Abstract
The hexameric membrane-spanning ATP-dependent metalloprotease FtsH is universally conserved in eubacteria, mitochondria, and chloroplasts, where it fulfills key functions in quality control and signaling. As a member of the self-compartmentalizing ATPases associated with various cellular activities (AAA+ proteases), FtsH converts the chemical energy stored in ATP via conformational rearrangements into a mechanical force that is used for substrate unfolding and translocation into the proteolytic chamber. The crystal structure of the ADP state of Thermotoga maritima FtsH showed a hexameric assembly consisting of a 6-fold symmetric protease disk and a 2-fold symmetric AAA ring. The 2.6 Å resolution structure of the cytosolic region of apo-FtsH presented here reveals a new arrangement where the ATPase ring shows perfect 6-fold symmetry with the crucial pore residues lining an open circular entrance. Triggered by this conformational change, a substrate-binding edge beta strand appears within the proteolytic domain. Comparison of the apo- and ADP-bound structure visualizes an inward movement of the aromatic pore residues and generates a model of substrate translocation by AAA+ proteases. Furthermore, we demonstrate that mutation of a conserved glycine in the linker region inactivates FtsH.
Keywords: AAA+ protease, conformational rearrangement, pore residue, FtsH, metalloprotease
The ATPases associated with various cellular activities (AAA+) family comprises a large group of proteins which share a common ATPase module, carrying the Walker A and B and other conserved sequence motifs (1, 2). They function as oligomers and usually form hexa- or heptameric rings, whereas in some cases oligomerization is ATP-dependent. AAA+ proteins are present in all kingdoms of life and participate in a wide range of different actions such as protein disaggregation, complex remodeling, and protein degradation (reviewed in refs. 3–5). In these processes, the energy of ATP is converted into a mechanical force by large conformational rearrangements. Until now, information on these immense structural changes is sparse; x-ray studies on HslU (6) and p97 (7) revealed only relatively small changes between the conformations of apo, ADP, and ATP state. In contrast, electron microscopic studies of p97 (8) and of the chaperone ClpB (9) displayed very large conformational rearrangements of the AAA domains.
The membrane-bound metalloprotease FtsH (10, 11) is a homohexameric complex that carries, contrary to most other AAA+ proteases, the AAA and proteolytic domain on the same polypeptide chain. The AAA domain bears the Walker A and B motifs necessary for nucleotide binding and hydrolysis; the second region of homology (SRH), which carries conserved arginine residues (“arginine fingers”) for oligomerization and nucleotide hydrolysis; and the conserved FGV pore motif required for substrate recognition and translocation. The protease domain exhibits the zinc-binding HEXXH finger print. The N-terminal AAA domain is preceded by 2 transmembrane helices flanking a small periplasmic region, thereby locating the largest part of the protein complex in the cytoplasm. FtsH is universally conserved in eubacteria, chloroplasts, and mitochondria as it has an essential function in protein turnover and processing. Of the 5 energy-dependent AAA+ proteases in Escherichia coli, namely Lon, HslVU, ClpXP, ClpAP, and FtsH, it is the only essential one, and malfunctioning of its human homolog paraplegin causes a drastic form of hereditary spastic paraplegia (12, 13). FtsH and its orthologs play a key role in quality and regulatory control within the cell by degrading a unique subset of substrates. On the one hand, the protease is able to identify and degrade nonfunctional or damaged membrane proteins by pulling them out of the lipid bilayer, followed by further substrate unfolding and translocation into the proteolytic chamber. On the other hand, certain cytosolic signaling proteins can be classified as regulatory control substrates. This group comprises proteins such as the heat-shock factor σ32, the lambda phage transcriptional activator CII (14), and LpxC (15), a key enzyme controlling the amount of membrane lipopolysaccharides.
