Abstract
In this paper, we systematically presented the orientation determination of protein helical secondary structures using vibrational spectroscopic methods, particularly the nonlinear Sum Frequency Generation (SFG) vibrational spectroscopy, along with linear vibrational spectroscopic techniques such as infrared spectroscopy and Raman scattering. SFG amide I signals can be collected using different polarization combinations of the input laser beams and output signal beam to measure the second order nonlinear optical susceptibility components of the helical amide I modes, which are related to their molecular hyperpolarizability elements through the orientation distribution of these helices. The molecular hyperpolarizability elements of amide I modes of a helix can be calculated based on the infrared transition dipole moment and Raman polarizability tensor of the helix; these quantities are determined by using the bond additivity model to sum over the individual infrared dipole transition moments and Raman polarizability tensors, respectively, of the peptide units (or the amino acid residues). The computed overall infrared transition dipole moment and Raman polarizability tensor of a helix can be validated by experimental data using polarized infrared and polarized Raman spectroscopy on samples with well-aligned helical structures.
From the deduced SFG hyperpolarizability elements and measured SFG second order nonlinear susceptibility components, orientation information regarding helical structures can be determined. Even though such orientation information can also be measured using polarized infrared or polarized Raman amide I signals, SFG has a much lower detection limit, which can be used to study the orientation of a helix when its surface coverage is much lower than a monolayer. In addition, the combination of different vibrational spectroscopic techniques, e.g., SFG and Attenuated Total Reflectance – Fourier Transform Infrared spectroscopy, provides more measured parameters for orientation determination, aiding in the deduction of more complicated orientation distributions. In this paper, we discussed two types of helices: the α-helix and 3–10 helix. However, the orientation determination method presented here is general, and thus can be applied to study other helices as well.
The calculations of SFG amide I hyperpolarizability components for α-helical and 3–10 helical structures with different chain lengths have also been performed. It was found that when the helices reach a certain length, the number of peptide units in the helix should not alter the data analysis substantially. It was shown in the calculation, however, that when the helix chain is short, the SFG hyperpolarizability component ratios can vary substantially when the chain length is changed. Because 3–10 helical structures can be quite short in proteins, the orientation determination for a short 3–10 helix needs to take into account the number of peptide units in the helix.
1. Introduction
Proteins and peptides play a crucial role in many biological functions in living organisms, from enzymatic reactions to ion transportation. Proteins are also widely used in many applications such as biosensing, food production, anti-biofouling and therapeutic agents for various diseases. Because of the importance and prevalence of proteins, their structures have been a subject of study in both science and engineering fields. The α-helix and β-sheet structures are the two most common protein secondary structures, which were proposed by Pauling based on the structural characteristics of amino acids and small peptides in 1951.1 In this paper we will focus on the study of helical secondary structures.
Helices, especially α-helices, are important structures in membrane-associated peptides and membrane proteins. Membrane peptides with α-helical structures play important roles in numerous biological processes. For instance, various natural and synthetic peptides, many of which adopt α-helical structures in cell membranes, have been proposed and tested as antibiotics to prevent bacterial drug resistance.2–6 The examination of the structural and orientation information of these helical peptides in cell membranes will aid in the rational design of antimicrobial peptides with improved activities. Membrane proteins with α-helical domains (e.g., potassium ion channels and G proteins) are crucial in cell biological functions such as ion transport and signal transduction, and elucidating relevant orientation information of these helices will lead to a more detailed understanding of their functions.7–10
Pauling’s idea of a non-integral α-helical structure was remarkably innovative. He came up with a model of such accuracy that it could not be surpassed for over 40 years. Surprisingly, Pauling came up with the model using a sheet of paper and in a couple of hours, while he was visiting Oxford sick with a cold.11 Three years after his visit to Oxford, Pauling published the two helical models that he called the α-helix (3.7–residue helix) and gamma-helix (5.1-residue helix).1,12 Even though the gamma-helical structure has never been discovered in any protein structures, the α-helical one was found to occur most frequently in nature and Pauling’s theoretical model was proved to match closely with the x-ray crystallography data of the actual structure. Even before the x-ray structure was elucidated, Bragg’s colleague, organic chemist Todd, personally admitted to Bragg that he preferred Pauling’s model over Bragg’s, which had been published a year earlier. Bragg’s model was simply proposed through the enumerated possible helical structures with integral numbers of amino acid residues per turn.13 Perutz also confirmed the α-helix model proposed by Pauling almost immediately after he came across Pauling’s paper on the model.11
