS-R deletions on each copy of NME4, we analyzed allele-specific expression in red blood cells, using a highly polymorphic, synonymous transcribed SNP in exon 4 of NME4 (A/G, rs14293). This SNP is contained in both the full-length transcript and the truncated erythroid-specific transcript (Fig. 1C). With one exception, six individuals with MCS-R deletions, who were informative for this SNP, displayed allele-specific expression of NME4 (Fig. 2B). Using somatic cell hybrids containing a single copy of human chromosome 16 derived from these individuals, we determined which SNP was in phase with the deletion and found that in all cases examined, deletion of the α-globin MCS-R occurred in cis to the eNME4 allele that was under-expressed. The reduced level of expression specifically from one allele in cis with the MCS-R deletion demonstrates that the erythroid-specific enhanced expression of NME4/eNME4 is regulated in cis by the α-globin major regulatory element (MCS-R2).
Identification of Long-Range Interactions Across the Terminal Region of Chromosome 16.
We hypothesized that this functional interaction between the MCS-Rs and NME4/eNME4 was mediated via a physical interaction. We have previously used chromosome conformation capture (3C) to demonstrate physical interactions between the α-globin genes and the MCS-Rs both in mouse (11) and human (12). Recently, we and others (13, 14), have developed this methodology into a modified circular 3C method (4C), which, after cross linking and ligation of chromosome loops, uses an inverse PCR protocol to detect all physical interactions with a genomic fragment of interest. Our assay has been modified to give extremely high sensitivity and is analyzed using a microarray-platform (see Methods). By using a DpnII fragment containing MCS-R2 (the major α-globin regulatory element) as the anchor fragment, we performed 4C analysis on nonerythroid and erythroid cells (Fig. 3A).
Fig. 3.
4C analysis from MCS-R2 identifies a physical interaction with NME4 in --/αα erythroid material. (A) 4C material hybridized to a tiled microarray. The dashed line represents the fixed fragment of MCS-R2. Shaded boxes show α-globin locus and NME4; nonerythroid, EBV-transformed B-lymphocyte cell line; erythroid, two-phase culture system for generation of erythroid cells. All tracks are representative of two biological replicates. Zoomed section shows signal from Lower. Actual enrichment of 4C-amplified material relative to genomic DNA (based on real-time PCR; QPCR) is shown for two amplicons (389776 and 396875). The y axis is a log scale. Arrows represent transcription of NME4 and eNME4. Primer sequences can be found in Table S6. (B) Pyrosequencing tracks from an --/αα individual informative for SNP rs14293 at NME4. (Upper) Genomic DNA; (Lower) 4C-amplified DNA. Peaks used for calculations are shaded; peak 4 corresponds to the G allele, peak 8 corresponds to the A allele; dispensation order of nucleotides is shown on the x axis; E, enzyme; S, substrate. Further information can be found in Tables S4 and S5.
In nonerythroid cells there are no interactions between MCS-R2 and genes in the α-globin locus (see Fig. 3A Top). In erythroid cells, where MCS-R2 becomes active and the α-globin genes are highly transcribed, there is a strong interaction between MCS-R2 and the α globin genes (see Fig. 3A Middle, erythroid controls 1 and 2). In these cells, although there is not a consistent interaction with NME4, we have observed rare interactions represented by small peaks of enrichment (for example, erythroid control 2) in approximately one in three 4C experiments (n = 9). Therefore, even though the functional data and some 4C data suggest an interaction does occur between MCS-R2 and NME4 in a normal, intact chromosome, it seems that when the α-globin genes are present, their interaction with the MCS-Rs may out-compete the much weaker and occasional interactions with NME4 (see below). Such infrequent interactions (MCS-R2/NME4) may often lie below the level of detection using this assay.
Deletion of the α-Globin Genes Increases Expression of NME4 via Competition for the Shared Regulatory Element (MCS-R2).
