Abstract
Arrhythmogenic right ventricular dysplasia (ARVD) is a genetic disorder resulting in fibrofatty replacement of right ventricular myocytes and consequent ventricular arrhythmias. Heterozygous mutations in PKP2 encoding plakophilin-2 have previously been reported to cause dominant ARVD with reduced penetrance. We report the first case of recessive ARVD caused by mutations in PKP2. Candidate gene analysis in a typical proband with this disorder identified a novel homozygous mutation in PKP2 (c.[2484C>T]+[2484C>T]), which is predicted to be translationally silent (p.Gly828). Analysis of the proband’s mRNA, however, shows that this mutation causes predominantly cryptic splicing, with a 7-nucleotide deletion in exon 12. The ensuing frame shift disrupts the last 54 amino acids of plakophilin-2 and extends the open reading frame by 145 nucleotides (48 amino acids) into the 3’ untranslated region. Haplotype analysis demonstrates the absence of remote consanguinity. Heterozygous family members produce approximately 60% of properly spliced PKP2 and do not have manifestations of ARVD. Further analysis of PKP2 mRNA sequence revealed two additional alternatively spliced transcripts. The possibility of cryptic or alternative splicing should be considered with identification of apparently synonymous nucleotide substitutions in this gene.
Keywords: PKP2, ARVD, alternate splicing, cryptic splicing
INTRODUCTION
Arrhythmogenic right ventricular dysplasia (ARVD; MIM# 107970, MIM# 609040) is a disease notable for prominent ventricular arrhythmias and replacement of right ventricular myocardium with fibrofatty tissue (Marcus et al., 1982). Patients often present with dizziness, palpitations, or syncope, but many experience sudden cardiac death as the first and only symptom (Dalal et al., 2005; Fontaine et al., 1999). Diagnosis is made according to task force criteria (TFC) established in 1994 (McKenna et al., 1994). Among families segregating ARVD, autosomal recessive inheritance has been demonstrated in both Naxos disease (MIM# 601214) (McKoy et al. 2000) and a similar disorder which results in ARVD with wooly hair and a pemphigous-like skin disorder (Alcalai et al., 2003). Autosomal dominant mutations in desmoplakin (DSP; MIM# 125647) (Bauce et al., 2005; Rampazzo et al., 2002), desmoglein-2 (DSG2; MIM# 125671) (Awad et al., 2006; Pilichou et al., 2006), transforming growth factor-beta 3 (TGFB3; MIM# 190230) (Beffagna et al., 2005), and the cardiac ryanodine receptor (RYR2; MIM# 180902) (Tiso et al., 2001) have also been identified in ARVD.
In 2004, Gerull and colleagues described mutations in the plakophilin-2 gene (PKP2; MIM# 602861) in 32 of 120 unrelated probands with ARVD (Gerull et al., 2004). All mutations identified were heterozygous and included insertion-deletions, nonsense, missense, and splice site alterations. Subsequent manuscripts have confirmed the high prevalence of dominant mutations in PKP2 in large series of ARVD probands (Dalal et al., 2006; Syrris et al., 2006; van Tintelen et al., 2006). To date, homozygous mutations in this gene have not been described.
Like plakoglobin, desmoplakin, and desmoglein-2, plakophilin-2 is a component of the desmosome, a complex structure of proteins involved in intercellular adhesion and signaling (Green and Gaudry, 2000). In addition, plakophilin-2 is also found in the nucleus, though its nuclear function is currently unknown (Schmidt and Jager, 2005). In mice, deletion of Pkp2 results in embryonic lethality due to impaired cardiac morphogenesis (Grossmann et al., 2004).
Here we describe a novel mutation in PKP2 in an individual diagnosed with ARVD. Genomic sequencing of PKP2 identified a homozygous mutation which is predicted to be translationally silent (c.2484C>T, p.Gly828Gly), but sequencing of the patient’s cDNA revealed a 7-base-pair deletion in roughly 80% of the transcripts (r.[2483_2489del]). The ensuing frame shift disrupts the C-terminus of plakophilin-2 and extends the open reading frame (ORF) into the 3’ untranslated region (UTR). Transcripts from all three of the proband’s heterozygous children undergo mutant splicing approximately 40% of the time. We also identify two previously unreported alternative splice forms of PKP2.
MATERIALS AND METHODS
Clinical Data
Written informed consent was obtained from all family members from whom blood was collected and analyzed. This work was reviewed and approved by the Johns Hopkins Medicine Institutional Review Board. The proband and family members were interviewed to determine family pedigree structure and family history of ARVD. The medical history of each subject was obtained by review of medical records, clinical evaluation, and patient interview. The diagnosis of ARVD was established based on the criteria set by the Task Force of the Working Group of Myocardial and Pericardial Disease of the European Society of Cardiology and of the Scientific Council on Cardiomyopathies of the International Society and Federation of Cardiology (McKenna et al., 1994).
Genomic Sequence Analysis
DNA was isolated from whole blood using standard methods (Qiagen, Valencia, CA, USA; www.qiagen.com). Mutation screening was performed by direct sequencing of genomic DNA. Polymerase chain reaction (PCR) products were generated for sequence analysis using intronic primers flanking each exon in the PKP2 gene, transcript variant 2b (GeneID: 5318; genomic contig: NC_000012; mRNA: NM_004572.2. The numbering of the coding sequence uses the A of the ATG start codon as position +1. Primer sequences were generously provided by Ludwig Thierfelder) (Gerull et al., 2004).