The mechanism of AAA+ proteases is currently the subject of intense investigation (reviewed in refs. 3, 4). However, due to the lack of structural data, there is a considerable gap in our understanding of the way of function. Currently, crystal structures of only one complete AAA+ protease (HslVU) are available, the very first such structure was the one from E. coli reported by Bochtler et al. (16), later those from Haemophilus influenzae (17) and another E. coli structure (18) were published (reviewed in ref. 19). Crystal structures of constructs spanning most of the cytosolic region, thus containing AAA and protease domains, of FtsH from Thermotoga maritima (20) and Thermus thermophilus (21) are also available. Many other AAA+ proteins have been crystallized only in the monomeric state and 6-fold-symmetric rings have been modeled frequently (22, 23). Recent biochemical data, however, have demonstrated that at least in some nucleotide states the symmetry has to be lower, e.g., only maximally 4 ATP molecules bind to 1 hexameric ClpX or PAN or HslU particle (24–26).
A conserved ΦVG motif (Φ indicating an aromatic amino acid) has been identified as mediator between ATP hydrolysis and substrate unfolding (27–33). The aromatic side chains of this motif are thought to grab a hydrophobic degradation tag and are implied to perform a power stroke inward movement, driven by conformational rearrangements of the AAA+ domains.
Earlier, we have reported the crystal structure of the cytosolic region of T. maritima FtsH, representing the ADP bound state and revealing a hexameric molecule composed of 2 rings (20). The protease ring shows the expected C6 symmetric assembly where the monomers possess a new fold consisting of 8 α-helices and a short 2-stranded antiparallel β-ribbon. Surprisingly, the AAA ring exhibited C2 symmetry only, leading to a model of protein unfolding and translocation where some of the crucial aromatic pore residues (Phe-234) move inwards. An alternative structure of a glycine-mutant of FtsH from T. thermophilus was published later, revealing a C3-symmetric AAA ring (21), suggesting a pair-wise mechanism of substrate translocation.
To gain more insight into the ATPase cycle and the unfolding and translocation mechanism, high-resolution structures of different nucleotide states are needed. Therefore, we have crystallized the apo-state and report the results here. The new structure reveals the large conformational rearrangements and pore residue movements necessary for protein unfolding and translocation and uncovers an active-site switch that is important for substrate fixation.
Results
The apo-Form of (Δtm)FtsH Is a C6 Symmetrical Hexamer.
To obtain nucleotide-free (Δtm)FtsH, we mutated the lysine in the Walker A motif to alanine (K207A). This mutation prevents nucleotide binding (2). The mutant crystallizes in spacegroup P622 with 3 monomers per asymmetric unit. Molecular replacement calculations failed to reveal the positions of the AAA domains, therefore, the structure was solved by selenomethionine single wavelength anomalous dispersion (SAD). Nineteen out of 27 seleniums were found, 6 of them in the 3 proteolytic domains (2 methionines each), the others belong to the AAA domains (Fig. S1). The resulting experimental electron density map allowed placement of all proteolytic and AAA domains. The latter have weaker density (Fig. S2), but the localization and arrangement is unambiguous.
The 3 monomers in the asymmetric unit belong to 2 hexameric particles, which are placed on the 6-fold and the 3-fold crystallographic axes. Both hexamers show no significant deviations from each other, i.e., both possess perfect 6-fold symmetry in both the AAA and the protease ring (Fig. 1). While the protease ring in the apo- and ADP-bound structures is essentially the same, very large conformational rearrangements can be observed within the AAA-ring. The transition from the ADP to the apo-form involves changes in the inter-domain angle between protease and AAA moieties by some 40 to 60° (Fig. 2), thus causing movements of the conserved pore residues Phe-234 of the FGV pore motif of more than 45 Å (Fig. 2A). These rearrangements of the AAA domains are largely of rigid body nature. A smaller movement takes place within the individual AAA domains itself. Due to the loss of the nucleotide, the angle between N-terminal “wedge” subdomain and C-terminal helical subdomain increases by about 20° (Fig. 2B). This change concurs with earlier studies (7, 34). This more open conformation of the AAA domain is incompatible with the C2-symmetric packing observed in the ADP-bound state.