The level of expression of eNME4 in erythroid cells is regulated by the α-globin MCS-Rs, and provisional evidence suggests that NME4 and MCS-R2 may physically interact, albeit rarely. Therefore, it seemed likely that NME4, even though it lies hundreds of kilobases from the α-globin cluster, may compete for the activity of the remote regulatory elements (MCS-R1 to -R4). To test this hypothesis, we examined the expression of eNME4 in the red cells of patients who have inherited chromosomes with either one (-α) or no (--) α genes rather than the normal duplicated pair of genes (αα). We observed that deletion of a single α-globin gene (-α) resulted in a small increase (factor of two) in expression of eNME4 (see Fig. 2A). However, deletions removing both α-globin genes (--) resulted in a dramatic increase (8-fold) in eNME4 expression when compared to normal controls (n = 22, P = 3.18 × 10−9) (see Fig. 2A). This group consisted of individuals each carrying mutations on one of three different chromosomes (--SEA, n = 18; --FIL, n = 3; --MED, n = 1).
As described before, we also determined whether up-regulation of eNME4 was caused by an increase in expression from one or both alleles. We found highly skewed patterns of NME4 expression in all informative individuals carrying these deletions (see Fig. 2B). Again, by generating somatic cell hybrids where material was available, we were able to link the nucleotide at the NME4 SNP to the deletion, and found that the up-regulated allele was always on the chromosome from which the α-globin gene deletion had occurred. This confirms that not only is the expression of eNME4 under the influence of the MCS-Rs lying ≈300 kb away, but the level of expression dramatically increases as the number of competing α-globin promoters in cis is reduced.
Identification of Long-Range Interactions Between NME4 and the α-Globin Regulatory Elements in the Absence of the α-Globin Genes.
Because expression of NME4 increases (from the affected “--” allele) in the absence of the α-globin promoters, it seemed possible that the previously noted rare interactions between MCS-R2 and NME4 in erythroid cells from nonthalassemic individuals (see above) might be increased in frequency, and therefore more readily detectable. To test this, we carried out 4C analysis on erythroid material from individuals heterozygous for deletions of both α-globin genes (--/αα), a common cause of α thalassemia in some regions. In this material, the interaction with the α-globin genes (from the intact [αα] chromosome) remains (see Fig. 3A Lower). However, now (observed in two independent experiments) there is clearly a more prominent interaction between MCS-R2 and NME4. This interaction is not restricted to the erythroid-specific promoter of NME4 (contained in intron 3) but is equally spread across the full length of the gene (see Fig. 3A, zoomed section). In addition to NME4, we observed a number of other interacting fragments associated with various genes along this region. Expression of these genes was analyzed in erythroid material, and they were found either not to be expressed in red blood cells (RHDBF1, MPG, MRPL28, DECR2) or did not show significantly different expression between controls and --/αα erythroid material (e.g., LUC7L) (Fig. S4). At present we do not fully understand the functional significance of these interactions; however, we hypothesize that they may represent structural interactions rather than having a functional effect on gene expression.
The interaction between MCS-R2 and NME4 identified by the 4C technique in --/αα erythroid material resulted in a ≈200-fold enrichment of NME4 DpnII-ligated DNA (see Fig. 3A, QPCR). If this interaction between MCS-R2 and NME4 occurs predominantly on the allele in cis with the deletion (as set out above), this allele should be overrepresented in the 4C-amplified material. To test this, we used the same technique as for the allele-specific expression (pyrosequencing of SNP rs14293 in exon 4 of NME4; the amplicon is contained entirely within a single DpnII fragment) on both genomic DNA and 4C-amplified DNA from an --/αα individual informative for this SNP (Fig. 3B). While the genomic DNA has equal representation of both NME4 alleles (see Fig. 3B Upper), DNA obtained from the MCS-R2 4C-amplified DNA is highly skewed toward one allele (97:3) (see Fig. 3B Lower). This is the NME4 allele in cis to the α-globin gene deletion, confirming that removal of the α-globin genes increases the interaction between MCS-R2 and NME4, which can now be readily detected by the 4C analysis.
Discussion
Here we have addressed the general question of how activation of a highly specialized gene cluster located within a gene-rich region of the genome affects expression of other genes in the shared chromosomal environment. By studying one such locus in detail, we have shown that although a region of conserved synteny, spanning ≈120 kb of the human α-globin cluster, is sufficient to obtain optimal tissue- and developmental stage-specific expression of the α-globin genes, this does not delimit the full extent over which globin gene activation exerts an effect. The process of α-globin activation results in significant effects on other apparently unrelated genes (C16orf35 and NME4). It is interesting to note that although C16orf35 lies adjacent to the α-globin cluster and contains the major erythroid-specific regulatory elements, NME4 lies 300 kb away from these elements. The α-globin regulatory elements appear to bypass, and therefore not affect expression, of at least five other genes contained within this chromosomal region, and yet specifically up-regulates NME4. This may be related to the chance appearance of an erythroid-specific element in this gene (see below).