Control DNA was obtained from NIGMS Human Genetic Cell Repository through the Coriell Institute for Medical Research (Camden, NJ, USA; ccr.coriell.org). Presence of this mutation was analyzed among 110 ethnically-matched and 100 ethnically-unmatched unaffected controls (420 control chromosomes) by creation of an Aar I restriction enzyme digest site (Fermentas Life Sciences, Hanover, MD, USA; www.fermentas.com). Analysis of 220 ethnically matched control chromosomes achieves at least 80% power to distinguish a missense mutation from a normal sequence variant.
Haplotype analysis for markers D12S345, D12S2080, and D12S1692 were performed as described in Gerull et al, 2004. Primer sequences for each marker were obtained from www.gdb.org.
cDNA Analysis
RNA was isolated from blood using the QIAamp RNA Blood Mini Kit (Qiagen) followed by clean-up and concentration with the RNeasy kit (Qiagen). RNA from cultured fibroblasts was isolated using Trizol (Invitrogen) followed by RNeasy clean-up (Qiagen). Reverse transcription was performed using an oligo-dT primer and Superscript III reverse transcriptase (Invitrogen, Carlsbad, CA, USA; www.invitrogen.com) according to manufacturer’s protocol. For cDNA generated from blood samples, PCR spanning the c.2484C> T mutation was performed using the following primers: #1 AGATGCTGCATGTTGGTGAC, #2 CAACCCAAGGGGATAAAAAC. For cDNA generated from fibroblasts, nested PCR was performed using the above primers for the first round of PCR, and the following nested primers for the second round of PCR: #3 CTCTCCCTGATTTGGTTTCC (exon 12), #4 GTGGCCGGTATCTACTGGTG (exon 14). Amplified cDNA was cloned into the pCR2.1 plasmid using the TOPO-TA cloning kit (Invitrogen). Plasmid DNA from 48 individual clones per individual was purified (Qiagen), and sequenced with M13 forward and reverse primers.
Statistical Analysis
Wild-type and mutant splicing tallies from each of the five clinical samples were compared pairwise using a two-tailed Fisher’s Exact Test. To compare control to proband and to children, tallies from the proband’s two clinical samples (lymphocytes and fibroblasts) were combined, and tallies from each of the heterozygous children were also combined before performing the Fisher’s Exact Test on the three groups.
RESULTS
Clinical Information
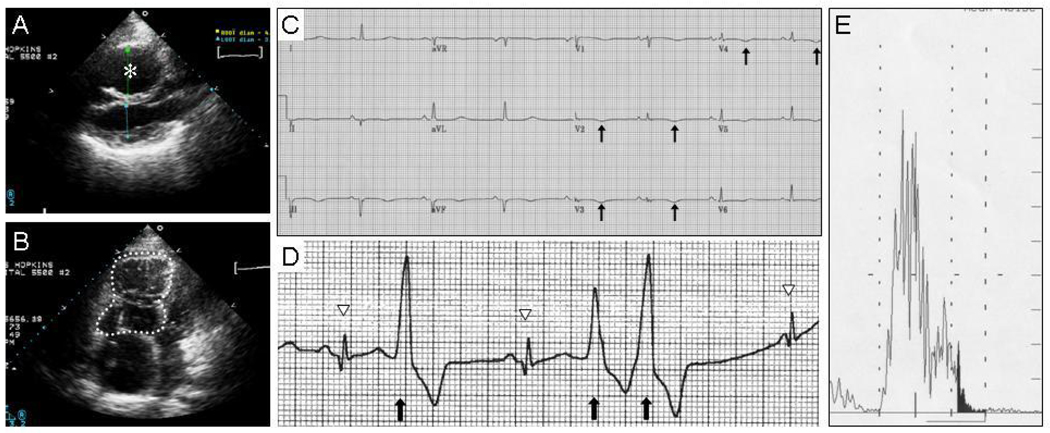
The proband is a 44-year-old woman of mixed Caucasian European ancestry. Her father is of German, English, and Scottish ancestry, and her mother is of Italian, French, and German ancestry. The proband was diagnosed with ARVD in her late teens. She meets consensus Task Force criteria for ARVD (McKenna et al., 1994) on the basis of one major and four minor diagnostic criteria (listed respectively): severe right ventricular (RV) dilatation and severely reduced RV ejection fraction with no left ventricular impairment (Fig. 1A, B), inverted T waves in right precordial leads (V2 and V3) in the absence of right bundle branch block (Fig. 1C); left bundle branch block-morphology ventricular tachycardia; greater than 1000 premature ventricular contractions (PVCs) in 24 hours on Holter monitoring (Fig. 1D); and late potentials on signal averaged ECG (Fig. 1E). The proband has no significant cutaneous abnormalities. She received an implantable cardiodefibrillator which has appropriately fired on multiple occasions. She is being treated with antiarrhythmic medications and has been arrhythmia-free for the past two years.
Figure 1.