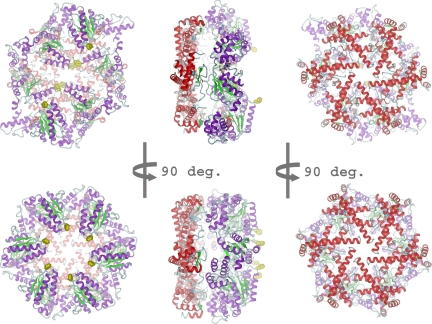
Fig. 1.
Cartoon representation of ADP-FtsH (Top) and apo-FtsH (Bottom) with view onto AAA ring (Left), side (Middle) and protease (Right). The protease ring is shown in red and the AAA ring in magenta and green. Zinc ions are displayed as cyan spheres, the pore residues Phe-234 in yellow space-filling mode. Carbon atoms of ADP and TAPI are shown in gray. (A) ADP-bound state, the AAA ring has C2 symmetry. (B) Apo-state, the AAA ring has C6 symmetry.
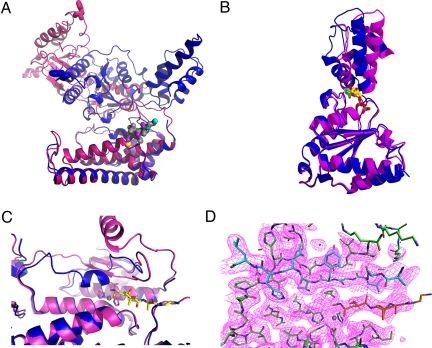
Fig. 2.
(A) Overlay of monomers in the ADP (blue) and apo-state (magenta). The pore phenylalanine Phe-234 is shown as sticks. The distance of these residues between ADP and apo-form is about 45 Å. Zn+ is shown as yellow sphere, the hydroxamate inhibitor as stick model with gray carbon atoms. The protease domain (bottom) was used as reference for the alignment. (B) Overlay of the AAA domain only. The angle between “wedge” subdomain and α-helical subdomain increases by about 20° in the apo form (red) compared to the ADP form (blue). (C) Overlay of the proteolytic active site in the ADP (blue) and apo-state (magenta). The active-size switch (residues 450 to 460) adopts a helical conformation in the ADP state and forms a β-strand in the apo-state. TAPI is displayed with yellow carbon atoms, Zn2+ as gray sphere. (D) Active site with electron density (1.0 σ) above mean. Zn2+ is shown as gray sphere, the active-site switch (residue 450 to 460) is depicted in cyan. The TAPI inhibitor is shown with orange carbon atoms.
An Active-Site Switch Formed by a Substrate-Binding β Strand.
A striking feature of all FtsH structures published so far is the small amount of β structure in the protease domain. The edge strands of β sheets in endo-proteinases, especially in zincins with their HEXXH motif that is also present in FtsH, are usually used for fixing the substrate polypeptide chain in an extended conformation around the scissile peptide bond by binding the substrate as an additional antiparallel β-strand to these edges. In the apo-form of FtsH described here, the stretch comprising amino acids 450 to 460 complements the short 2-stranded β-ribbon already present in the ADP form by an additional antiparallel strand. This segment is disordered in 4 of the 6 independent monomers of the ADP state, and an α-helical conformation is observed in the remaining 2 (Fig. 2C). The cocrystallized hydroxamic acid inhibitor TAPI binds in an extended parallel conformation to this newly formed edge strand (Fig. 2 C and D). The formation of this edge strand is independent of the presence of inhibitors or substrates, because it is also observed in crystals of the K207A mutant obtained in the absence of these (e.g., the selenium-labeled crystal use for structure determination).
The G404L Mutant Is Monomeric and Inactive.
To improve crystal quality by restricting conformational flexibility, Suno and coworkers mutated a glycine residue (G399 in T. thermophilus) in the linker between AAA and protease domain to leucine (21). This glycine is completely conserved among all FtsH homologs (Fig. S3). To examine the effects of this mutation more closely, we introduced the same mutation in T. maritima FtsH, replacing Gly-404 by a leucine. Contrary to the wild type, this mutant elutes exclusively as a monomer from the gel filtration column (Fig. 3C). Activity measurements show that the T. maritima G404L mutant is inactive in the ATPase assay and possesses only very low proteolytic activity (Fig. 3 A and B).
