Abstract
Tumor cells are metabolically reprogrammed to fuel cell proliferation. Most transformed cells take up high levels of glucose and produce ATP through aerobic glycolysis. In cells exhibiting aerobic glycolysis, a significant fraction of glucose carbon is also directed into de novo lipogenesis and nucleotide biosynthesis. The glucose-responsive transcription factor carbohydrate responsive element binding protein (ChREBP) was previously shown to be important for redirecting glucose metabolism in support of lipogenesis in nonproliferating hepatocytes. However, whether it plays a more generalized role in reprogramming metabolism during cell proliferation has not been examined. Here, we demonstrated that the expression of ChREBP can be induced in response to mitogenic stimulation and that the induction of ChREBP is required for efficient cell proliferation. Suppression of ChREBP resulted in diminished aerobic glycolysis, de novo lipogenesis, and nucleotide biosynthesis, but stimulated mitochondrial respiration, suggesting a metabolic switch from aerobic glycolysis to oxidative phosphorylation. Cells in which ChREBP was suppressed by RNAi exhibited p53 activation and cell cycle arrest. In vivo, suppression of ChREBP led to a p53-dependent reduction in tumor growth. These results demonstrate that ChREBP plays a key role both in redirecting glucose metabolism to anabolic pathways and suppressing p53 activity.
Keywords: cancer biology, cell biology, metabolism
Many human tumors display a high rate of aerobic glycolysis, de novo fatty acid synthesis, and nucleotide biosynthesis (1, 2). Previous findings suggest that the increased glucose metabolism promotes lipogenesis and nucleotide biosynthesis, and enhances tumor cell growth and proliferation by providing essential synthetic and bioenergetic requirements (3–7). Although the metabolic alterations might not be initiating events in oncogenesis, recent success in blocking carcinogenesis by targeting tumor metabolism suggests that aerobic glycolysis plays an important role in sustaining tumor growth (5–11). Thus, understanding the genes that are required to support anabolic metabolism in normal and tumor cells may provide us with strategies for cancer therapy and prevention.
The basic helix–loop–helix leucine zipper (bHLH-LZ) transcription factor carbohydrate responsive element binding protein (ChREBP) is a critical mediator of glucose-dependent induction of glycolytic and lipogenic enzyme genes in metabolic tissues (12–17). Cellular level of nutrients (such as glucose and fatty acid) regulates the level and activity of ChREBP in hepatocytes and adipocytes (18, 19). In adults, expression of ChREBP is detectable in most tissues but is at its highest levels in liver (20). ChREBP-null mice show decreased glycolysis and lipogenesis as well as intolerance to dietary carbohydrate (20). Loss of ChREBP in leptin-null ob/ob mice results in alleviation of obesity (21). Although the function of ChREBP in hepatocytes has been extensively investigated, little is known about the role of ChREBP in proliferating or transformed cells.
In this study, the role of ChREBP in cancer cell proliferation and metabolism was investigated. We report that ChREBP was required for cell proliferation in HCT116 colorectal cancer cells and HepG2 hepatoblastoma cells. Suppression of ChREBP resulted in diminished aerobic glycolysis, de novo lipogenesis, and nucleotide biosynthesis, and was accompanied by stimulation of mitochondrial respiration. Thus, ChREBP is required to maintain high levels of aerobic glycolysis in these cells. Attenuation of ChREBP activated p53 and induced the expression of p53 target genes. Consistent with this, ChREBP inhibition reduced tumor growth in vivo via a p53-dependent mechanism. However, the loss of p53 resulted in only a partial rescue of the metabolic defect in ChREBP-deficient cells, suggesting that p53 is a partial mediator of the metabolic phenotype induced by ChREBP suppression in cancer cells. These results demonstrate that in addition to its role in organismal metabolism, ChREBP plays a key role in regulating the metabolism of proliferating cells.
Results
ChREBP Expression Is Modulated by Growth Factor Stimulation.
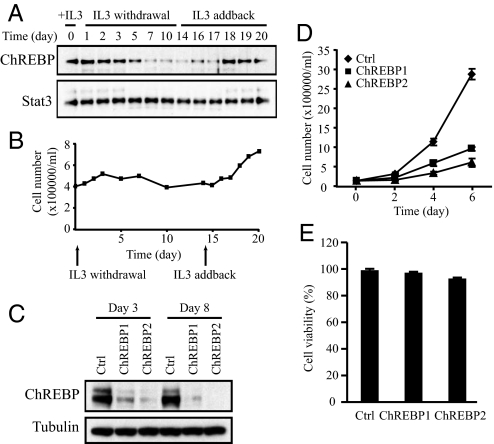
The expression of ChREBP in response to growth factor withdrawal and stimulation was examined in IL-3-dependent immortalized hematopoietic cells. IL-3-dependent cells exit the cell cycle upon IL-3 withdrawal and re-enter cell proliferation following IL-3 readdition (22). Cells cultured in the presence of IL-3 (day 0) stopped proliferation 3 days after IL-3 withdrawal and remained quiescent in the absence of IL-3 for 10 more days (Fig. 1B). At day 14, IL-3 was added back to the medium and the cells resumed proliferation on day 18 (Fig. 1B). ChREBP protein was highly expressed in the presence of IL-3 and declined progressively with growth factor withdrawal (Fig. 1A). Restimulation with IL-3 did not immediately restore ChREBP expression. Instead ChREBP levels restored several days after IL-3 readdition as the cells resumed exponential growth (Fig. 1A).