Proband meets diagnostic criteria for ARVD. A, B: Right ventricular dilatation on echocardiogram, shown from a parasternal long axis view (A), and from an apical four-chamber view (B). The right ventricle is marked with an asterisk in (A) and is traced with a dotted line in (B). C: T-wave inversions (arrows) in precordial leads V2 to V4) on 12-lead electrocardiogram (ECG). D: Premature ventricular contractions (PVCs, arrows) on Holter monitoring. Normal sinus beats are denoted with arrowheads. E: Late potentials (shaded region) on signal averaged ECG.
Genomic Sequence Analysis
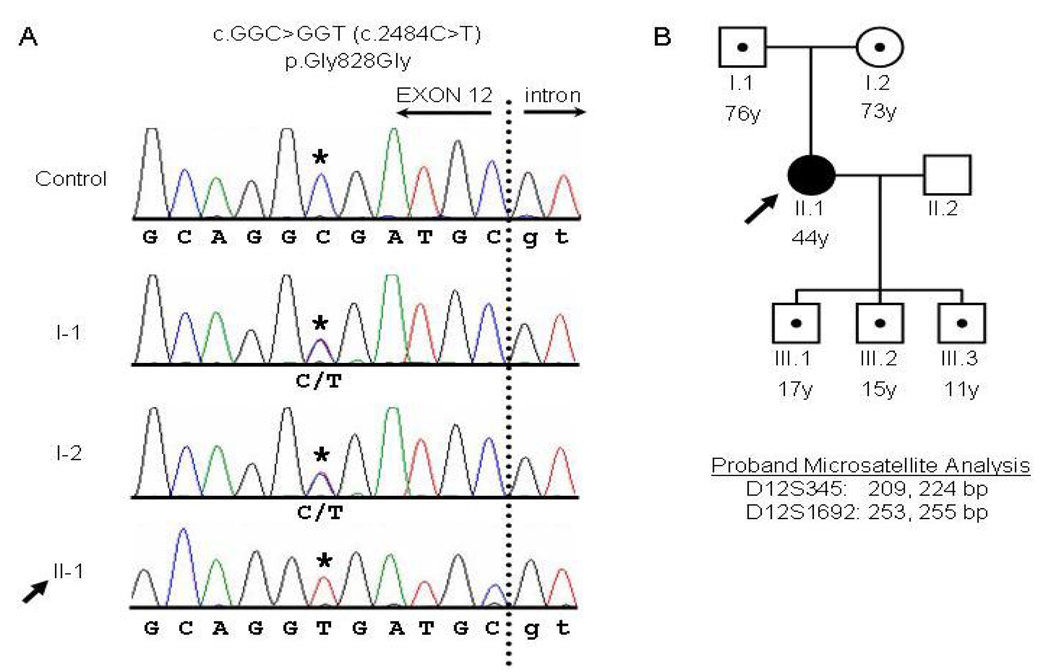
Sequencing of the 14 exons and flanking intronic regions of PKP2 revealed a homozygous C-to-T point mutation at position 2484 (c.[2484C>T]+[2484C>T], numbered relative to the start of the PKP2b coding sequence NM_004572.2; Fig. 2A). This mutation is not predicted to alter the amino acid sequence as it results in a synonymous glycine-to-glycine substitution at residue 828 (p.Gly828Gly) (Fig. 2B). This sequence variant was not present in 110 ethnically-matched controls or 100 ethnically-mismatched controls (420 chromosomes). Sequence analysis of DSG2 and DSP identified no mutations in these genes.
Figure 2.
PKP2 genomic sequencing and pedigree analysis. A: PKP2 genomic sequence analysis of the end of exon 12 and the downstream 5’ intronic splice site for a control individual, the proband (II-1), and the proband’s parents (I-1 and I-2). The dotted line indicates the boundary between the exon (capital letters) and intron (lowercase). The c.2484C>T mutation (NM_004572.2) is indicated with an asterisk. The proband’s parents are each heterozygous (C/T) at position c.2484, and the proband (arrow) is homozygous (T/T) at this position. B: Pedigree of the proband’s family. Squares, male; circles, female; black shading, affected; white shading, unaffected; dots indicate heterozygous individuals as described in (A). For the proband, the lengths of the microsatellite markers D12S345 and D12S1692 are indicated below the pedigree.
Pedigree Analysis
The proband’s parents and three children (Fig. 2B) underwent screening for ARVD. At ages greater than 70 years, individuals I-1 and I-2 each had normal electrocardiograms (ECGs), and exercise stress testing, and I-1 had a normal echocardiogram. These 2 individuals did not consent to additional cardiac evaluation. Individuals III-1, III-2, and III-3 were evaluated by ECG, echocardiogram, cardiac MRI, signal-averaged ECG, exercise stress testing, and Holter monitoring, and none met any major or minor diagnostic criteria. These family members are all heterozygous for the c.2484C>T sequence variant (Fig. 2A, 2B). Two polymorphic microsatellite markers within or near the PKP2 locus on either side of the mutation (D12S1692 and D12S345) were sized for haplotype analysis. Sizes for marker D12S1692 (within the PKP2 intron 7, telomeric to the mutation) were 253 and 255 bp, and sizes for marker D12S345 (within 1 cM centromeric to the PKP2 locus) were 209 and 224 bp, indicating that the proband is heterozygous at these two markers. Marker D12S2080, which is further telomeric beyond marker D12S1692, was not informative, and no single nucleotide polymorphisms within the PKP2 coding sequence or immediate flanking introns were heterozygous for the proband.