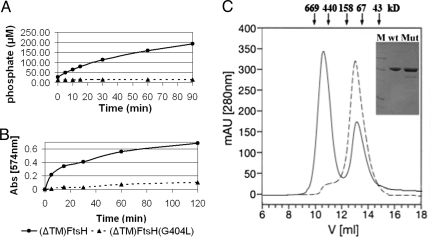
Fig. 3.
Activity assays and oligomeric state for the G404L mutant. (A) ATPase activities of wild-type Δ(tm)FtsH (solid line, ●) and Δ(tm)G404L (dashed line, ▴). (B) Resorufin casein proteolytic assay, symbols as in (A). (C) Gel filtration profile of wild-type Δ(tm)FtsH (solid line) and G404L (dashed line). The insert shows the SDS/PAGE with molecular weight markers in lane M (66, 45, 24, 14, and 12 kDa), wild-type Δ(tm)FtsH (wt) and G404L (Mut).
Discussion
The mechanism through which AAA+ proteases unfold and translocate their substrates into the proteolytic chambers is currently a topic of intense discussion. A gap in structural information has been a hindrance to achieve a deeper understanding. The 2 crystal structures of T. maritima FtsH in its ADP- and apo-state provide a basis for the movements occurring between the different nucleotide states. The protease ring is the stationary component and nucleotide-driven movements of the AAA domains accomplish substrate unfolding and translocation by pulling the polypeptide chain against and through the central pore. The 2 structures available now illuminate the transition from the ADP- to the apo-state. The ATP-loaded hexamer might look roughly similar as the nucleotide-free, but is presumably not 6-fold symmetric anymore, because it is unlikely that 6 ATP bind simultaneously and ATPs are bound with different affinities (24–26). Furthermore, the nucleotide induced subdomain closure within the AAA moiety would lead to a more compact, closed appearance. However, the location of the pore residues could still be similar to the apo-state and the inward movement of 4 of the 6 phenylalanines, as seen in the ADP-bound conformation (Fig. 4) (20), describes the actual “pulling” on the substrate polypeptide chain. The currently most accepted hypothesis is that the energy-stroke of ATP hydrolysis is converted into a mechanical force by conformational changes involving the pore residues. Atomic force microscopy experiments show that typically a force of a few hundred pico-Newtons (pN) is needed to unfold many proteins. Estimating the energy of ATP hydrolysis under cellular conditions as 45 kJ/mol, a movement of 50 Å can generate a force of about 150 pN. While some cryo-EM reconstructions of different nucleotide states can be interpreted in this way, x-ray structures have failed so far to demonstrate conformational changes in this order of magnitude. The structure presented here reveals for the first time nucleotide-dependent changes in this order of magnitude (see Movie S1), whereby hydrolysis does not take place simultaneously in all subunits (24, 35). Our model is in agreement with the 1:1 enzyme:substrate stoichiometry that has been observed in at least 2 examples of AAA+ proteins (24, 25). On the other hand, the proposed alternative model (21) would principally allow the simultaneous binding of 3 substrate molecules. We could also provide evidence here that the low-resolution model proposed by Suno et al. (21) suffers from an underlying nonconservative mutation that, at least for T. maritima FtsH, inactivates the protein.
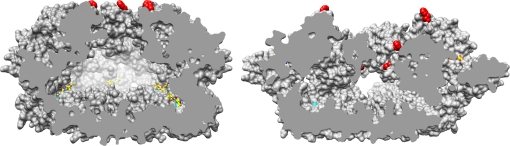
Fig. 4.
Cut-away surface representation of apo (Left) and ADP-bound state (Right). Pore residues Phe-234 are shown in red. The inward movement and staggered arrangement of these residues in the ADP state, after power stroke, is clearly visible. Zinc ions are indicated as cyan spheres, ADP and TAPI carbon atoms are shown in yellow.