Fig. 1.
ChREBP protein level is regulated by growth factor signaling and is required for cell proliferation. Data in (A–C) are representative of at least three experiments. Data in (D) and (E) are presented as the mean ± SD of triplicate samples. (A) Western blot analysis of protein extracts harvested at the times indicated using antibodies to ChREBP and Stat3. The Stat3 blot serves as a loading control. IL-3-dependent bax−/−bak−/− mouse hematopoietic cells were grown in IL-3 and subjected to IL-3 withdrawal for 2 weeks, followed by restimulation with IL-3 at day 14. (B) Cell numbers of bax−/−bak−/− hematopoietic cultures that were grown in the presence or absence of IL-3 as described in A. (C) Western blot analysis of protein extracts of HCT116 cells transiently transfected with siRNA for control (Ctrl) and ChREBP (ChREBP1 and 2) using antibodies to ChREBP and tubulin at days 3 and 8 posttransfection. Human ChREBP protein displayed as a doublet whereas mouse ChREBP protein was a single band (A), which correlated with the presence of different ChREBP transcription variants in human. (D) Cell proliferation of HCT116 cells transfected with siRNA oligonucleotides for control (Ctrl) and ChREBP (ChREBP1 and 2). (E) Cell viability of HCT116 cells transfected with the indicated siRNA oligonucleotides at day 5 post-transfection.
Proliferative Cells Depend on ChREBP to Maintain Proliferative Expansion.
To assess the effects of ChREBP suppression on continuously proliferating cells, RNA interference (RNAi) was used to reduce ChREBP levels in the human HCT116 colorectal cancer cell line. Transient transfection of HCT116 cells with two independent ChREBP siRNA oligonucleotides (ChREBP1 and ChREBP2) led to a reproducible decline in ChREBP mRNA and protein levels compared to a siRNA control at days 3 and 8 after transfection (Fig. 1C).
To observe the effects of ChREBP suppression on cell proliferation, cells were replated at equal numbers 48 h after control and ChREBP siRNA transfection. Cell numbers were counted at days 2, 4, and 6 after replating. During this period, ChREBP protein remained at very low levels in ChREBP siRNA-transfected cells (Fig. 1C). Cell proliferation was severely impaired in ChREBP knockdown cells compared with control cells (Fig. 1D). ChREBP2 siRNA was more potent than ChREBP1 siRNA in reducing ChREBP protein levels and rate of proliferation (Fig. 1 C and D). Cell viability remained >90% in control and ChREBP knockdown cells (Fig. 1E).
To generalize our findings that suppression of ChREBP resulted in decreased proliferation, we performed the RNAi experiment using another cancer cell line-human hepatocellular carcinoma HepG2 cells. Similarly, transient transfection of HepG2 cells with ChREBP1 and ChREBP2 siRNAs led to a decline in ChREBP protein levels and cell proliferation compared to a siRNA control (Fig. S1 A and B).
Suppression of ChREBP Leads to a Metabolic Switch from Aerobic Glycolysis to Mitochondrial Respiration.
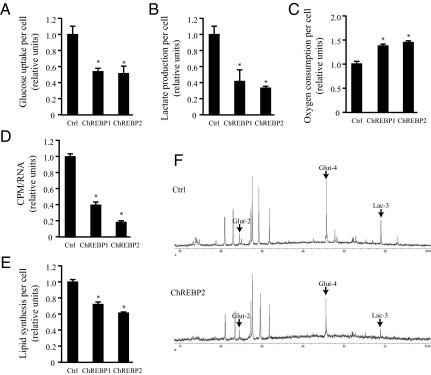
To investigate the metabolic effects of ChREBP suppression on tumor cells, the glycolytic activity of HCT116 cells transfected with either control or ChREBP siRNA were compared by quantifying glucose uptake and lactate production per cell at day 3 after transfection (Fig. 2 A and B). ChREBP1 and 2 siRNA transfected cells exhibited reduced glucose uptake and lactate production, indicating a loss of aerobic glycolysis. Oxygen consumption rates indicated that ChREBP-deficient cells displayed increased mitochondrial respiration (Fig. 2C). Furthermore, ChREBP-deficient cells displayed reduced glucose flux through the pentose phosphate pathway for RNA biosynthesis (Fig. 2D). The de novo lipid biosynthesis pathway was also impaired by ChREBP suppression (Fig. 2E).
Fig. 2.