cDNA Sequence Analysis
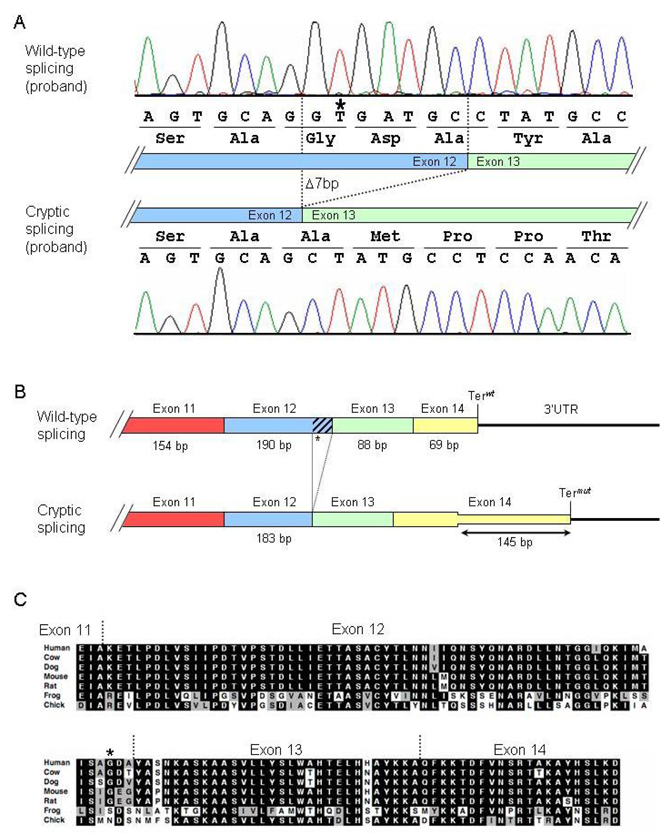
The c.2484C>T mutation changes the dinucleotide sequence starting at position c.2483 from GC to GT and is located 7 base pairs upstream of the end of exon 12. Because of this close proximity to the downstream wild-type 5’ splice site, we reasoned that this GT sequence could create a cryptic splice donor and cause aberrant splicing of the PKP2 transcript. Indeed, we observed that most of the proband’s transcripts were alternatively spliced to the mutant splice donor (Fig. 3A). Alternative splicing results in a 7-nucleotide deletion at the end of exon 12 (r.[2483_2489del]), and the ensuing frame-shift disrupts the last 54 amino acids of plakophilin-2 and extends the open reading frame by 145 nucleotides (48 amino acids) into the 3’UTR (Fig. 3B). The C-terminal 54 residues are well conserved among vertebrate species (Fig. 3C).
Figure 3.
Cryptic splicing of the PKP2 transcript caused by the 2484C→T mutation. A: Chromatograms of cloned RT-PCR products. Top panel: wild-type splicing in the proband. Bottom panel: mutant splicing results in deletion of the last seven nucleotides in exon 12 and causes a subsequent frame shift. B: Diagram illustrating consequences of cryptic splicing on translation. Exons are indicated by boxes, and the 3’UTR is drawn as a line. Terwt and Termut indicate the wild-type and mutant termination codons, respectively. C: ClustalW sequence alignment of plakophilin-2 orthologs demonstrating conservation of the C-terminal residues that are disrupted in the mutant splice forms. The G828G mutation is indicated with an asterisk (*). Black shading, residue identity; gray shading, conservation of residue character.
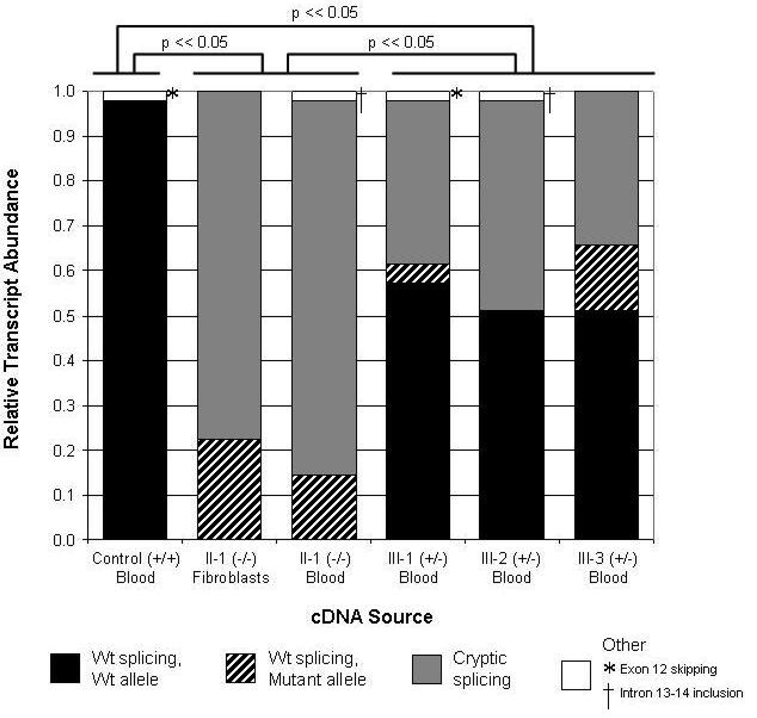
In order to quantify the relative frequency of wild-type and mutant splicing, cloned RT-PCR products generated from the proband’s RNA were analyzed by sequencing (Fig. 4). In lymphocytes, wild-type splicing occurred only 17% of the time (8 of 48 clones), whereas mutant splicing occurred 83% of the time (40 of 48 clones). To determine whether aberrant splicing was recapitulated in another tissue, the proband’s cultured dermal fibroblasts were also analyzed. Similar to lymphocytes, wild-type splicing occurred 22% of the time (8 of 36 clones), and mutant splicing occurred 78% of the time (28 of 36 clones). Combining these data, the ratio of mutant to wild-type splicing is roughly 4 to 1. There was no significant difference in mutant splicing between the proband’s lymphocytes and fibroblasts (p = 0.58). The 7-bp deletion was not observed in any of 47 clones from a control sample (Fig. 4). The difference in splicing ratio between the proband’s two samples and the control sample was highly significant (p < 10−20).
