Abstract
Based on the X-ray structure of the enzyme strictosidine synthase, the glucose moiety of the seco-iridoid glucoside, secologanin, appears to be the key for orienting the substrate. We hypothesized that removing the glucose moiety would allow alternate stereoisomers of secologanin to be turned over. A convenient synthesis to prepare stereoisomers of des-vinyl secologanin is presented. The choice of protective group was the key to access this series of compounds. The analogs were assayed with strictosidine synthase and, interestingly, both the natural 2,4-trans diastereomer and the unnatural 2,4-cis diastereomer are turned over. The trans/cis selectivity increases with increased acetal substituent size. The results add to our understanding of how strictosidine synthase discriminates among stereoisomers.
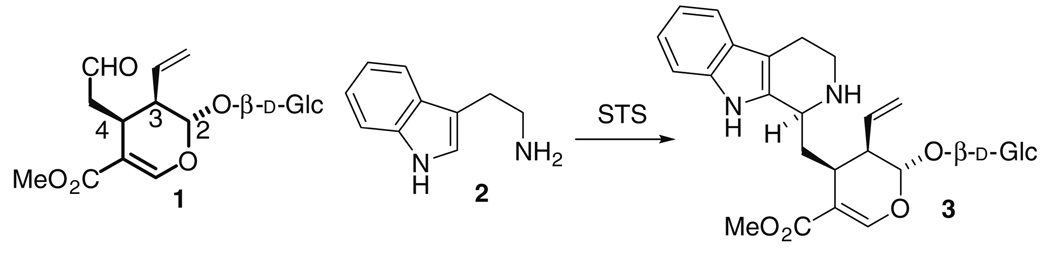
Many biologically active natural products are found in plants; for example, the monoterpene indole alkaloids, vinblastine and vincristine are produced in Catharanthus roseus, ajmaline in Rauvolfia serpentina and camptothecin in Ophiorrhiza pumila.1 The monoterpene-derived seco-iridoid β-d-glucoside, secologanin 1, is the precursor for all monoterpene indole alkaloids. Secologanin 1 is a densely functionalized molecule that contains three stereogenic centers on its dihydropyran core [2(S)3(R)4(S)] (Scheme 1). Only one stereoisomer of 1 is produced biosynthetically. Herein we describe the synthesis of alternate stereoisomers of des-vinyl secologanin using Tietze’s tandem Knoevenagel-hetero-Diels–Alder reaction.2 These analogs were used to assess the stereochemical preference of strictosidine synthase, the first committed enzyme in the biosynthetic pathways leading to monoterpene indole alkaloids. It is demonstrated for the first time that strictosidine synthase accepts more than one aldehyde stereoisomer.
Scheme 1.
Pictet-Spengler reaction catalyzed by strictosidine synthase (STS).
The first step in the biosynthesis of monoterpene indole alkaloids is the Pictet-Spengler reaction between secologanin 1 and tryptamine 2 to form the tetrahydro-β-carboline, strictosidine 3, a reaction catalyzed by the enzyme strictosidine synthase (Scheme 1).3 Strictosidine synthase acts as a gate-keeping enzyme because it has a restrictive substrate scope; only substrates with minor perturbations to the structure of 1 and 2 are recognized by the enzyme, and thereby allowed to enter this alkaloid pathway.4,5