It is also of general interest that this 500-kb region contains numerous potential enhancer blocker or boundary elements (DNaseI hypersensitive sites associated with binding of CTCF) (Fig. S5), which clearly do not act as such, at least in erythroid cells. These observations (and others, for example refs. 15 and 16) question the models of the genome in which genes are thought to be compartmentalized and insulated from activation or repression by the activity of nearby, unrelated cis elements and genes.
The mechanism by which expression of a nearby gene (C16orf35, which contains the α-globin MCS-R elements) is up-regulated is not clear. Others have suggested that such “bystander” activation simply results from location of a gene within an active chromatin domain (17), although the details of this mechanism have not been addressed. By contrast, we have shown that activation of NME4 (300 kb away) results from a direct physical interaction between multiprotein complexes assembled at an erythroid-like element at NME4 and at the α-globin MCS-R elements, as observed for other long-range enhancer/promoter interactions (11, 18).
It is interesting that the influence of MCS-R2 on NME4 expression is modulated by the number of α-globin genes in cis. This finding was most rigorously tested by the fact that the expression of eNME4 was up-regulated in individuals, each carrying one of three independent deletions that remove both α-globin genes. As the only genetic feature these individuals share is the deletion of the α-globin genes, it is compelling evidence that this is indeed the causative factor in this up-regulation.
Previous studies have suggested that closely linked promoters may compete for the activity of a shared enhancer (19, 20). This principle also seems to apply to the α-globin cluster where, from chromosomes containing between one (-α) to five (ααααα) identical α genes, the α-globin output does not increase in a linear fashion but appears to be limited by the available interaction with a single MCS-R (or complex formed by more than one MCS element): the most proximal gene competing most efficiently and the most distal gene competing least (reviewed in ref. 21). Also consistent with this competitive model, we recently demonstrated that a regulatory SNP lying between MCS-R2 and the normal α promoters (αα), which creates a new erythroid promoter, appears to out-compete the more distal α-globin promoters for access to MCS-R2, thereby causing α-thalassemia (22). These principles also seem to apply to the interaction between the α-globin regulatory elements and NME4, the observations being most readily explained by competition between this gene and the α globin promoters for the MCS elements. This leads to a situation in which polymorphic variation in the number of α-globin genes radically affects expression of an unrelated gene located 300 kb away.
An important question is whether the apparently unrelated genes have any significant biological function. It has been previously argued that activated bystander genes have arisen by chance, and have no known biological function (17, 23). Although the role of C16orf35 is currently unknown, a promoter knockout model of this gene has no obvious additional effects on erythropoiesis. In the case of NME4, in the mouse this gene is located on a separate chromosome from the α-globin cluster, and its expression is not up-regulated in murine erythroid cells (Fig. S3C). This suggests that NME4 does not play a general role in erythropoiesis, but is an example of a gene that (in humans) has become activated by the MCS-R elements by chance. Clearly this type of mechanism, in general, could play an important role in the recruitment of novel genes to existing biological circuits during evolution.
It seems likely that the principles established at this well-characterized locus will apply to many other regions of the genome. The adventitious activation of apparently unrelated genes clearly provides a potential pitfall in the interpretation of global gene expression studies and expression quantitative trait loci data, as changes in the expression of some genes may play no role in the processes being studied. In particular, our findings are relevant to the interpretation of CNV data. Recent genome wide studies have shown that not only do CNVs account for a large proportion of heritable variation in gene expression, but surprisingly up to 50% of this variation is because of genes lying beyond the CNV interval (24). Variation in the number of α-globin genes, generated by frequent homologous recombination between duplicated sequences, provided one of the first examples of a polymorphic CNV in human populations (21). Here we have shown how such CNVs may alter the expression of coordinately regulated genes, outside the deletions or insertions, across hundreds of kilobases of a chromosome, almost certainly by altering competition between promoters for a shared regulatory element (Fig. 4). It seems likely that this mechanism will explain some elusive diseases and phenotypes that are not currently explained by the gain or loss of genes that physically lie within associated CNVs.