Suppression of ChREBP leads to reduced aerobic glycolysis and anabolic metabolism, accompanied by increased mitochondrial oxygen consumption in HCT116 cells. In (A–E), data are presented as the mean ± SD of triplicate samples. Data in (F) are representative of at least three experiments. (A) Glucose uptake and (B) lactate production of HCT116 cells transfected with the indicated siRNA at day 3 post-transfection. The data were normalized by cell number. *, P < 0.01. (C) Oxygen consumption of HCT116 cells transfected with the indicated siRNA at day 3 post-transfection. The data were normalized by cell number. *, P < 0.005. (D) Measurement of RNA synthesis from D-[U-14C6] glucose in HCT116 cells transfected with the indicated siRNA at day 3 post-transfection. The data were normalized by RNA amount. *, P < 0.0001. (E) Measurement of lipid produced from D-[6-14C] glucose in HCT116 cells transfected with the indicated siRNA at day 3 post-transfection. The data were normalized by cell number. *, P < 0.001. (F) Spectra of HCT116 cells transfected with control (Ctrl) and ChREBP2 siRNA and incubated with D-[1,6-13C2] glucose at day 3 post-transfection. Glut-2, Glut-4, and Lac-3 represent [2-13C] glutamate, [4-13C] glutamate and [3-13C] lactate, respectively.
13C NMR (NMR) spectroscopy was used to confirm the above effects of ChREBP suppression on glucose metabolism. At day 3 after control or ChREBP2 siRNA transfection, HCT116 cells were cultured with [1,6-13C2]glucose for 5 h and glucose-derived metabolic intermediates were detected by 13C NMR spectra. The level of [3-13C]lactate produced from [1,6-13C2]glucose was lower in ChREBP2 siRNA-transfected cells compared with control cells (Fig. 2F), which was consistent with the measurement of lactate production per cell and indicated the cells had undergone a reduction in their glycolytic rate (Fig. 2B). The steady-state ratio of [2-13C]glutamate to [4-13C]glutamate increased from 0.23 ± 0.04 in control cells to 0.37 ± 0.07 in ChREBP knockdown cells (P < 0.05) (Fig. 2F). The increase in this ratio suggests the ChREBP-deficient cells have undergone an increase in the rate at which pyruvate enters the TCA cycle in relationship to the anaplerotic flux into the TCA cycle from amino acids as pyruvate carboxylase activity was found to be negligible in HCT116 cells. Taken together, these data indicate that ChREBP induces a transcriptional program that is required to maintain aerobic glycolysis in cancer cells and that cells revert to greater glucose catabolism in the mitochondria in the absence of ChREBP.
Suppression of ChREBP Induces p53 Activation and Cell Cycle Arrest.
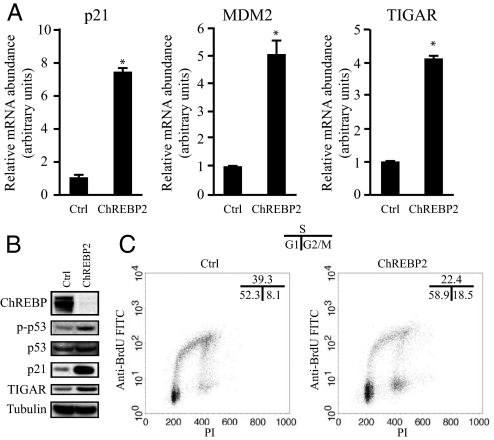
To examine the transcriptional changes that occur after ChREBP inhibition, RNA microarray analysis was performed with RNA from HCT116 cells 3 days after transfection with control or ChREBP2 siRNA. Isolated RNA was hybridized to Affymetrix human U133 plus 2.0 expression microarrays containing >47,000 transcripts and variants. Pairwise analysis of the hybridization profiles revealed that among the genes whose expression was highly increased in the ChREBP2 siRNA-transfected sample relative to the control-transfected sample, three were established p53-dependent targets: p21, MDM2 and TIGAR (23, 24). The microarray results were verified with real-time quantitative PCR for p21, MDM2 and TIGAR (Fig. 3A). In addition to the accumulation of p21, MDM2 and TIGAR mRNA transcripts in ChREBP knockdown cells, the level of p21 and TIGAR protein was also increased compared to that of control cells (Fig. 3 A and B). No difference in the level of total p53 protein was observed in ChREBP-deficient cells, but the level of p53 that was phosphorylated on Ser-15 increased as ChREBP expression declined (Fig. 3B).
Fig. 3.
Attenuation of ChREBP activates p53 and induces cell cycle arrest. Data in (A) are presented as the mean ± SD of triplicate samples. Data in (B) and (C) are representative of at least three experiments. (A) Quantitative PCR analysis of p21, MDM2, and TIGAR in HCT116 cells transfected with either control or ChREBP2 siRNA at day 3 post-transfection. *, P < 0.01. (B) Western blot analysis of HCT116 cells transfected with either control or ChREBP2 siRNA using indicated antibodies at day 3 post-transfection. (C) FACS analysis for BrdU incorporation and DNA content (PI) of HCT116 cells transfected with either control or ChREBP2 siRNA and pulse labeled with BrdU at day 3 post-transfection. Numbers indicate the percentage of cells in the G1, S, and G2/M phases.