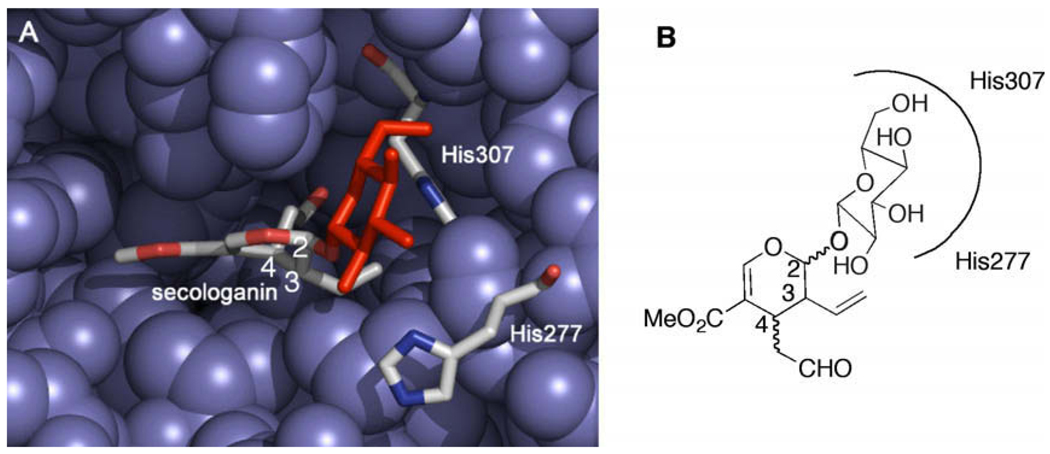
The X-ray structure of strictosidine synthase (Rauvolfia serpentina) in complex with 1 (PDB ID: 2FPC)6 suggests that the glucose moiety appears to form hydrogen bonds with histidine residues 277 and 307 (Fig. 1). These interactions of glucose with the enzyme suggest that the glucose moiety plays a key role in recognition of the substrate. We hypothesized that removing the glucose would enable the enzyme to turnover a greater number of secologanin analogs.
Figure 1.
(A) X-ray structure of strictosidine synthase in complex with secologanin (PDB ID: 2FPC). The glucose moiety (red) is positioned to hydrogen bond to two histidine residues. (B) Without the glucose group (gray) the secologanin-binding pocket could potentially accommodate substrates with different geometries.
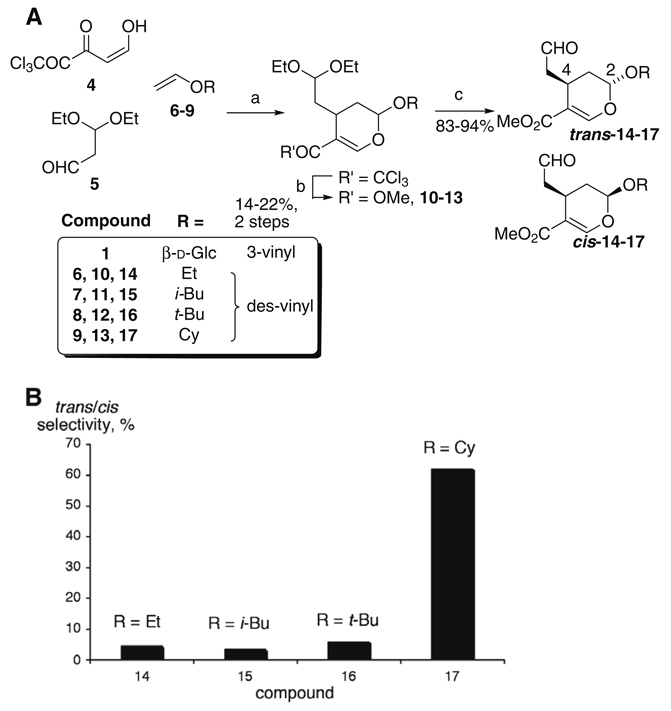
Only a limited number of secologanin 1 analogs are accessible by semi-synthesis,7 and there have been no prior reports of synthesis of analogs of 1 with alternate stereochemistry. To this end, we used total synthesis to obtain des-vinyl aglucone O-analogs of 1. Tietze’s tandem Knoevenagel-hetero-Diels–Alder reaction allowed rapid access to acetal-protected des-vinyl O-analogs as previously described.2 However, some of the analogs reported previously could not be deprotected (step c, Fig. 2). After testing several protecting groups to overcome the challenges associated with accessing the final aldehyde product, we found that the acyclic diethoxyacetal provided a convenient solution. Briefly, trichloromethyl ketone 4 was reacted with monoprotected malondialdehyde 58 and vinyl ethers 6–9 in the presence of potassium fluoride (Fig. 2A). The resulting cycloadduct was filtered through deactivated aluminum oxide and partially purified by silica gel chromatography as described previously.2 Trichloromethyl ketones were subjected to methanolysis in the presence of 1,8-diazabicycloundec-7-ene to form methyl esters 10–13.9 Finally, deprotection with aqueous oxalic acid and silica gel afforded aldehydes 14–17 (Fig. 2A).10 Purification by silica gel chromatography separated the two sets of enantiomers, which were each characterized by proton NMR. The trans versus cis configuration is distinguished by the splitting of the acetal proton signal ~5 ppm: cis: (t, J = 2–3 Hz), trans: (dd, J = 2–3, 7–8 Hz).2
Figure 2.