Figure 4.
Quantification of wild-type and mutant cryptic splicing of the PKP2 transcript in six clinical samples. Relative transcript abundance in two sources of RNA from the proband is compared to transcript abundance in RNA derived from an unaffected control and from the proband’s three heterozygous offspring. Proportion of wild-type splicing derived from the wild-type allele (c.2484C) is shaded in black. Proportion of wild-type splicing derived from the mutant allele (c.2484T) is shaded with black diagonal bars. Proportion of cryptic splicing with the 7-bp deletion at the end of exon 12 is shaded in gray. Minor splice forms (see Fig. 5), shaded in white, include exon 12 skipping (*) and intron 13–14 retention (†). Statistical differences in the relative abundance of wild-type and mutant splice forms are indicated above the graph, comparing control, combined proband samples, and combined heterozygous samples.
We next sought to determine the extent to which alternative splicing occurs in the proband’s three children (III-1, III-2, III-3), who are all heterozygous for the c.2484C>T mutation (Fig. 4). As expected, transcripts derived from the wild-type allele (c.2484C) were spliced normally and contributed to at least half of the total number of transcripts (59%, 52%, and 51% for III-1, III-2, and III-3, respectively). Transcripts with the 7-bp deletion comprised 37% (III-1), 48% (III-2), and 34% (III-3) of total transcripts. Normally spliced transcripts derived from the mutant allele comprised 4% (III-1), 0% (III-2), and 15% (III-3) of total transcripts. As with the proband, most of the transcripts derived from the mutant allele (c.2484T) were abnormally spliced. The combined ratio of mutant to wild-type splicing from their mutant allele is about 6 to 1. There was no significant difference in the fraction of wild-type transcripts between any two of the three siblings (p = 0.30 between III-1 and III-2; p = 0.21 between III-2 and III-3; p = 0.83 between III-1 and III-3). The difference in splicing ratio between the children’s samples and either the control sample or the proband sample was highly significant (p < 10−8).
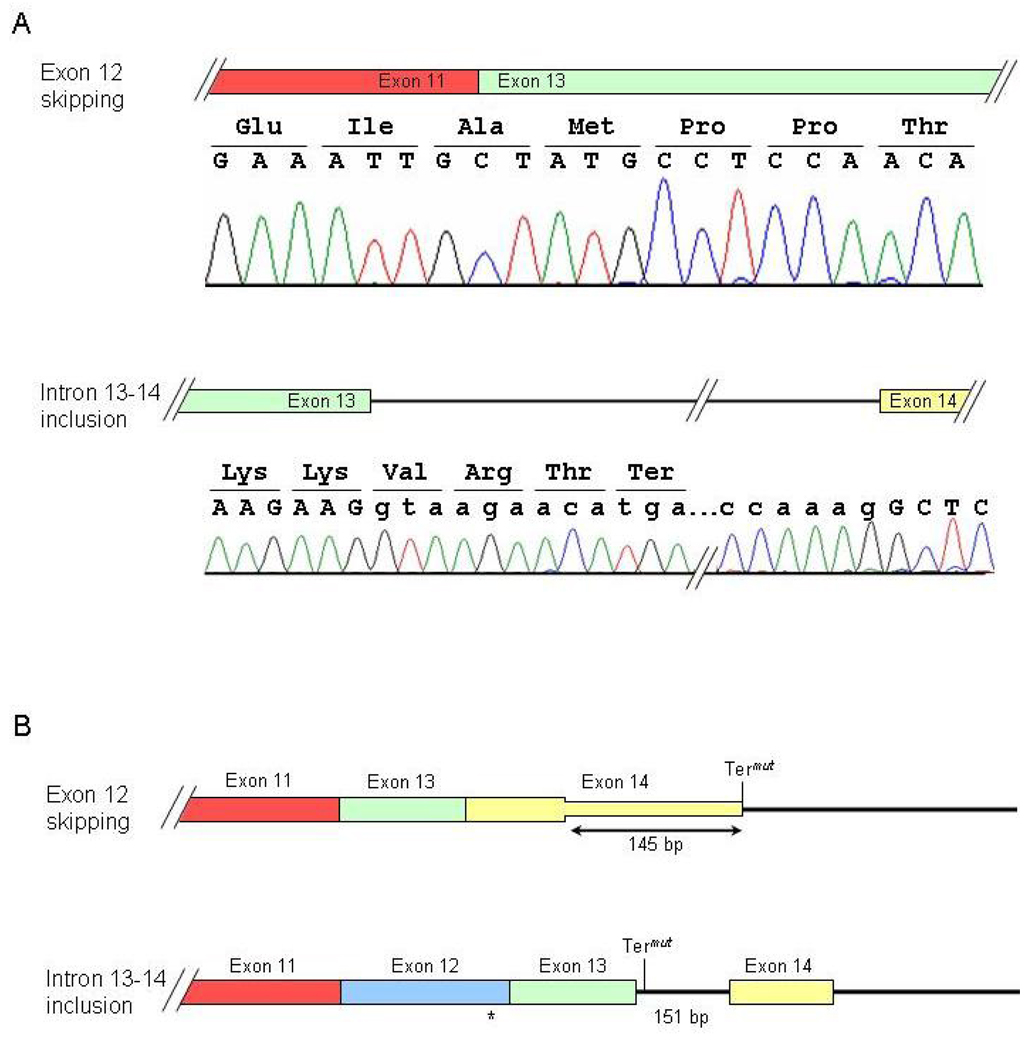
During our analysis, we observed two alternative splice forms of PKP2 that were previously unreported (Fig. 5). One clone from the control sample, and one from individual III-1 showed complete skipping of exon 12, r[2361_2550del]. This results in the loss of 61 well-conserved amino acids encoded by exon 12 and causes a frame shift that extends the open reading frame by 145 bp into the 3’UTR. In addition, one clone from the proband and one clone from individual III-2 displayed retention of intron 13–14 with proper splicing at other exon junctions, r2638_2639ins2638+1_2639−1. This results in the addition of three residues after exon 13 followed by a premature termination codon (PTC) and the loss of 21 amino acids encoded by exon 14. As this PTC resides in what is now the final exon (comprised of exon 13, intron 13–14, and exon 14), it would not be expected to render the transcript susceptible to nonsense-mediated mRNA decay (NMD).
Figure 5.