The structure of apo-FtsH presented here together with the fully ADP-loaded structure presented earlier (20) provides for the first time a high-resolution image of the movement of the crucial pore residues. We propose that this mechanism is universal for all AAA+ proteins—substrate processing is achieved by pulling it through the central pore.
Experimental Procedures
Cloning, Protein Expression, and Purification.
(Δtm)FtsH (amino acids 147–610 of FtsH from T. maritima with K410L-K415A surface mutations) and its mutants were expressed and purified as described previously (20). Point mutations were introduced by a modified overlap-extension protocol (36). Mutation K207A was generated employing the primers K207A-for (5′-gtagggcctcccgggaccggtgcaacgctccttgcgagggc-3′) and K207A-rev (5′-gccctcgcaaggagcgttgcaccggtcccgggaggccctac-3′), while the G404L mutation was introduced with primers G404L-for (5′-gggtgatagccttgccggcaagaaagtcgaagctgatcagc-3′) and G404L-rev (5′-cgactttcttgccggcaaggctatcaccctgtctatggcttcc-3′).
Crystallization.
Crystals were obtained at 20 °C using the microbatch method by mixing equal volumes of protein (20 mg/ml) and crystallization buffer: 30% (wt/vol) PEG 400, 200 mM CaCl2, 100 mM Hepes (pH 7.5), 200 mM glycine, and 0.1–0.2% (wt/vol) low-melt agarose (FMC BioProduct). Crystals belong to the spacegroup P622 (compare Table 1). For crystallization of (Δtm)FtsH(K207A) bound to TAPI-0, 1 μl of 30 mM TAPI-0 (Calbiochem) was added to the crystallization set-up. For cryoprotection, crystals were transferred to crystallization buffer containing 5% (vol/vol) glycerol for about 1 min before flash-cooling in liquid nitrogen. Selenomethionine-incorporated protein was produced by the methionine biosynthesis inhibition method (37) and purified as the wild-type protein.
Table 1.
Data collection and refinement statistics
| Crystal | TAPI | SeMet |
|---|---|---|
| Unit cell | ||
| a, b, c (Å) | 190.64, 190.64, 152.21 | 191.20, 191.20, 152.63 |
| α, β, γ (degrees) | 90.0, 90.0, 120.0 | 90.0, 90.0, 120.0 |
| Data collection | ||
| Beamline | ID-29, ESRF, Grenoble | ID-29, ESRF, Grenoble |
| Wavelength (Å) | 0.987 | 0.987 |
| Resolution range (Å) | 34.0–2.60 (2.74–2.60) | 50–3.20 (3.37–3.20) |
| No. unique reflections | 50,229 | 27,700 |
| Redundancy | 10.9 (4.2) | 13.7 (14.3) |
| Completeness (%) | 99.7 (98.2) | 100.0 (100.0) |
| Rsym (%) | 7.9 (91.1) | 9.5 (44.2) |
| I/σ (I) | 20.6 (1.9) | 24.3 (6.5) |
| Refinement | ||
| Resolution range (Å) | 45.78–2.60 | |
| No. reflections work/test set | 48,222/1,986 | |
| No. atoms | ||
| Total | 9,999 | |
| Water molecules/Zn2+ | 0/3 | |
| TAPI | 87 | |
| R/R-free (%) | 25.0 (31.7)/30.1 (40.1) | |
| RMS bonds (Å) | 0.006 | |
| RMS angles (deg.) | 0.94 | |
| Ramachandran outliers* (%) | 4.2 |
*Ramachandran outliers according to ref. 44.
X-Ray Data Collection, Phasing, and Refinement.
X-ray data were collected at 100 K at beamlines X06SA (Swiss Light Source, Paul-Scherer-Institute, Villigen, Switzerland), and ID29 (European Synchrotron Radiation Facility, Grenoble, France) as well as at the National Light Source, Brookhaven National Laboratory, Upton, NY. Data collection statistics are given in Table 1. Diffraction intensities were processed and scaled with XDS (38) and MOSFLM/SCALA (39, 40). Selenium positions were determined employing SHELXD (41) and phases were computed by SHARP (42). Refinement was carried out using PHENIX (43). Ramachandran outliers were determined according to (44).