Loss of ChREBP led to a switch from aerobic glycolysis to oxidative phosphorylation. Mitochondrial respiration is a major source for reactive oxygen species (ROS) generation (25). ROS-mediated oxidative stress can result in phosphorylation of p53 at Serine 15 and induce p21 transcription (26–28). To explore the possible mechanism for loss of ChREBP leading to p53 phosphorylation, the cellular ROS content was measured, which was quantified by flow cytometry using CM-DCFDA as a fluorescent probe. Compared to control cells, the ChREBP siRNA-transfected cells exhibited an increased ROS level (Fig. S2A). Moreover, the cellular glutathione level was lower in ChREBP-deficient cells compared with control cells (Fig. S2B). As expected, the increased ROS exhibited by cells transfected with ChREBP siRNA was suppressed by the addition of N-acetyl cysteine (NAC) (Fig. S2C). Treatment with NAC also eliminated the induction of p53 phosphorylation and p21 expression (Fig. S2D).
p53 activation can lead to apoptosis and/or cell cycle arrest (29). To characterize the effects of ChREBP suppression on cell cycle progression, control and ChREBP2 siRNA-transfected HCT116 cells were assessed for propidium iodide (PI) staining and BrdU incorporation. A 17% decrease in the number of cells in S phase was observed, accompanied by an increase in the percentage of cells in both G1 and G2/M phases in ChREBP2 siRNA-transfected cells compared to control cells (Fig. 3C).
p53 Contributes to the Metabolic and Growth Phenotype Induced by ChREBP Knockdown in HCT116 Colorectal Cancer Cells.
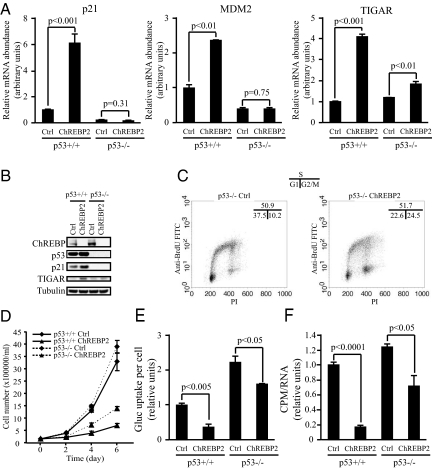
To test if p53 was required for the up-regulation of p21, MDM2, and TIGAR mRNA in ChREBP-deficient cells, p53−/− HCT116 cells were analyzed. The basal ChREBP level was higher in p53−/− HCT116 cells compared with wild-type counterparts (Fig. 4B). Transient transfection of ChREBP2 siRNA led to effective reduction in ChREBP protein level in both p53+/+ and p53−/− HCT116 cells (Fig. 4B). The basal mRNA level of p21 and MDM2 was lower in p53−/− cells compared with p53+/+ cells (Fig. 4A). Furthermore, the up-regulation of p21 and MDM2 mRNA after ChREBP2 siRNA transfection was blocked by loss of p53 (Fig. 4A). The basal mRNA and protein level of TIGAR was slightly higher in p53−/− cells compared with p53+/+ cells (Fig. 4 A and B). The up-regulation of TIGAR mRNA and protein in ChREBP2 siRNA transfected cells was partially blocked by loss of p53 (Fig. 4 A and B).
Fig. 4.
p53 is an important mediator of the growth and metabolic phenotype induced by ChREBP suppression. Data in (A) and (D–F) are presented as the mean ± SD of triplicate samples. Data in (B) and (C) are representative of at least three experiments. (A) Quantitative PCR analysis of p21, MDM2 and TIGAR in p53+/+ and p53−/− HCT116 cells transfected with either control or ChREBP2 siRNA at day 3 post-transfection. (B) Western blot analysis of p53+/+ and p53−/− HCT116 cells transfected with either control or ChREBP2 siRNA using indicated antibodies at day 3 post-transfection. (C) FACS analysis for BrdU incorporation and DNA content (PI) of p53+/+ and p53−/− HCT116 cells transfected with either control or ChREBP2 siRNA and pulse labeled with BrdU at day 3 post-transfection. Numbers indicate the percentage of cells in the G1, S, and G2/M phases. (D) Cell proliferation of p53+/+ and p53−/− HCT116 cells transfected with siRNA for control and ChREBP2. (E) Glucose uptake of p53+/+ and p53−/− HCT116 cells transfected with either control or ChREBP2 siRNA at day 3 post-transfection. The data were normalized by cell number. (F) Measurement of RNA synthesis from D-[U-14C6] glucose in p53+/+ and p53−/− HCT116 cells transfected with either control or ChREBP2 siRNA at day 3 post-transfection. The data were normalized by RNA amount.
Suppression of ChREBP resulted in G1 and G2/M arrest in p53+/+ HCT116 cells (Fig. 3C). To assess the role of p53 in ChREBP-dependent G1 and G2/M arrest, cell cycle analysis was performed using PI and BrdU staining for p53−/− HCT116 cells transfected with either control or ChREBP2 siRNA. In contrast to their wild-type counterparts, p53−/− cells transfected with ChREBP2 siRNA readily entered S phase (Fig. 4C). However, a greater proportion of p53−/− ChREBP-deficient cells had accumulated in G2/M in comparison to p53−/− control cells (Fig. 4C).
Growth kinetics of HCT116 cells with different genotypes of p53 and ChREBP were compared by counting cell numbers at days 2, 4, and 6 after replating equal number of cells at 48 h post-siRNA transfection. p53+/+ and p53−/− cells transfected with control siRNA showed similar growth curves (Fig. 4D). Both p53+/+ and p53−/− cells exhibited a pronounced reduction in growth when treated with ChREBP RNAi. However, the degree of impairment was reproducibly less in the p53−/− cells as compared to p53+/+ cells.