(A) Synthesis of des-vinyl secologanin O-analogs. (a) Postassium fluoride (50 mg mmol−1 6–9), dichloromethane; (b) 1,8-diazabicycloundec-7-ene (0.01 equiv), methanol; (c) oxalic acid and silica gel (each 10% w/v) tetrahydrofuran/water (4:1). (B) Diastereoselectivity of strictosidine synthase. The initial rates of product formation were obtained (<15% conversion, <20% error) and the average and normalized rate of three experiments is reported for trans and cis diastereomers of the same aldehyde substrate (14–17).
No secologanin aglycones had previously been tested biochemically. Aldehydes 14–17 were therefore each assayed with strictosidine synthase in the presence of tryptamine 2 and monitored for appearance of the product by high-resolution electrospray ionization (ESI)-MS. Aldehydes 14–17 each proved to be competent substrates. Surprisingly, both the 2,4-trans configuration—found in the natural substrate secologanin—and the unnatural 2,4-cis configuration were accepted by strictosidine synthase. For smaller aldehydes 14–16, the trans/cis selectivity was low (de = 5–10%), whereas for 17 the selectivity for the trans configuration was significantly higher >60% (Fig. 2B). Therefore, increased acetal substituent size appears to be correlated with a substrate preference for the natural trans stereochemistry.
We reported the successful synthesis of a series of des-vinyl secologanin aglycones. For the first time, it is shown that strictosidine synthase accepts alternate stereoisomers. Notably, turnover of ‘unnatural’ cis stereoisomers occurs when the acetal substituent is small. This study demonstrates that a central alkaloid biosynthetic enzyme can recognize a variety of stereoisomers.
Acknowledgment
We acknowledge financial support from the NIH (GM074820).
References and notes
- 1.O’Connor SE, Maresh JJ. Nat. Prod. Rep. 2006;23:532–547. doi: 10.1039/b512615k. [DOI] [PubMed] [Google Scholar]
- 2.Tietze LF, Meier H, Nutt H. Chem. Ber. 1989;122:643–650. [Google Scholar]; Tietze LF, Meier H, Nutt H. Liebigs Ann. Chem. 1990:253–260. [Google Scholar]; Tietze LF. Angew. Chem. 1983;95:840–853. [Google Scholar]
- 3.Maresh JJ, Giddings LA, Friedrich A, Loris EA, Panjikar S, Stöckigt J, Peters B, O’Connor SE. J. Am. Chem. Soc. 2008;130:710–723. doi: 10.1021/ja077190z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McCoy EA, Galan MC, O’Connor SE. Bioorg. Med. Chem. Lett. 2006;16:2475–2478. doi: 10.1016/j.bmcl.2006.01.098. [DOI] [PubMed] [Google Scholar]; Treimer JF, Zenk MH. Eur. J. Biochem. 1979;101:225–233. doi: 10.1111/j.1432-1033.1979.tb04235.x. [DOI] [PubMed] [Google Scholar]
- 5.Loris EA, Panjikar S, Ruppert M, Barleben L, Unger M, Schubel H, Stöckigt J. Chem. Biol. 2007;14:979–985. doi: 10.1016/j.chembiol.2007.08.009. [DOI] [PubMed] [Google Scholar]
- 6.Ma X, Panjikar S, Koepke J, Loris E, Stöckigt J. Plant Cell. 2006;23:532–547. doi: 10.1105/tpc.105.038018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galan MC, O’Connor SE. Tetrahedron Lett. 2005;47:1563–1565. [Google Scholar]
- 8.The aldehyde was prepared by Dess–Martin periodinane mediated oxidation of 3,3-diethoxy-1-propanol. 1H NMR (300 MHz, CDCl3): δ 9.72 (t, 1H, J = 2.3), 4.93 (t, 1H, J = 5.5), 3.65 (qd, 1H, J = 7.1, 9.3), 3.52 (qd, 1H, J = 7.1, 9.3), 2.70 (dd, 1H, J = 2.3, 5.5), 1.78 (t, 1H, J = 7.1); 13C NMR (75 MHz, CDCl3): δ 200.17, 98.85, 62.30, 48.22, 15.40.