Two novel splice forms of PKP2. A: Chromatograms of RT-PCR products demonstrating skipping of exon 12 (top panel) and retention of intron 13–14 (bottom panel). B: Diagram illustrating consequences of alternative splice forms on translation. Exons are indicated by boxes; the intron and 3’UTR is drawn as lines. Termut indicates the mutant termination codon.
DISCUSSION
Both dominant and recessive forms of ARVD have previously been described, though recessive mutations in PKP2 manifesting as ARVD have not yet been reported. However, assuming a dominant model, Syrris et al described that independent of family history of ARVD, 4 of 32 heterozygous mutation carriers met task force criteria (TFC), 22/32 heterozygous mutation carriers had sub-diagnostic manifestations of disease, and 5 heterozygous mutation carriers had no ARVD criteria other than family history of ARVD (Syrris et al., 2006). Low penetrance in this disorder suggests that additional genetic and environmental factors may contribute to this condition.
Mutations may be defined on the basis of a nucleotide change resulting in alteration of a functional domain, conservation at that site, and absence of this nucleotide change in a population of unrelated unaffected controls. Initial inspection of the c.2484C>T mutation suggests that this translationally silent, glycine-to-glycine substitution, would not likely result in ARVD. However, the possibility of creation of a 5’ splice donor on the basis of this alteration led to our inspection of the mRNA transcribed with this variant. Several algorithm-based splice site prediction programs either failed to predict cryptic splicing (GeneSplicer, BDGP) or predicted that wild-type splicing would occur more frequently than alternative splicing (NatGene2). However, experimental testing of the effect of the c.2484C>T mutation on PKP2 splicing demonstrates that the mutation creates a splice donor that is utilized four to six times more frequently than the wild-type splice donor. This finding emphasizes the importance of examining other translationally-silent nucleotide changes in PKP2 that may create a cryptic splice site or disrupt an exonic splicing enhancer (ESE) or silencer (ESS), and thus contribute to development of ARVD.
The c.2484C>T transition is seen on both of the proband’s PKP2 alleles, suggesting that remote consanguinity may have contributed to her condition. However, haplotype analysis of markers on either side of the mutation indicates that she has two different PKP2 alleles. One alternate possibility is that the mutation occurred on a common ancestral allele and that the number of repeats in the closest informative markers changed size over several generations. In addition, the c.2484C>T transition occurs at the site of a CpG dinucleotide. Transitions at CpG dinucleotides are frequent mutations, as a methylated C can be deaminated to T and miscopied. This suggests that the two mutations may have arisen independently. Among sixty other probands with ARVD screened for mutations in PKP2, no other individual was found to have this mutation, nor was it seen among 420 control chromosomes screened, but its occurrence in homozygosity in this proband indicates that this mutation may be present in others.