To examine whether p53 contributes to the metabolic defects resulting from suppression of ChREBP, both p53+/+ and p53−/− HCT116 cells were transiently transfected with control and ChREBP2 siRNA and assayed for aerobic glycolysis, lipid biosynthesis and nucleotide biosynthesis. The basal level of glucose uptake and lactate production was significantly higher in p53−/− cells compared with that in p53+/+ cells (Fig. 4E), as reported by others (30). Suppression of ChREBP led to a more modest inhibition of glucose uptake and lactate production in p53−/− cells compared with p53+/+ cells (Fig. 4E). Similarly, suppression of ChREBP led to less inhibition of lipid biosynthesis and glucose flux for the ribose production required for nucleotide biosynthesis in p53−/− cells compared with p53+/+ cells (Fig. 4F).
Attenuation of ChREBP Can Reduce Tumor Growth in Vivo.
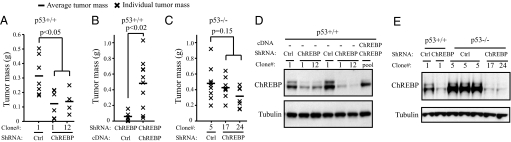
To examine the effects of stable ChREBP gene knockdown on the in vivo tumorigenicity of HCT116 cells, stable clones of HCT116 with short hairpin RNA (shRNA)-mediated suppression of ChREBP were established. Two clones (ChREBP-1 and ChREBP-12) stably transfected with pSM2C-ChREBP which displayed reduced ChREBP levels were chosen for further study. To determine their tumorigenicity in vivo, vector control (Ctrl-1) and ChREBP knockdown clones (ChREBP-1 and ChREBP-12) were injected s.c. into nude mice and examined for tumor formation. Each mouse was injected with a vector control inoculation in one flank and a ChREBP shRNA clone in the other, so that tumor comparisons were controlled for each mouse. The growth of tumors was monitored every 3 days, and tumors were excised and weighed 18 days post-injection. The results demonstrated that ChREBP knockdown cells formed smaller tumors in vivo compared with control cells (Fig. 5A). Western blot analysis of protein lysates from the excised tumors confirmed maintenance of the ChREBP knockdown phenotype by the tumor cells (Fig. 5D).
Fig. 5.
ChREBP suppression reduced tumor growth in vivo via a p53-dependent mechanism. (A–C) Charts depicting the mass of s.c. tumors formed in nude mice 18 days (A and B) and 10 days (C) after injection of indicated HCT116 stable transfectants. (A) Clones of p53+/+ HCT116 cells: ctrl-1 was generated by stable transfection of control pSM2C vector, while independent clones ChREBP-1 and ChREBP-12 were generated using the pSM2C-ChREBP shRNA plasmid. (B) Pools of p53+/+ HCT116 ChREBP-1 cells stably transfected with either empty pcDNA3 vector (Ctrl) or with a pcDNA3-mutant ChREBP plasmid. (C) Clones of p53−/− HCT116 cells: ctrl-5 was generated by stable transfection of control pSM2C vector, while independent clones ChREBP-17 and ChREBP-24 were generated using the pSM2C-ChREBP shRNA plasmid. (D and E) Western blot analysis of tumor protein extracts demonstrating efficient suppression of ChREBP levels in p53+/+ and p53−/− HCT116 cells stably transfected with ChREBP shRNA constructs as described in A–C. (D) Lanes 1–3 and 4–7 represent protein samples from two different animals, respectively. (E) Lanes 1–2, 3, and 4–7 represent protein samples from three different animals, respectively.
To determine if decreased tumor growth was due to loss of ChREBP expression, we generated stable cells which overexpressed either vector control or mutant ChREBP cDNA in the ChREBP-1 knockdown clone. A nonsuppressible ChREBP cDNA containing several mutations in the region targeted by the double-stranded RNA was created. Introducing the nonsuppressible ChREBP cDNA in ChREBP shRNA knockdown cells increased the tumor growth and rescued the tumorigenicity of ChREBP knockdown cells to levels similar to those of control cells (Fig. 5B). This finding demonstrates that the phenotype of reduced tumorigenicity in ChREBP knockdown HCT116 cells was due to the suppression of the ChREBP gene and not as a nonspecific effect of RNAi.
The contribution of p53 to the reduced tumor growth due to loss of ChREBP was investigated by assessing stable clones of HCT116 p53−/− cells with shRNA-mediated suppression of ChREBP. Two clones (ChREBP-17 and ChREBP-24) stably transfected with ChREBP shRNA which displayed reduced ChREBP levels were chosen and assessed for tumorigenicity. Because xenografts of p53−/− HCT116 cells in nude mice exhibited a shorter latency and increase in tumor growth kinetics compared with their p53+/+ counterparts, p53−/− tumors were excised and weighed 10 days post-injection. The results demonstrated that p53−/− control (Ctrl-5) and ChREBP knockdown cells (ChREBP-17 and ChREBP-24) formed similar sized tumors (Fig. 5C). Western blot analysis of protein lysates from the excised tumors showed that ChREBP shRNA efficiently suppressed the expression of ChREBP in p53−/− HCT116 cells (Fig. 5E).