- 9.cis-10: 1H NMR (600 MHz, CDCl3): δ ppm 7.43 (s, 1H), 5.10–5.07 (m, 1H), 4.64 (dd, 1H, J = 5.1, 7.0), 3.93 (dq, 1H, J = 7.1, 14.3), 3.70–3.40 (m, 5H), 3.66 (s, 3H), 2.66–2.59 (m, 1H), 2.10–2.03 (m, 1H), 2.03–1.89 (m, 2H), 1.80 (ddd, 1H, J = 2.9), 1.21 (t, 3H, J = 7.1), 1.19–1.13 (m, 6H); 13C NMR (200 MHz, CDCl3): δ ppm 168.01b, 167.85a, 153.44b, 152.25a, 110.63a, 109.80b, 101.70b, 101.52a, 98.44a,b, 65.30b, 64.74a, 62.41b, 61.78a, 59.85b, 59.25a, 51.32a,b, 51.30a,b, 38.53b, 36.15a, 31.46b, 29.60a, 26.60b, 24.54a, 15.52–15.29; trans-10: 1H NMR (600 MHz, CDCl3): δ ppm 7.43 (s, 1H), 4.97 (dd, 1H, J = 1.7, 9.1), 4.58 (t, 1H, J = 5.7), 3.79 (dq, 1H, J = 7.1, 14.2), 3.70–3.40 (m, 5H), 3.66 (s, 3H), 2.75–2.70 (m, 1H), 2.10–2.03 (m, 1H), 2.03–1.89 (m, 2H), 1.72 (ddd, 1H, J = 5.9, 9.1, 14.3), 1.47 (ddd, 1H, J = 5.4, 9.8, 14.5, 1.21), 1.19–1.13 (m, 9H, b); ESI-MS(C15H26O6) m/z calcd: 325.1622 [M+Na]+, found: 325.1623 [M+Na]+. cis-11: 1H NMR (300 MHz, CDCl3): δ 7.42 (s, 1H), 5.07 (t, 1H, J = 2.7), 4.65 (dd, 1H, J = 5.6, 6.6), 3.65 (s, 3H), 3.68–3.40 (m, 4H), 3.52 (dd, 1H, J = 6.5, 9.1), 3.19 (dd, 1H, J = 6.4, 9.1), 2.66–2.57 (m, 1H), 2.11 (td, 1H, J = 2.3, 14.2), 2.02–1.70 (m, 4H), 1.19–1.10 (m, 6H), 0.84 (dd, 6H, J = 1.4, 6.7); 13C NMR (125 MHz, CDCl3): δ 167.99, 152.20, 110.60, 101.77, 98.81, 96.09, 61.93, 58.77, 51.28, 36.05, 29.11, 28.62, 24.29, 19.41, 15.47, 15.54; trans-11: 1H NMR (300 MHz, CDCl3): δ 7.42 (s, 1H), 4.94 (dd, 1H, J = 2.2, 9.0), 4.57 (t, 1H, J = 5.7), 3.65 (s, 3H), 3.68–3.40 (m, 5H), 3.25 (dd, 1H, J = 6.9, 9.3), 2.72 (dq, 1H, J = 3.5, 6.4), 2.02–1.70 (m, 4H), 1.46 (ddd, 1H, J = 5.4, 9.7, 14.3), 1.19–1.10 (m, 6H), 0.87 (dd, 6H, J = 2.2, 6.7); 13C NMR (125 MHz, CDCl3): δ 167.84, 153.46, 109.78, 101.77, 98.81, 76.44, 62.33, 59.97, 51.28, 38.46, 31.40, 26.35, 19.35, 15.54, 