Vibrational spectroscopic techniques have been widely used in the studies of proteins, including the orientation determination of helical structures. Widely used vibrational spectroscopic techniques include polarized Attenuated Total Reflectance-Fourier Transform Infrared Spectroscopy (ATR-FTIR),14–28 polarized Raman spectroscopy,19,20,29–38 and, recently, Sum Frequency Generation (SFG) vibrational spectroscopy.39–55 The orientation analyses of helical structures using these techniques require knowledge of the detailed structure of the helices at the atomic level. For example, for polarized ATR-FTIR studies, in order to measure the order parameter, S, of the amide I vibrational mode of a helix, which is necessary to determine its overall orientation, it is essential to know the detailed structure of the helix, as well as the relative angular position between the transition dipole moment and the helical axis.15,21,23,28 For Raman studies, atomic structural details of helices are also required to correlate the polarized Raman results to the orientation of the helix by projecting the Raman tensor onto the molecular frame of the helix. For SFG, which can be regarded as a combination of infrared (IR) absorption and Raman scattering, to determine the orientation of a helix using polarized SFG spectra, knowledge of the detailed structure of the helix is required. Due to the accuracy of Pauling’s proposed α-helical structure, it has been used extensively in the orientation determination of α-helices using vibrational spectroscopic techniques.
ATR-FTIR and polarized Raman have been applied to investigate the orientation of protein and peptide structures, including numerous helical structures.14,15,23,28,31,33,38 More details about such research will be discussed in this article later in sections 2.3 and 2.6. SFG has several advantages over ATR-FTIR and polarized Raman in studying the orientation of proteins and peptides at interfaces and in cell membrane environments. Details regarding these advantages have been discussed in a recent review paper56 and will not be repeated here. The combination of different vibrational spectroscopic techniques in the study provides more measured parameters for the orientation determination of peptides and proteins, aiding in the deduction of more complicated orientation distributions, which will also be discussed in detail later in section 2.6.
SFG amide I signal of an interfacial α-helical structure was successfully observed in 2005,57 and subsequently, SFG methodologies were developed to determine the orientation of α-helical structures. First, group theory and the projection operator method were applied to calculate qualitatively the SFG hyperpolarizability tensor and its relation to the SFG measured susceptibility tensor.58 The orientation of fibrinogen at a polymer/protein solution interface (based on its α-helical coiled coils),59 α-helical melittin in a lipid bilayer,60 and Gβγ associated with a lipid bilayer (based on its α-helical domains), were then experimentally measured.61 We found that fibrinogen molecules adopt a broad orientation distribution on the polymer surface; melittin molecules exhibit two well-defined orientations: one parallel to the bilayer surface and another one perpendicular to it; and that the Gβγ orientation is influenced by the lipid composition in the bilayer. In this paper, we systematically present the detailed methodology to determine the orientation of α-helical structures using SFG. The majority of these details have not been reported previously. We also validate various parameters needed to develop and refine the methodology. This paper further extends the method to determine the orientation of 3–10 helices and discusses whether the number of peptide units in a helix can alter the methodology to determine the helix orientation.
2. Orientation Determination of an α-Helix
2.1. Introduction of Pauling’s α-helix
The first and most important assumption that Pauling and his coworkers made in their model of the α-helix was that each peptide bond is planar due to the resonance structure between the carbonyl C=O bond and the amide C–N bond.1 Based on this assumption, two helical models were constructed and proposed: a gamma-helix and an α-helix, with 5.1 residues per turn and 3.7 residues (later refined to 3.6 residues according to X-ray diffraction results) per turn, respectively.
In a Pauling α-helix, the structure repeats itself every 5.4 Å along the helical axis. Alpha-helices have 3.6 amino acid residues per turn. Each residue is related to the next one by a translation of 1.5 Å along the helical axis and a rotation of 100° (Figure 1). One important aspect of this structure is the intra-molecular hydrogen bonding scheme that renders the structure very stable. Every backbone carbonyl C=O and N-H group on a peptide unit is hydrogen-bonded to another N-H and C=O, respectively, on another unit four residues away. Additionally, the backbone C=O groups point in the same direction, while the N-H groups point in the opposite direction.