Fig. 4.
Schematic representation of the effect of activated α-globin MCS-Rs, and structural polymorphisms, on the expression of surrounding genes. (A) In erythroid cells the α-globin MCS-Rs (black bars) up-regulate the α globin genes (red boxes), and also C16orf35 (yellow box; mechanism unknown) and NME4 (blue box; via a physical interaction in cis). (B) Variation in the number of α-globin genes (shown in this example as deletion of both adult α-globin genes) results in variation in the expression of NME4 through competition for the shared enhancer element. Boxes represent genes as shown in Fig. 1A; gray boxes are unaffected genes. The light gray area represents the region of conserved synteny across the α-globin locus. Arrowhead lines indicate interactions, thickness of the line represents frequency of the interaction.
Methods
Patients.
The patients studied were ascertained by reduced hematological indices. Deletions were confirmed with either Multiplex Ligation-dependent Probe Amplification or Southern blotting. Controls are individuals with no evidence of hematological defects. Consent was obtained in accordance with standard ethics approval guidelines. All individuals are homozygous for the common haplotype surrounding NME4.
Cell Types.
Erythroid cells were obtained using a two-phase culture system as previously described (25). Red blood cells were purified from whole blood (according to ref. 26), for expression analysis. hES cells were obtained from the H1 HES cell line, grown according to manufacturer's instructions. Nonerythroid cells were either primary T lymphocytes (ChIP, Northern analysis), or EBV-transformed B-lymphocyte cell lines (expression analysis, 4C analysis).
Expression Analysis.
For all cell types, RNA was extracted with Tri reagent as per manufacturer's instructions (Sigma). For Northern blots, 20 μg of total RNA were assayed, using the NorthernMax-Gly kit as per manufacturer's instructions (Ambion). For real-time expression analysis, RNA was DNaseI treated (Ambion) and cDNA was generated with SuperScript III (Invitrogen) as per manufacturer's instructions. Real-time PCR assays were obtained from either Applied Biosystem's Assay-On-Demand resource, or designed with Primer Express software. Both +RT and −RT templates were analyzed to detect genomic contamination. Expression in hES and erythroid cells was calculated relative to a control sequence in the 18S ribosomal RNA gene (Eurogentec RT-CKFT-18S). Expression in red blood cells was calculated relative to CD71, to correct for stage of erythropoiesis. For the latter, the mean of expression of each gene in the control samples is set to 100%, and expression in the deletion patient samples is expressed relative to this mean for each gene analyzed. For details of assays, real time primers and probes, and Northern probes see Tables S1–S3.
Statistical Analysis.
Significance of differences in expression between control and deletion samples was calculated with a two-tailed Student's t test assuming nonequal variance. Significance in variation of allele-specific expression was calculated with an f test.
ChIP and ChIP-chip.
ChIP was performed as previously described (5). Briefly, for one immunoprecipitation, 1 × 107 cultured primary human erythroblasts or T lymphocytes were cross-linked with 1% formaldehyde for 10 min. DNA was sheared by sonication to fragments under 500 base pairs. Antibodies used were H3Ac (06–599, Upstate), H4ac (06–866, Upstate), H3K4me2 (07–030, Upstate), H3K4me3 (ab8580, Abcam), CTCF (07–729, Upstate), GATA1 (sc1234, Santa Cruz), and SCL (gifted by C. Porcher). ChIP DNA was analyzed by real time PCR, calculated relative to input, and normalized to β-actin promoter. For details of primers and probes see Table S6. ChIP-chip was performed by hybridization to a custom α-globin tiled microarray as previously described (5), and enrichment was validated with real-time PCR.
Modified Circular Chromosome Conformation Capture.