15.47. ESI-MS (C17H30O6+) calcd m/z 353.1940 [M+Na]+, obsd: m/z 353.1957 [M+Na]+. cis-12: 1H NMR (300 MHz, CDCl3): δ 7.42 (d, 1H, J = 0.8), 5.32 (t, 1H, J = 3.1), 4.64 (dd, 1H, J = 4.7, 7.4), 3.65 (s, 3H), 3.67–3.57 (m, 2H), 3.50–3.41 (m, 2H), 2.03 (ddd, 1H, J = 4.1, 7.4, 13.8), 1.99–1.85 (m, 4H), 1.77 (ddd, 1H, J = 3.0, 6.6, 14.0), 1.20 (s, 9H), 1.19–1.13 (m, 6H); 13C NMR (125 MHz, CDCl3): δ 152.8, 110.2, 101.4, 93.3, 75.8, 61.8, 58.9, 51.2, 38.9, 36.1, 30.9, 28.7, 25.0, 15.5. trans-12: 1H NMR (300 MHz, CDCl3): δ 7.42–7.41 (m, 1H), 5.20 (dd, 1H, J = 2.4, 9.5), 4.55 (t, 1H, J = 5.5), 3.64 (s, 3H), 3.69–3.57 (m, 2H), 3.49–3.41 (m, 2H), 2.74–2.68 (m, 1H), 2.48 (ddd, 1H, J = 1.5, 5.6, 7.2), 1.98–1.85 (m, 2H), 1.49 (ddd, 1H, J = 5.7, 9.8, 14.2), 1.73–1.67 (m, 1H), 1.23 (s, 9H), 1.18–1.13 (m, 6H); 13C NMR (125 MHz, CDCl3): δ 154.0, 109.3, 102.2, 93.1, 76.3, 62.4, 60.4, 51.2, 39.8, 32.7, 28.7, 26.9, 15.5; ESI-MS (C17H31O6+) calcd m/z 353.1935 [M+Na]+, obsd: m/z 353.1941 [M+Na]+. cis-13: 1H NMR (400 MHz, CDCl3): δ 7.42 (s, 1H), 5.23 (t, 1H, J = 2.8), 4.66 (dd, 1H, J = 5.0, 7.2), 3.66 (s, 3H), 3.70–3.40 (m, 4H), 2.63 (ddd, 2H, J = 4.5, 7.2, 9.9), 2.05 (td, 1H, J = 2.8, 14.1), 2.00–1.20 (m, 13H), 1.18–1.13 (m, 6H); 13C NMR (75 MHz, CDCl3): δ 152.42, 110.53, 101.47, 96.34, 76.29, 61.82, 58.96, 51.27, 36.16, 33.45, 31.61, 29.88, 25.80, 24.61, 23.65, 23.62, 15.49; trans-13: 1H NMR (300 MHz, CDCl3): δ 7.42 (s, 1H), 5.11 (dd, 1H, J = 2.3, 9.3), 4.57 (t, 1H, J = 5.6), 3.66 (s, 3H), 3.70–3.40 (m, 4H), 2.72 (ddd, 1H, J = 3.2, 5.9, 12.6), 2.00–1.20 (m, 14H), 1.18–1.13 (m, 6H); 13C NMR (125 MHz, CDCl3): δ 153.69, 109.62, 101.96, 96.80, 77.64, 62.38, 60.15, 51.29, 38.68, 33.67, 31.94, 30.43, 26.65, 25.69, 24.32, 24.19, 15.56; ESI-MS (C19H32O6), calcd m/z 379.2097 [M+Na]+, obsd m/z 379.2097 [M+Na]+.