Discussion
ChREBP has been shown to play critical roles in glycolysis and lipogenesis in hepatocytes and adipocytes (12–17). The present findings suggest that ChREBP also plays a role in redirecting glucose metabolism for lipid biosynthesis during cell proliferation. In addition, our findings suggest that ChREBP also plays a role in the stimulation of glucose-dependent de novo nucleotide biosynthesis. In our microarray analysis, several genes were identified which were involved in nucleotide metabolism and underwent about a 2-fold decrease in ChREBP siRNA-transfected HCT116 cells compared to control cells. Among these genes, the microarray results for dihydrofolate reductase (DHFR) and ribonucleoside-diphosphate reductase subunit M1 (RRM1) were verified with real-time quantitative PCR. A direct role of ChREBP in directing the transcription of any of these genes could potentially account for the reduction in nucleotide synthesis we observed in ChREBP-deficient cells. Alternatively, the expression of these genes may decline as a reaction to the failure to up-regulate established ChREBP target genes that reprogram glucose metabolism. Our data add to a growing literature that ChREBP contributes to the programming in redirecting glucose into anabolic processes and away from catabolism in the mitochondria.
The results presented here have provided evidence that ChREBP plays a critical role in the intracellular reprogramming of glucose in response to growth factor and/or oncogene induced changes in glucose uptake. In the absence of ChREBP, the failure to reprogram glucose metabolism to fuel growth-induced consumption of synthetic precursors results in redox stress, glutathione depletion and p53 induction. The elevated ROS generation and decreased GSH level in ChREBP-depleted cells were observed at as early as day 2 after siRNA transfection compared with control cells, which was followed by p53 activation at day 3. The findings of increased oxygen consumption in ChREBP knockdown cells compared with control cells is consistent with reported effects of p53 on mitochondrial oxidative phosphorylation.
Our data suggest that p53 was required for the G1 arrest that resulted from suppression of ChREBP. In vitro although, ChREBP-deficient p53−/− cells still display an altered metabolic phenotype, a reduced growth curve, and evidence of a G2/M arrest. These effects may result not from direct effects on cell cycle control genes but the reduced rate of anabolic synthesis of macromolecules. Impaired synthesis of macromolecules has been shown to lead to a prolonged G2/M in model organisms (31).
In conclusion, our results demonstrate a critical role of ChREBP in directing glucose metabolism into anabolic pathways such as lipid and nucleotide biosynthesis during cell growth. ChREBP suppression in cancer cell lines resulted in decreased glycolysis and increased oxygen consumption, as well as diminished proliferative and tumorigenic potential of cancer cells. These data demonstrate that ChREBP contributes to the glycolytic phenotype exhibited by many cancer cells. Whether ChREBP can directly serve as an oncogene in transforming cells remains to be determined. Although these cancer cells are capable of carrying out oxidative phosphorylation, they undergo the glycolytic shift because aerobic glycolysis is advantageous for cell proliferation and tumorigenicity. One of the most obvious explanations is that glycolytic metabolites are required for macromolecular biosynthesis of fatty acids, nucleotides, and nonessential amino acids required for anabolic cell growth. Therefore, our results add to the growing evidence that aerobic glycolysis contributes to cancer cell proliferation and tumorigenicity.
Materials and Methods
The SI Materials and Methods provides detailed information about cell culture reagents and conditions, RNA and protein analysis, analysis of cellular ROS and GSH and NAC treatment, and statistical analysis.
Cell Proliferation, Cell Viability, and Cell Cycle Analysis.
Cell proliferation and cell viability were determined by counting cells using trypan-blue exclusion assay. For cell proliferation analysis, 48 h after control and ChREBP siRNA transfection, cells were replated at 150,000 cells per well of 6-well plate. Cell numbers were counted at days 2, 4, and 6 after replating. For cell cycle analysis, cells were pulsed with 10 μM BrdU (Sigma) for 1 h, then trypsinized, fixed with ice-cold 70% ethanol, labeled with an anti-BrdU antibody (PharMingen) and propidium iodide (PI, Sigma), and analyzed by flow cytometry using an FACSCalibur flow cytometer (BD Biosciences).
siRNA Transfection.
Control siRNA (#12935–100) and siRNA oligonucleotides targeting ChREBP were obtained from Invitrogen. The ChREBP1 siRNA sequences are 5′-CCAGAUGCGAGACAUGUUUUU-3′ and 5′-AAACAUGUCUCGCAUCUGGUU-3′. The ChREBP2 siRNA sequences are 5′-GCACCCUUGGCAAACCUUUUU-3′ and 5′-AAAGGUUUGCCAAGGGUGCUU-3′. siRNA transfections were performed using Lipofectamine™ RNAiMAX (Invitrogen) according to the manufacturer's protocol.
Metabolism Measurements.