Complete knock out of the Pkp2 gene in mice causes embryonic lethality, indicating a critical function for this gene in early development (Grossmann et al., 2004), but the threshold of minimal plakophilin-2 required for proper development in not clear. The proband in our study properly splices PKP2 in two separate tissues only ~20% of the time, which suggests that this level of wild-type plakophilin-2 is sufficient for human embryonic development. The remaining ~80% of mutant plakophilin-2 that is frame-shifted at the C-terminus may also retain partial function. The N-terminus of plakophilin-2 is sufficient for its localization to the cell border, and this region is also responsible for most of its interactions to other desmosomal components (Chen et al., 2002). However, other ARVD-causing C-terminal mutations have been identified in PKP2 (Dalal et al., 2006; Gerull et al., 2004; Syrris et al., 2006; van Tintelen et al., 2006), suggesting an important role for this region in proper plakophilin-2 structure and function.
In our study, the absence of phenotypic features of ARVD among heterozygous family members may be due to an insufficient dominant negative effect imposed by the ~40% improperly spliced plakophilin-2 on the ~60% wild-type protein. Alternatively, haploinsufficiency for wild-type plakophilin-2 may be responsible in the more common dominant form of PKP2-associated ARVD. In that model, achieving greater than a required 50% of wild-type plakophilin-2 may result in the absence of phenotypic features of ARVD. The consequence of heterozygosity for this mutation remains unknown at this point for the children of the proband in the setting of an age-dependent phenotype with average age at presentation of 29 ± 13 years (Dalal et al., 2005).
We describe two additional splice isoforms of plakophilin-2 that were identified in our analyses. Skipping of exon 12 occurs in lymphocytes obtained from both an unrelated unaffected control individual and a heterozygous mutation carrier. Inclusion of intron 13 occurs in both the proband and a heterozygous mutation carrier. The role of these splice isoforms in human disease or in alternate functions of plakophilin-2 remains unknown, as does the frequency of these splice isoforms in cardiac tissue. However, their presence alerts us to the possibility that additional intronic variants may influence splicing in this region.
To our knowledge, this represents the first documented case of ARVD being caused by a recessive mutation in PKP2. Additional analyses of other conserved nucleotide alterations in this gene may lead to increased recognition of pathogenic mutations.
ACKNOWLEDGMENTS
We thank the proband and her family for participation in this study. This work is supported by grants from the W.W. Smith Charitable Trust (DPJ) and the D.W. Reynolds Foundation. We would also like to acknowledge the Johns Hopkins ARVD Program (www.arvd.com) which is supported by the Bogle Foundation, the Campanella family, and the Wilmerding Endowments.
REFERENCES
- Alcalai R, Metzger S, Rosenheck S, Meiner V, Chajek-Shaul T. A recessive mutation in desmoplakin causes arrhythmogenic right ventricular dysplasia, skin disorder, and woolly hair. J Am Coll Cardiol. 2003;42(2):319–327. doi: 10.1016/s0735-1097(03)00628-4. [DOI] [PubMed] [Google Scholar]
- Awad MM, Dalal D, Cho E, Amat-Alarcon N, James C, Tichnell C, Tucker A, Russell SD, Bluemke DA, Dietz HC, et al. DSG2 Mutations Contribute to Arrhythmogenic Right Ventricular Dysplasia/Cardiomyopathy. Am J Hum Genet. 2006;79:136–142. doi: 10.1086/504393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauce B, Basso C, Rampazzo A, Beffagna G, Daliento L, Frigo G, Malacrida S, Settimo L, Danieli G, Thiene G, et al. Clinical profile of four families with arrhythmogenic right ventricular cardiomyopathy caused by dominant desmoplakin mutations. Eur Heart J. 2005;26(16):1666–1675. doi: 10.1093/eurheartj/ehi341. [DOI] [PubMed] [Google Scholar]
- Beffagna G, Occhi G, Nava A, Vitiello L, Ditadi A, Basso C, Bauce B, Carraro G, Thiene G, Towbin JA, et al. Regulatory mutations in transforming growth factor-beta3 gene cause arrhythmogenic right ventricular cardiomyopathy type 1. Cardiovasc Res. 2005;65(2):366–373. doi: 10.1016/j.cardiores.2004.10.005. [DOI] [PubMed] [Google Scholar]
- Chen X, Bonne S, Hatzfeld M, van Roy F, Green KJ. Protein binding and functional characterization of plakophilin 2. Evidence for its diverse roles in desmosomes and beta -catenin signaling. J Biol Chem. 2002;277(12):10512–10522. doi: 10.1074/jbc.M108765200. [DOI] [PubMed] [Google Scholar]