- 10.cis-14: 1H NMR (CDCl3, 400 MHz): δ 9.74 (t, 1H, J = 1.2), 7.50 (d, 1H, J = 0.5), 5.14 (t, 1H, J = 2.5), 3.78 (dq, 1H, J = 7.1, 9.5), 3.68 (s, 3H), 3.50 (dq, 1H, J = 7.1, 9.5), 3.11–3.05 (m, 1H), 2.99 (ddd, 1H, J = 1.3, 9.6, 17.7), 2.75 (ddd, 1H, J = 1.0, 3.4, 17.4), 2.00 (td, 1H, J = 2.2, 14.4), 1.89 (ddd, 1H, J = 2.8, 6.7, 14.4), 1.16 (t, 3H, J = 7.1; 13C NMR (CDCl3, 100 MHz): δ 202.43, 167.67, 153.10, 108.81, 97.96, 64.87, 51.46, 47.87, 30.06, 22.34, 15.29. trans-14: 1H NMR (CDCl3, 500 MHz): δ 9.74 (t, 1H, J = 1.2), 7.49 (d, 1H, J = 1.1), 4.97 (dd, 1H, J = 2.4, 7.5), 3.90 (dq, 1H, J = 7.1, 9.5), 3.68 (s, 3H), 3.58 (dq, 1H, J = 7.1, 9.5), 3.24–3.18 (m, 1H), 2.87 (ddd, 1H, J = 1.2, 4.2, 16.9), 2.38 (ddd, 1H, J = 2.2, 9.0, 16.9), 1.93 (ddd, 1H, J = 6.2, 7.3, 13.7), 1.77 (ddd, 1H, J = 2.4, 5.2, 13.9), 1.21 (t, 3H, J = 7.1); ESI-MS (C11H16O5), m/z calcd m/z 251.0890 [M+Na]+, obsd m/z 251.0884 [M+Na]+. cis-15: 1H NMR (500 MHz, CDCl3): δ 9.75 (t, 1H, J = 1.3), 7.49 (d, 1H, J = 0.7), 5.11 (t, 1H, J = 2.5), 3.68 (s, 3H), 3.51 (dd, 1H, J = 6.7, 9.2), 3.21 (dd, 1H, J = 6.4, 9.2), 3.14–3.05 (m, 1H), 2.99 (ddd, 1H, J = 1.4, 9.6, 17.4), 2.80–2.70 (m, 1H), 2.03 (td, 1H, J = 2.1, 14.5), 1.93–1.84 (m, 1H), 1.80 (dd, 1H, J = 6.6, 13.4), 0.85 (d, 6H, J = 6.6); 13C NMR (125 MHz, CDCl3): δ 202.32, 167.65, 153.04, 108.84, 98.39, 76.26, 51.43, 48.00, 29.83, 28.58, 22.24, 19.46, 19.44. trans-15: 1H NMR (500 MHz, CDCl3): δ 9.74 (t, 1H, J = 1.8), 7.49 (d, 1H, J = 1.1), 4.95 (dd, 1H, J = 2.4 7.2), 3.67 (s, 3H), 3.61 (dd, 1H, J = 6.6, 9.2), 2.27 (dd, 1H, J = 6.8, 9.2), 3.24–3.18 (m, 1H), 2.88 (ddd, 1H, J = 1.5, 4.2, 17.0), 2.38 (ddd, 1H, J = 2.1, 9.0, 17.0), 1.96 (td, 1H, J = 6.4, 13.7), 1.84 (td, 1H, J = 6.7, 13.4), 1.76 (ddd, 1H, J = 2.4, 5.5, 13.9), 0.88 (dd, 6H, J = 3.4, 6.7); 13C NMR (125 MHz, CDCl3): δ 201.28, 167.51, 154.15, 108.29, 98.17, 76.32, 51.45, 48.65, 32.49, 28.62, 23.79, 19.42, 19.38; ESI-MS (C13H20O5+), calcd m/z 279.1208 [M+Na]+, obsd