Figure 1.
Correlation between the direction of the amide I transition dipole moment in one peptide unit and the molecular axis of an α-helix. The first peptide unit in Pauling’s α-helix is also illustrated.
Extensive research has been done to analyze the three amide I vibrational modes of α-helices19,33–35,37,62–68: A, E1 and E2. The two modes A and E1 are IR active, while all three modes are Raman active. Because an SFG active mode needs to be both IR and Raman active, only the A and E1 modes are SFG active. The A and E1 modes are parallel and perpendicular to the helical axis, respectively. The SFG hyperpolarizability tensor can be expressed as a tensor product of the IR transition dipole moment and the Raman polarizability tensor:
| (1) |
where (l,m,n) are the molecular coordinate indices, and the superscript “*” denotes the complex conjugate,andare the IR dipole moment and the Raman polarizability derivatives with respect to the normal coordinate of the qth vibrational mode, respectively (in this paper, we refer to these derivatives as components of the IR transition dipole moment and components of Raman polarizability tensor, respectively). As Eq. (1) indicates, if both the IR transition dipole moment and Raman polarizability tensor are known, the SFG hyperpolarizability tensor of that vibrational mode can be deduced. In this paper, the IR transition dipole moment and Raman polarizability tensor for a helix are calculated from the IR transition dipole moment and the Raman polarizability tensor of a peptide unit using the bond additivity model according to the α-helix symmetry and structure (Sections 2.2 and 2.3). These calculated quantities are compared to and validated by the experimentally measured quantities acquired by polarized IR and Raman spectroscopic techniques in the literature. The SFG amide I hyperpolarizability tensor for an α-helix is then deduced by incorporating these values into Eq. (1).
2.2. IR transition dipole moment of an α-helix amide I mode
Higgs successfully applied group theory to characterize and derive the selection rules of the amide I modes of an α-helix in 1953.19 According to Higgs, the components of the dipole moments (M+, M−, Mo) of the nth peptide unit are:
| (2) |
| (3) |
| (4) |
where ψ indicates the angular distance around the helical axis between two adjacent peptide units; M+(n), M−(n) and Mo(n) are the dipole moment components of the nth peptide unit that are involved in the absorption of right circularly polarized, left circularly polarized, and linearly parallel polarized IR radiation, respectively; and the terms einψ and e−inψ perform the translations from the first peptide unit to the nth peptide unit. The two types of linearly polarized light, parallel and perpendicular, are referenced to the principal axis of the helix.
The IR absorption intensities of each dipole moment component, for the entire α-helix, can be determined by summing over all the peptide units. For the IR absorption using the right/left circularly polarized or linearly perpendicular polarized light,
| (5) |
where ϕk is the angle between the transition dipole moment Mk and the helical axis, and fk is a proportional factor.
For the linearly parallel polarized absorption,
| (6) |
Therefore, the absorption intensity ratio between the perpendicular and the parallel modes is:
| (7) |
The angle ϕk can be deduced by measuring the dichroic ratio of amide I modes of perfectly aligned α-helical structures. After ϕk is deduced, polarized IR measurements can be used to determine the orientation of α-helical structures.
There have been numerous efforts to deduce the angle ϕk using IR dichroism from polarized FTIR experiments. In the early 1960’s, a series of findings reported ϕk values ranging from 30 to 40 degrees. Among these studies, both Miyazawa and Blout (ϕk= 29–34°)24 and Tsuboi (ϕk= 39°)27 reported their measurements on the α-helix of poly-γ-benzyl-L-glutamate. Bradbury et al. studied the structure of the ω-form of poly-β-benzyl-L-aspartate (ϕk=40°).17 This type of work has also been done more recently. In 1995, ϕk was determined by Axelsen from his studies on the peptide L24 (ϕk<34°),14 and in 2000 by Marsh from studies on poly(γ-methyl-L-glutamate)x-co-(γ-n-octadecyl-L-glutamate)y (ϕk= 38°).23 As discussed by Bradbury et al., Miyazawa and Blout assumed a planar (instead of uniaxial) orientation in the model, which caused his result to be slightly different from others. If a uniaxial model had been applied in Miyazawa and Blout’s analysis, a range of 33°–37.5° would be deduced from their data, which would correlate better with other studies.17
Since the α-helices used in the research mentioned above may not be ideal and can vary from one sample to another, the amide I signals may not be simple and/or straightforward for analysis. The amide I signal can be affected by the hydrogen bonding scheme and the dipole-dipole coupling among the neighboring groups within a particular α-helical structure. This could explain the discrepancies among the reported ϕk values of α-helical structures with a varying number of peptide units, but such discrepancies are not substantial.