ATPase Assay.
ATPase activity measurements were performed using the EnzChek Phosphate Assay Kit from Invitrogen according to the method of Webb (45). 50 μl 20x reaction buffer (1M Tris-HCl (7.5), 20 mM MgCl2, 2 mM sodium azide), 200 μl 2-amino-6-mercapto-7-methylpurine riboside substrate solution (MESG), 10 μl purine nucleoside phosphorylase (1 unit), and 3 nmol of purified FtsH protein were mixed in a total volume of 1 ml. The mixture was incubated for 10 min at room temperature; 1 mM ATP was then added as experimental substrate and change of absorbance at 360 nm was recorded. Absolute ATPase activity was calculated following the manufacturer's instructions.
Protease Activity Assay.
Protease activity of purified FtsH proteins was determined as described before (20) using resorufin-labeled casein (Roche) as substrate. Samples were incubated at 50 °C. Reactions were terminated after 0, 5, 15, 30, 60, and 120 min by adding 5% trichloroacetic acid (wt/vol) and further incubation at 50 °C for 10 min. Samples were centrifuged at 12′000g for 5 min before 400 μl of supernatant were mixed with 600 μl assay buffer (0.5M Tris-HCl, pH 8.8). Protease activity was determined by measuring the absorbance at 574 nm.
Supplementary Material
Acknowledgments.
We thank Christoph Muller-Dieckmann and Gordon Leonard for their help during data collection at European Synchrotron Radiation Facility, Grenoble, France; Clemens Schulze-Briese at Swiss Light Source, Villigen, Switzerland; and Robert Sweet and the RapiData2006 course team, Brookhaven National Laboratory, Upton, NY. This workwas supported by the Swiss National Science Foundation, the Berner Hochschulstiftung, and the University of Bern.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3KDS).
This article contains supporting information online at www.pnas.org/cgi/content/full/0910708106/DCSupplemental.
References
- 1.Erzberger JP, Berger JM. Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu Rev Biophys Biomol Struct. 2006;35:93–114. doi: 10.1146/annurev.biophys.35.040405.101933. [DOI] [PubMed] [Google Scholar]
- 2.Hanson PI, Whiteheart SW. AAA+ proteins: Have engine, will work. Nat Rev Mol Cell Biol. 2005;6:519–529. doi: 10.1038/nrm1684. [DOI] [PubMed] [Google Scholar]
- 3.Baker TA, Sauer RT. ATP-dependent proteases of bacteria: Recognition logic and operating principles. Trends Biochem Sci. 2006;31:647–653. doi: 10.1016/j.tibs.2006.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sauer RT, et al. Sculpting the proteome with AAA(+) proteases and disassembly machines. Cell. 2004;119:9–18. doi: 10.1016/j.cell.2004.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Striebel F, Kress W, Weber-Ban E. Controlled destruction: AAA+ ATPases in protein degradation from bacteria to eukaryotes. Curr Opin Struct Biol. 2009;19:209–217. doi: 10.1016/j.sbi.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 6.Wang J. Nucleotide-dependent domain motions within rings of the RecA/AAA(+) superfamily. J Struct Biol. 2004;148:259–267. doi: 10.1016/j.jsb.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 7.DeLaBarre B, Brunger AT. Nucleotide dependent motion and mechanism of action of p97/VCP. J Mol Biol. 2005;347:437–452. doi: 10.1016/j.jmb.2005.01.060. [DOI] [PubMed] [Google Scholar]
- 8.Rouiller I, et al. Conformational changes of the multifunction p97 AAA ATPase during its ATPase cycle. Nat Struct Biol. 2002;9:950–957. doi: 10.1038/nsb872. [DOI] [PubMed] [Google Scholar]