For 4C, 1 × 107 cells were fixed with 2% formaldehyde in medium for 10 min at room temperature with agitation. Following quenching with glycine, cells were lysed (10 mM Tris pH8.0, 10 mM NaCl, 0.2% Nonidet P-40, 1× proteinase inhibitor) and resuspended in 1× DpnII digestion buffer (NEB), 0.3% SDS, 2% Triton X-100, and 500U DpnII at 37 °C overnight with shaking. The enzyme was inactivated at 65 °C for 25 min with shaking, and the total volume resuspended in 7 ml 1× ligation buffer with 1% Triton X-100. Following incubation at 37 °C for 1 h, the samples were cooled on ice for 2 min and 240 units of high-concentration T4 DNA ligase (Fermentas) was added. Following incubation overnight at 16 °C, 300 μg of proteinase K was added and cross-links reversed at 65 °C overnight with rotation. The samples were treated with 15 μg RNase (Roche) at 37 °C for 30 min. Following phenol/chloroform extraction and ethanol precipitation, DNA was resuspended in 500 μl 1× ligation buffer and 60 units high-concentration T4 DNA ligase (Fermentas) and incubated for 2 h at 16 °C with shaking. Following phenol/chloroform extraction and ethanol precipitation, DNA was resuspended in 100 μl water, of which 10 μl was used as template in Advantage-GC PCR (Clontech) as per manufacturer's instructions. Primer sequences can be found in Table S6. The resultant amplified DNA was ethanol precipitated and resuspended in 20 μl water, of which 5 μl was hybridized to a customized α globin tiled microarray using sonicated genomic DNA as input, as previously described (5). Enrichment of NME4 in 4C material was quantitated by real-time PCR, and normalized to an unenriched amplicon.
Pyrosequencing.
The ratio of expression of allele-specific transcripts and 4C-amplified DNA of NME4 was ascertained by pyrosequencing. Primer and dispensation information is contained in Table S4 and S5. Peak height is directly proportional to the amount of nucleotide incorporated. Analysis was performed in duplicate and an average obtained.
Sequence Information.
All human sequence positions correspond to the International Human Genome Sequencing Consortium Human March 2006 (hg18) Assembly sequence. The NME4 gene corresponds to sequence position chr16:387193–390755; eNME4 corresponds to sequence position chr16:389609–390755.
Supplementary Material
Acknowledgments.
We thank the clinicians and the members of the families studied for their participation, particularly Dr. C. L. Harteveld (Leiden University Medical Center, The Netherlands), Dr. D. Rund (Hadassah University Hospital, Israel), Dr. D. Filon (Hadassah Medical Center, Israel), Dr. H. Frischknecht (Institute for Medical & Molecular Diagnostics Ltd, Switzerland), Dr. S. L. Thein (King's College Hospital, United Kingdom) Dr. N. Gattermann (Heinrich-Heine University, Germany), Dr. J. Finlayson (QEII Medical Centre, Western Australia), and Dr. R. Hutch (Cork, Ireland). We thank Dr. C. Porcher for the kind gift of the SCL antibody, Dr. I. Dunham for assistance with microarrays, the Computational Biology Research Group, Oxford University, for bioinformatic support, and Prof. W. Wood for critical reading of the manuscript. This work was supported by the Medical Research Council, the Wellcome Trust and the National Institute for Health Research Biomedical Research Centre Program. K.M.L. was supported by an Oxford Nuffield Medical Fellowship, Oxford University.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0909331106/DCSupplemental.
References
- 1.Kim TH, et al. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kleinjan DA, van Heyningen V. Long-range control of gene expression: Emerging mechanisms and disruption in disease. Am J Hum Genet. 2005;76:8–32. doi: 10.1086/426833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dean A. On a chromosome far, far away: LCRs and gene expression. Trends Genet. 2006;22:38–45. doi: 10.1016/j.tig.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 4.Hughes JR, et al. Annotation of cis-regulatory elements by identification, subclassification, and functional assessment of multispecies conserved sequences. Proc Natl Acad Sci USA. 2005;102:9830–9835. doi: 10.1073/pnas.0503401102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Gobbi M, et al. Tissue-specific histone modification and transcription factor binding in alpha globin gene expression. Blood. 2007;110:4503–4510. doi: 10.1182/blood-2007-06-097964. [DOI] [PubMed] [Google Scholar]