Glucose uptake and lactate production was measured using the Nova Biomedical Flex Analyzer. Oxygen consumption was measured using a Clark-type oxygen electrode (Hansatech Instruments). RNA biosynthesis was assayed by measuring incorporation of 14C into RNA after culture of cells with universally labeled D-[U-14C6] glucose (PerkinElmer). RNA was then extracted using RNeasy columns (Qiagen) and 14C incorporation was assayed using a Beckman LS 6500 scintillation counter. 14C cpm for each sample were normalized by the amount of RNA. Lipid biosynthesis was assayed after incubation of cells with D-[6-14C] glucose (Sigma) as described in ref. 4. Additional metabolites were analyzed by incubating cells with 7 mM D-[1,6-13C2] glucose (Sigma) at 37 °C for 5 h. Cells were then extracted with 10% perchloric acid and soluble metabolites were analyzed by 13C NMR spectroscopy as described in ref. 32.
ShRNA and cDNA Transfection.
The control and ChREBP shRNA (#RHS1764–97192923) constructs were obtained from Open Biosystems. The ChREBP shRNA sequence (sense) is 5′-CTGAGTACATCCTTATGCT-3′. The cDNA encoding the complete coding region of human ChREBP cDNA was purchased from Invitrogen and then subcloned into the pcDNA3 vector. The ChREBP cDNA refractory to shRNA silencing was designed by introducing the following mutations highlighted by letters in bold type: 5′-CT GAG TAT CTT TTA ATG CT-3′ using the QuikChange Site-Directed Mutagenesis Kit (Stratagene). These mutations do not affect the amino acid sequence except for a substitution from Ile (ATC) to Leu (CTT), and produce the mutant ChREBP mRNA refractory to RNAi. ShRNAs and cDNAs were transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions.
Xenograft Experiments.
All studies involving animals were performed according to approved IACUC protocols at the University of Pennsylvania. In brief, 6 × 106 HCT116 p53 +/+ or −/− cells resuspended in 200 μL PBS were s.c. injected into the flanks of athymic nude male mice (Charles River). The tumor size was measured every 3 days, and tumors were excised for weight measurements, histological examination 18 and 10 days after p53+/+ and p53−/− cell injection, respectively.
Supplementary Material
Acknowledgments.
We thank Georgia Hatzivassiliou, Justin Cross, Thi Bui, and Uma Sachdeva for technical assistance with the manuscript and members of the Thompson laboratory for discussions and comments on the manuscript. This work was supported by National Institutes of Health grants.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0911316106/DCSupplemental.
References
- 1.Kim JW, Dang CV. Cancer's molecular sweet tooth and the Warburg effect. Cancer Res. 2006;66:8927–8930. doi: 10.1158/0008-5472.CAN-06-1501. [DOI] [PubMed] [Google Scholar]
- 2.Tong X, Zhao F, Thompson CB. The molecular determinants of de novo nucleotide biosynthesis in cancer cells. Curr Opin Genet Dev. 2009;19:32–37. doi: 10.1016/j.gde.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boros LG, et al. Transforming growth factor beta2 promotes glucose carbon incorporation into nucleic acid ribose through the nonoxidative pentose cycle in lung epithelial carcinoma cells. Cancer Res. 2000;60:1183–1185. [PubMed] [Google Scholar]
- 4.Hatzivassiliou G, et al. ATP citrate lyase inhibition can suppress tumor cell growth. Cancer Cell. 2005;8:311–321. doi: 10.1016/j.ccr.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 5.Christofk HR, et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumor growth. Nature. 2008;452:230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- 6.Liu YC, et al. Global regulation of nucleotide biosynthetic genes by c-Myc. PLoS ONE. 2008;3:e2722. doi: 10.1371/journal.pone.0002722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mannava S, et al. Direct role of nucleotide metabolism in C-MYC-dependent proliferation of melanoma cells. Cell Cycle. 2008;7:2392–2400. doi: 10.4161/cc.6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ko YH, et al. Advanced cancers: Eradication in all cases using 3-bromopyruvate therapy to deplete ATP. Biochem Biophys Res Commun. 2004;324:269–275. doi: 10.1016/j.bbrc.2004.09.047. [DOI] [PubMed] [Google Scholar]
- 9.Kim JW, Gao P, Liu YC, Semenza GL, Dang CV. Hypoxia-inducible factor 1 and dysregulated c-Myc cooperatively induce vascular endothelial growth factor and metabolic switches hexokinase 2 and pyruvate dehydrogenase kinase 1. Mol Cell Biol. 2007;27:7381–7393. doi: 10.1128/MCB.00440-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fantin VR, St-Pierre J, Leder P. Attenuation of LDH-A expression uncovers a link between glycolysis, mitochondrial physiology, and tumor maintenance. Cancer Cell. 2006;9:425–434. doi: 10.1016/j.ccr.2006.04.023. [DOI] [PubMed] [Google Scholar]
- 11.Telang S, et al. Ras transformation requires metabolic control by 6-phosphofructo-2-kinase. Oncogene. 2006;25:7225–7234. doi: 10.1038/sj.onc.1209709. [DOI] [PubMed] [Google Scholar]