- Dalal D, Molin LH, Piccini JP, Tichnell C, James C, Bomma C, Prakasa K, Towbin JA, Marcus FI, Spevak PJ, et al. Clinical features of arrhythmogenic right ventricular dysplasia/cardiomyopathy associated with mutations in plakophilin-2. Circulation. 2006 doi: 10.1161/CIRCULATIONAHA.105.568642. in press. [DOI] [PubMed] [Google Scholar]
- Dalal D, Nasir K, Bomma C, Prakasa K, Tandri H, Piccini J, Roguin A, Tichnell C, James C, Russell SD, et al. Arrhythmogenic right ventricular dysplasia: a United States experience. Circulation. 2005;112(25):3823–3832. doi: 10.1161/CIRCULATIONAHA.105.542266. [DOI] [PubMed] [Google Scholar]
- Fontaine G, Fontaliran F, Hebert JL, Chemla D, Zenati O, Lecarpentier Y, Frank R. Arrhythmogenic right ventricular dysplasia. Annu Rev Med. 1999;50:17–35. doi: 10.1146/annurev.med.50.1.17. [DOI] [PubMed] [Google Scholar]
- Gerull B, Heuser A, Wichter T, Paul M, Basson CT, McDermottz DA, Lerman BB, Markowitz SM, Ellinor PT, MacRae CA, et al. Mutations in the desmosomal protein plakophilin-2 are common in arrhythmogenic right ventricular cardiomyopathy. Nat Genet. 2004;36(11):1162–1164. doi: 10.1038/ng1461. [DOI] [PubMed] [Google Scholar]
- Green KJ, Gaudry CA. Are desmosomes more than tethers for intermediate filaments? Nat Rev Mol Cell Biol. 2000;1(3):208–216. doi: 10.1038/35043032. [DOI] [PubMed] [Google Scholar]
- Grossmann KS, Grund C, Huelsken J, Behrend M, Erdmann B, Franke WW, Birchmeier W. Requirement of plakophilin 2 for heart morphogenesis and cardiac junction formation. J Cell Biol. 2004;167(1):149–160. doi: 10.1083/jcb.200402096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus FI, Fontaine GH, Guiraudon G, Frank R, Laurenceau JL, Malergue C, Grosgogeat Y. Right ventricular dysplasia: a report of 24 adult cases. Circulation. 1982;65(2):384–398. doi: 10.1161/01.cir.65.2.384. [DOI] [PubMed] [Google Scholar]
- McKenna WJ, Thiene G, Nava A, Fontaliran F, Blomstrom-Lundqvist C, Fontaine G, Camerini F. Diagnosis of arrhythmogenic right ventricular dysplasia/cardiomyopathy. Task Force of the Working Group Myocardial and Pericardial Disease of the European Society of Cardiology and of the Scientific Council on Cardiomyopathies of the International Society and Federation of Cardiology. Br Heart J. 1994;71(3):215–218. doi: 10.1136/hrt.71.3.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKoy G, Protonotarios N, Crosby A, Tsatsopoulou A, Anastasakis A, Coonar A, Norman M, Baboonian C, Jeffery S, McKenna WJ. Identification of a deletion in plakoglobin in arrhythmogenic right ventricular cardiomyopathy with palmoplantar keratoderma and woolly hair (Naxos disease) The Lancet. 2000;355(9221):2119–2124. doi: 10.1016/S0140-6736(00)02379-5. [DOI] [PubMed] [Google Scholar]
- Pilichou K, Nava A, Basso C, Beffagna G, Bauce B, Lorenzon A, Frigo G, Vettori A, Valente M, Towbin J, et al. Mutations in desmoglein-2 gene are associated with arrhythmogenic right ventricular cardiomyopathy. Circulation. 2006;113(9):1171–1179. doi: 10.1161/CIRCULATIONAHA.105.583674. [DOI] [PubMed] [Google Scholar]
- Rampazzo A, Nava A, Malacrida S, Beffagna G, Bauce B, Rossi V, Zimbello R, Simionati B, Basso C, Thiene G, et al. Mutation in human desmoplakin domain binding to plakoglobin causes a dominant form of arrhythmogenic right ventricular cardiomyopathy. Am J Hum Genet. 2002;71(5):1200–1206. doi: 10.1086/344208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A, Jager S. Plakophilins--hard work in the desmosome, recreation in the nucleus? Eur J Cell Biol. 2005;84(2–3):189–204. doi: 10.1016/j.ejcb.2004.12.020. [DOI] [PubMed] [Google Scholar]
- Syrris P, Ward D, Asimaki A, Sen-Chowdhry S, Ebrahim HY, Evans A, Hitomi N, Norman M, Pantazis A, Shaw AL, et al. Clinical expression of plakophilin-2 mutations in familial arrhythmogenic right ventricular cardiomyopathy. Circulation. 2006;113(3):356–364. doi: 10.1161/CIRCULATIONAHA.105.561654. [DOI] [PubMed] [Google Scholar]
- Tiso N, Stephan DA, Nava A, Bagattin A, Devaney JM, Stanchi F, Larderet G, Brahmbhatt B, Brown K, Bauce B, et al. Identification of mutations in the cardiac ryanodine receptor gene in families affected with arrhythmogenic right ventricular cardiomyopathy type 2 (ARVD2) Hum Mol Genet. 2001;10(3):189–194. doi: 10.1093/hmg/10.3.189. [DOI] [PubMed] [Google Scholar]
- van Tintelen JP, Entius MM, Bhuiyan ZA, Jongbloed R, Wiesfeld AC, Wilde AA, van der Smagt J, Boven LG, Mannens MM, van Langen IM, et al. Plakophilin-2 mutations are the major determinant of familial arrhythmogenic right ventricular dysplasia/cardiomyopathy. Circulation. 2006;113(13):1650–1658. doi: 10.1161/CIRCULATIONAHA.105.609719. [DOI] [PubMed] [Google Scholar]