m/z 279.1209 [M+Na]+. cis-16: 1H NMR (500 MHz, CDCl3): δ 9.74 (t, 1H, J = 1.3), 7.49 (d, 1H, J = 0.6), 5.38 (t, 1H, J = 2.8), 3.67 (s, 3H), 3.09 (qd, 1H, J = 4.4, 9.0), 2.99 (ddd, 1H, J = 1.4, 9.5, 17.7), 2.77 (ddd, 1H, J = 1.3, 4.0, 17.7), 1.87 (dd, 2H, J = 3.0, 4.4), 1.20 (s, 9H); 13C NMR (125 MHz, CDCl3): δ 202.6, 167.8, 153.6, 108.3, 92.9, 76.2, 51.4, 48.0, 31.4, 28.7, 22.8. trans-16: 1H NMR (500 MHz, CDCl3): δ 9.74 (dd, 1H, J = 1.8, 2.2), 7.49 (d, 1H, J = 1.0), 5.20 (dd, 1H, J = 2.5, 7.8), 3.67 (s, 3H), 3.24–3.18 (m, 1H), 2.86 (ddd, 1H, J = 1.5, 5.4, 16.7), 2.38 (ddd, 1H, J = 2.3, 8.9, 16.7), 1.89 (ddd, 1H, J = 5.9, 7.9, 13.8), 1.67 (ddd, 1H, J = 2.5, 3.7, 13.9), 1.24 (s, 9H); 13C NMR (125 MHz, CDCl3): δ 201.4, 167.6, 154.7, 101.7, 92.6, 76.5, 51.4, 49.0, 33.7, 28.7, 24.3; ESI-MS (C13H20O5), calcd m/z 279.1208 [M+Na]+, obsd m/z 279.1200 [M+Na]+. cis-17: 1H NMR (500 MHz, CDCl3): δ 9.74 (t, 1H, J = 1.3), 7.49 (d, 1H, J = 0.6), 5.27 (t, 1H, J = 2.6), 3.67 (s, 3H), 3.65–3.59 (m, 1H), 3.10–3.05 (m, 1H), 3.02 (ddd, 1H, J = 1.3, 9.6, 17.5), 2.78–2.72 (m, 1H), 1.97 (td, 1H, J = 2.3, 14.4), 1.88 (ddd, 1H, J = 2.8, 6.5, 14.4), 1.82–1.72 (m, 2H), 1.68–1.58 (m, 2H), 1.48–1.41 (m, 1H), 1.40–1.13 (m, 5H); 13C NMR (125 MHz, CDCl3): δ 202.53, 167.73, 153.26, 108.70, 96.10, 76.55, 51.40, 48.02, 33.45, 31.59, 30.43, 25.72, 23.91, 23.74, 22.45; trans-17: 1H NMR (500 MHz, CDCl3): δ 9.74 (t, 1H, J = 1.8), 7.48 (d, 1H, J = 1.1), 5.10 (dd, 1H, J = 2.4, 7.5), 3.67 (s, 3H), 3.68–3.62 (m, 1H), 3.21 (td, 1H, J = 5.1, 14.6), 2.86 (ddd, 1H, J = 1.5, 4.2, 16.9), 2.38 (ddd, 1H, J = 2.2, 8.9, 16.9), 1.93 (ddd, 1H, J = 6.1, 7.4, 13.7), 1.89–1.82 (m, 2H), 1.74 (ddd, 1H, J = 2.4, 5.2, 13.9), 1.72–1.67 (m, 2H), 1.53–1.47 (m, 1H), 1.42–1.15 (m, 5H); 13C NMR (125 MHz, CDCl3): δ 201.36, 167.58, 154.37, 108.07, 96.15, 51.44, 48.80, 39.78, 33.61, 32.86, 32.03, 25.67, 24.26, 24.14, 24.00; ESI-MS (C15H22O5+), calcd m/z 283.1545 [M+H]+, obsd m/z 283.1544 [M+H]+.