- 12.Yamashita H, et al. A glucose-responsive transcription factor that regulates carbohydrate metabolism in the liver. Proc Natl Acad Sci USA. 2001;98:9116–9121. doi: 10.1073/pnas.161284298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang H, Wollheim CB. ChREBP rather than USF2 regulates glucose stimulation of endogenous L-pyruvate kinase expression in insulin-secreting cells. J Biol Chem. 2002;277:32746–32752. doi: 10.1074/jbc.M201635200. [DOI] [PubMed] [Google Scholar]
- 14.Collier JJ, et al. c-Myc and ChREBP regulate glucose-mediated expression of the L-type pyruvate kinase gene in INS-1-derived 832/13 cells. Am J Physiol Endocrinol Metab. 2007;293:E48–56. doi: 10.1152/ajpendo.00357.2006. [DOI] [PubMed] [Google Scholar]
- 15.da Silva Xavier G, Rutter GA, Diraison F, Andreolas C, Leclerc I. ChREBP binding to fatty acid synthase and L-type pyruvate kinase genes is stimulated by glucose in pancreatic beta-cells. J Lipid Res. 2006;47:2482–2491. doi: 10.1194/jlr.M600289-JLR200. [DOI] [PubMed] [Google Scholar]
- 16.Dentin R, et al. Hepatic glucokinase is required for the synergistic action of ChREBP and SREBP-1c on glycolytic and lipogenic gene expression. J Biol Chem. 2004;279:20314–20326. doi: 10.1074/jbc.M312475200. [DOI] [PubMed] [Google Scholar]
- 17.Billin AN, Ayer DE. The Mlx network: Evidence for a parallel Max-like transcriptional network that regulates energy metabolism. Curr Top Microbiol Immunol. 2006;302:255–278. doi: 10.1007/3-540-32952-8_10. [DOI] [PubMed] [Google Scholar]
- 18.Kawaguchi T, Osatomi K, Yamashita H, Kabashima T, Uyeda K. Mechanism for fatty acid “sparing” effect on glucose-induced transcription: Regulation of carbohydrate-responsive element-binding protein by AMP-activated protein kinase. J Biol Chem. 2002;277:3829–3835. doi: 10.1074/jbc.M107895200. [DOI] [PubMed] [Google Scholar]
- 19.He Z, Jiang T, Wang Z, Levi M, Li J. Modulation of carbohydrate response element-binding protein gene expression in 3T3–L1 adipocytes and rat adipose tissue. Am J Physiol Endocrinol Metab. 2004;287:E424–430. doi: 10.1152/ajpendo.00568.2003. [DOI] [PubMed] [Google Scholar]
- 20.Iizuka K, Bruick RK, Liang G, Horton JD, Uyeda K. Deficiency of carbohydrate response element-binding protein (ChREBP) reduces lipogenesis as well as glycolysis. Proc Natl Acad Sci USA. 2004;101:7281–7286. doi: 10.1073/pnas.0401516101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iizuka K, Miller B, Uyeda K. Deficiency of carbohydrate-activated transcription factor ChREBP prevents obesity and improves plasma glucose control in leptin-deficient (ob/ob) mice. Am J Physiol Endocrinol Metab. 2006;291:E358–364. doi: 10.1152/ajpendo.00027.2006. [DOI] [PubMed] [Google Scholar]
- 22.Lum JJ, et al. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell. 2005;120:237–248. doi: 10.1016/j.cell.2004.11.046. [DOI] [PubMed] [Google Scholar]
- 23.Bensaad K, et al. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126:107–120. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 24.Smeenk L, et al. Characterization of genome-wide p53-binding sites upon stress response. Nucleic Acids Res. 2008;36:3639–3654. doi: 10.1093/nar/gkn232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Balaban RS, Nemoto S, Finkel T. Mitochondria, oxidants, and aging. Cell. 2005;120:483–495. doi: 10.1016/j.cell.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 26.Ito K, et al. Induction of apoptosis in leukemic cells by homovanillic acid derivative, capsaicin, through oxidative stress: Implication of phosphorylation of p53 at Ser-15 residue by reactive oxygen species. Cancer Res. 2004;64:1071–1078. doi: 10.1158/0008-5472.can-03-1670. [DOI] [PubMed] [Google Scholar]
- 27.Catalano A, Rodilossi S, Caprari P, Coppola V, Procopio A. 5-Lipoxygenase regulates senescence-like growth arrest by promoting ROS-dependent p53 activation. EMBO J. 2005;24:170–179. doi: 10.1038/sj.emboj.7600502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hammond EM, Dorie MJ, Giaccia AJ. ATR/ATM targets are phosphorylated by ATR in response to hypoxia and ATM in response to reoxygenation. J Biol Chem. 2003;278:12207–12213. doi: 10.1074/jbc.M212360200. [DOI] [PubMed] [Google Scholar]
- 29.Vousden KH, Prives C. Blinded by the light: The growing complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 30.Matoba S, et al. p53 regulates mitochondrial respiration. Science. 2006;312:1650–1653. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- 31.Moseley JB, Mayeux A, Paoletti A, Nurse P. A spatial gradient coordinates cell size and mitotic entry in fission yeast. Nature. 2009;459:857–860. doi: 10.1038/nature08074. [DOI] [PubMed] [Google Scholar]
- 32.DeBerardinis RJ, et al. Beyond aerobic glycolysis: Transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci USA. 2007;104:19345–19350. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.