- 6.Wallace HA, et al. Manipulating the mouse genome to engineer precise functional syntenic replacements with human sequence. Cell. 2007;128:197–209. doi: 10.1016/j.cell.2006.11.044. [DOI] [PubMed] [Google Scholar]
- 7.Milon L, et al. The human nm23–H4 gene product is a mitochondrial nucleoside diphosphate kinase. J Biol Chem. 2000;275:14264–14272. doi: 10.1074/jbc.275.19.14264. [DOI] [PubMed] [Google Scholar]
- 8.Milon L, et al. nm23–H4, a new member of the family of human nm23/nucleoside diphosphate kinase genes localised on chromosome 16p13. Hum Genet. 1997;99:550–557. doi: 10.1007/s004390050405. [DOI] [PubMed] [Google Scholar]
- 9.Higgs DR, Wood WG. Long-range regulation of alpha globin gene expression during erythropoiesis. Curr Opin Hematol. 2008;15:176–183. doi: 10.1097/MOH.0b013e3282f734c4. [DOI] [PubMed] [Google Scholar]
- 10.Sexton T, Bantignies F, Cavalli G. Genomic interactions: Chromatin loops and gene meeting points in transcriptional regulation. Semin Cell Dev Biol. 2009;20:849–855. doi: 10.1016/j.semcdb.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 11.Vernimmen D, De Gobbi M, Sloane-Stanley JA, Wood WG, Higgs DR. Long-range chromosomal interactions regulate the timing of the transition between poised and active gene expression. EMBO J. 2007;26:2041–2051. doi: 10.1038/sj.emboj.7601654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vernimmen D, et al. Chromosome looping at the alpha globin locus is mediated via the major upstream regulatory element (HS-40) Blood. 2009;114:4253–4260. doi: 10.1182/blood-2009-03-213439. [DOI] [PubMed] [Google Scholar]
- 13.Zhao Z, et al. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat Genet. 2006;38:1341–1347. doi: 10.1038/ng1891. [DOI] [PubMed] [Google Scholar]
- 14.Simonis M, et al. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C) Nat Genet. 2006;38:1348–1354. doi: 10.1038/ng1896. [DOI] [PubMed] [Google Scholar]
- 15.Kokubu C, et al. A transposon-based chromosomal engineering method to survey a large cis-regulatory landscape in mice. Nat Genet. 2009;41:946–952. doi: 10.1038/ng.397. [DOI] [PubMed] [Google Scholar]
- 16.Bender MA, et al. Flanking HS-62.5 and 3′ HS1, and regions upstream of the LCR, are not required for beta-globin transcription. Blood. 2006;108:1395–1401. doi: 10.1182/blood-2006-04-014431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cajiao I, Zhang A, Yoo EJ, Cooke NE, Liebhaber SA. Bystander gene activation by a locus control region. EMBO J. 2004;23:3854–3863. doi: 10.1038/sj.emboj.7600365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Laat W, Grosveld F. Spatial organization of gene expression: The active chromatin hub. Chromosome Res. 2003;11:447–459. doi: 10.1023/a:1024922626726. [DOI] [PubMed] [Google Scholar]
- 19.Choi OR, Engel JD. Developmental regulation of beta-globin gene switching. Cell. 1988;55:17–26. doi: 10.1016/0092-8674(88)90005-0. [DOI] [PubMed] [Google Scholar]
- 20.Dillon N, Trimborn T, Strouboulis J, Fraser P, Grosveld F. The effect of distance on long-range chromatin interactions. Mol Cell. 1997;1:131–139. doi: 10.1016/s1097-2765(00)80014-3. [DOI] [PubMed] [Google Scholar]
- 21.Higgs DR, et al. A review of the molecular genetics of the human alpha-globin gene cluster. Blood. 1989;73:1081–1104. [PubMed] [Google Scholar]
- 22.De Gobbi M, et al. A regulatory SNP causes a human genetic disease by creating a new transcriptional promoter. Science. 2006;312:1215–1217. doi: 10.1126/science.1126431. [DOI] [PubMed] [Google Scholar]
- 23.Spitz F, Gonzalez F, Duboule D. A global control region defines a chromosomal regulatory landscape containing the HoxD cluster. Cell. 2003;113:405–417. doi: 10.1016/s0092-8674(03)00310-6. [DOI] [PubMed] [Google Scholar]
- 24.Stranger BE, et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–853. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pope SH, Fibach E, Sun J, Chin K, Rodgers GP. Two-phase liquid culture system models normal human adult erythropoiesis at the molecular level. Eur J Haematol. 2000;64:292–303. doi: 10.1034/j.1600-0609.2000.90032.x. [DOI] [PubMed] [Google Scholar]
- 26.Beutler E, West C, Blume KG. The removal of leukocytes and platelets from whole blood. J Lab Clin Med. 1976;88:328–333. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.