- 9.Lee S, Choi JM, Tsai FT. Visualizing the ATPase cycle in a protein disaggregating machine: Structural basis for substrate binding by ClpB. Mol Cell. 2007;25:261–271. doi: 10.1016/j.molcel.2007.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Akiyama Y, Yoshihisa T, Ito K. FtsH, a membrane-bound ATPase, forms a complex in the cytoplasmic membrane of Escherichia coli. J Biol Chem. 1995;270:23485–23490. doi: 10.1074/jbc.270.40.23485. [DOI] [PubMed] [Google Scholar]
- 11.Ito K, Akiyama Y. Cellular functions, mechanism of action, and regulation of ftsh protease. Annu Rev Microbiol. 2005;59:211–231. doi: 10.1146/annurev.micro.59.030804.121316. [DOI] [PubMed] [Google Scholar]
- 12.Nolden M, et al. The m-AAA protease defective in hereditary spastic paraplegia controls ribosome assembly in mitochondria. Cell. 2005;123:277–289. doi: 10.1016/j.cell.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 13.Rugarli EI, Langer T. Translating m-AAA protease function in mitochondria to hereditary spastic paraplegia. Trends Mol Med. 2006;12:262–269. doi: 10.1016/j.molmed.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 14.Herman C, Thevenet D, D'Ari R, Bouloc P. The HflB protease of Escherichia coli degrades its inhibitor lambda cIII. J Bacteriol. 1997;179:358–363. doi: 10.1128/jb.179.2.358-363.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Inagawa T, et al. Defective plasmid partition in ftsH mutants of Escherichia coli. Mol Genet Genomics. 2001;265:755–762. doi: 10.1007/s004380100488. [DOI] [PubMed] [Google Scholar]
- 16.Bochtler M, et al. The structures of HsIU and the ATP-dependent protease HsIU-HsIV. Nature. 2000;403:800–805. doi: 10.1038/35001629. [DOI] [PubMed] [Google Scholar]
- 17.Sousa MC, et al. Crystal and solution structures of an HslUV protease-chaperone complex. Cell. 2000;103:633–643. doi: 10.1016/s0092-8674(00)00166-5. [DOI] [PubMed] [Google Scholar]
- 18.Wang J, et al. Crystal structures of the HslVU peptidase-ATPase complex reveal an ATP-dependent proteolysis mechanism. Structure. 2001;9:177–184. doi: 10.1016/s0969-2126(01)00570-6. [DOI] [PubMed] [Google Scholar]
- 19.Groll M, et al. Molecular machines for protein degradation. ChemBioChem. 2005;6:222–256. doi: 10.1002/cbic.200400313. [DOI] [PubMed] [Google Scholar]
- 20.Bieniossek C, et al. The molecular architecture of the metalloprotease FtsH. Proc Natl Acad Sci USA. 2006;103:3066–3071. doi: 10.1073/pnas.0600031103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suno R, et al. Structure of the whole cytosolic region of ATP-dependent protease FtsH. Mol Cell. 2006;22:575–585. doi: 10.1016/j.molcel.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 22.Lee S, et al. The structure of ClpB: A molecular chaperone that rescues proteins from an aggregated state. Cell. 2003;115:229–240. doi: 10.1016/s0092-8674(03)00807-9. [DOI] [PubMed] [Google Scholar]
- 23.Niwa H, et al. Hexameric ring structure of the ATPase domain of the membrane-integrated metalloprotease FtsH from Thermus thermophilus HB8. Structure. 2002;10:1415–1423. doi: 10.1016/s0969-2126(02)00855-9. [DOI] [PubMed] [Google Scholar]
- 24.Hersch GL, et al. Asymmetric interactions of ATP with the AAA+ ClpX6 unfoldase: Allosteric control of a protein machine. Cell. 2005;121:1017–1027. doi: 10.1016/j.cell.2005.05.024. [DOI] [PubMed] [Google Scholar]
- 25.Horwitz AA, et al. ATP-induced structural transitions in PAN, the proteasome-regulatory ATPase complex in Archaea. J Biol Chem. 2007;282:22921–22929. doi: 10.1074/jbc.M702846200. [DOI] [PubMed] [Google Scholar]
- 26.Yakamavich JA, Baker TA, Sauer RT. Asymmetric nucleotide transactions of the HslUV protease. J Mol Biol. 2008;380:946–957. doi: 10.1016/j.jmb.2008.05.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DeLaBarre B, Christianson JC, Kopito RR, Brunger AT. Central pore residues mediate the p97/VCP activity required for ERAD. Mol Cell. 2006;22:451–462. doi: 10.1016/j.molcel.2006.03.036. [DOI] [PubMed] [Google Scholar]
- 28.Erbse AH, et al. Conserved residues in the N-domain of the AAA+ chaperone ClpA regulate substrate recognition and unfolding. FEBS J. 2008;275:1400–1410. doi: 10.1111/j.1742-4658.2008.06304.x. [DOI] [PubMed] [Google Scholar]
- 29.Farbman ME, Gershenson A, Licht S. Role of a conserved pore residue in the formation of a prehydrolytic high substrate affinity state in the AAA+ chaperone ClpA. Biochemistry. 2008;47:13497–13505. doi: 10.1021/bi801140y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Graef M, Langer T. Substrate specific consequences of central pore mutations in the i-AAA protease Yme1 on substrate engagement. J Struct Biol. 2006;156:101–108. doi: 10.1016/j.jsb.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 31.Martin A, Baker TA, Sauer RT. Pore loops of the AAA+ ClpX machine grip substrates to drive translocation and unfolding. Nat Struct Mol Biol. 2008;15:1147–1151. doi: 10.1038/nsmb.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park E, et al. Role of the GYVG pore motif of HslU ATPase in protein unfolding and translocation for degradation by HslV peptidase. J Biol Chem. 2005;280:22892–22898. doi: 10.1074/jbc.M500035200. [DOI] [PubMed] [Google Scholar]
- 33.Schlieker C, et al. Substrate recognition by the AAA+ chaperone ClpB. Nat Struct Mol Biol. 2004;11:607–615. doi: 10.1038/nsmb787. [DOI] [PubMed] [Google Scholar]
- 34.Wang J, et al. Nucleotide-dependent conformational changes in a protease-associated ATPase HsIU. Structure. 2001;9:1107–1116. doi: 10.1016/s0969-2126(01)00670-0. [DOI] [PubMed] [Google Scholar]
- 35.Martin A, Baker TA, Sauer RT. Rebuilt AAA + motors reveal operating principles for ATP-fuelled machines. Nature. 2005;437:1115–1120. doi: 10.1038/nature04031. [DOI] [PubMed] [Google Scholar]
- 36.Zheng L, Baumann U, Reymond JL. An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res. 2004;32:e115. doi: 10.1093/nar/gnh110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van Duyne GD, et al. Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J Mol Biol. 1993;229:105–124. doi: 10.1006/jmbi.1993.1012. [DOI] [PubMed] [Google Scholar]
- 38.Kabsch W. In: International Tables for Crystallography. Rossmann MG, Arnold E, editors. Vol F. Dordrecht, The Netherlands: Kluwer Academic Publisher; 2001. pp. 730–734. [Google Scholar]
- 39.Evans P. Scaling and assessment of data quality. Acta Crystallogr D. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 40.Leslie AG. The integration of macromolecular diffraction data. Acta Crystallogr D. 2006;62:48–57. doi: 10.1107/S0907444905039107. [DOI] [PubMed] [Google Scholar]
- 41.Schneider TR, Sheldrick GM. Substructure solution with SHELXD. Acta Crystallogr D. 2002;58:1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- 42.Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated structure solution with autoSHARP. Methods Mol Biol. 2007;364:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- 43.Adams PD, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 44.Kleywegt GJ, Jones TA. Phi/psi-chology: Ramachandran revisited. Structure. 1996;4:1395–1400. doi: 10.1016/s0969-2126(96)00147-5. [DOI] [PubMed] [Google Scholar]
- 45.Webb MR. A continuous spectrophotometric assay for inorganic phosphate and for measuring phosphate release kinetics in biological systems. Proc Natl Acad Sci USA. 1992;89:4884–4887. doi: 10.1073/pnas.89.11.4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.