Here, a methodology is implemented, similar to that proposed by Suzuki, to calculate the ratio between the dipole moment projections perpendicular and parallel to the principal axis of an α-helix.26 Wang’s corrected ϕk angle of 42° is used in these calculations.59 It is interesting to note that Marsh also found through transmission FTIR that ϕk should be 42°.23 The results from the calculations are then cross-checked with the calculations from Choi69 and the experimental data obtained from α-Poly(L-alanine)37 and PG3023, which possess well-defined right-handed α-helical structures (see below for more details).
The parallel and perpendicular components (relative to the principal axis) of the amide I IR transition dipole moment of an α-helix are calculated using the bond additivity model. In this model, the dipole moment of each peptide unit in the helix is projected onto the parallel and perpendicular directions in the helix’s molecular frame. The projections of these dipole moments onto the parallel and perpendicular axes in the molecular frame are then integrated to obtain the perpendicular and parallel dipole moment components of the helix. The calculated IR transition dipole moments (in the unit of the IR transition dipole moment of each peptide unit) for the two IR allowed amide I modes of an α-helix are:
For the A mode (the parallel mode):
| (8) |
For the E1 mode (the perpendicular mode):
| (9) |
The angles in the parentheses on the left hand side of Eqs. (8) and (9) are the vibrational phase differences of the adjacent peptide units. From these results, the ratio of M(y+x)/Mz can be deduced to be 0.64. This calculated ratio matches closely with the value of 0.62 obtained from polarized IR measurements using well-aligned α-helical α-Poly(L-alanine) by Lee et al.20 This value also falls within the experimental range of 0.5 to 1.0 determined by polarized IR spectra of α-helical PG30 by Marsh et al.23 Moreover, it is in good agreement with the calculated result from Choi that suggests a M(y+x)/Mz ratio of 0.62.69 The bond additivity method used to calculate the IR transition dipole moment of an α-helix appears to yield a reasonably accurate M(y+x)/Mz ratio of 0.64.
2.3. Raman polarizability tensor of an α-helix amide I mode
The Raman tensors of vibrational modes of various functional groups, such as the ester C=O stretch, the amide I and III modes, and the C-Cphenyl stretch, have been successfully described by Tsuboi by investigating Raman spectra of a uniaxial tetragonal aspartame crystal.30 The Raman tensor for the amide I mode (Figure 2) takes the following form:
| (10) |
Figure 2.
A. Principal axes of a single peptide group for the amide I Raman tensor.30
For a regular helical structure with infinite length, according to its symmetry, the three tensors of the vibrational modes A, E1 and E2 can be written as:34
| (11) |
| (12) |
| (13) |
In the above equations, α11, α12, α13 etc. are the components of the Raman polarizability tensor when the axes are chosen such that the z-axis corresponds to the coordinate index 3; the y-axis corresponds to the coordinate index 2; and the x-axis corresponds to the coordinate index 1. Because the E2 mode is not IR allowed, it is not necessary to consider this mode while calculating the SFG hyperpolarizability, and will thus be ignored in the remainder of this discussion.
In the case of the α-helical structure, the Stokes Raman polarizability tensors of the vibrational modes A and E1 can be written as the following:
For the A mode:
| (14) |
where .
For the E1 mode:
| (15) |
| (16) |
where ,ΘEo and ΘAo are only generic phase terms. While all amide I groups in different peptide units vibrate in phase in the A mode, there is a phase difference of 100° in the E1 mode between two adjacent peptide units.
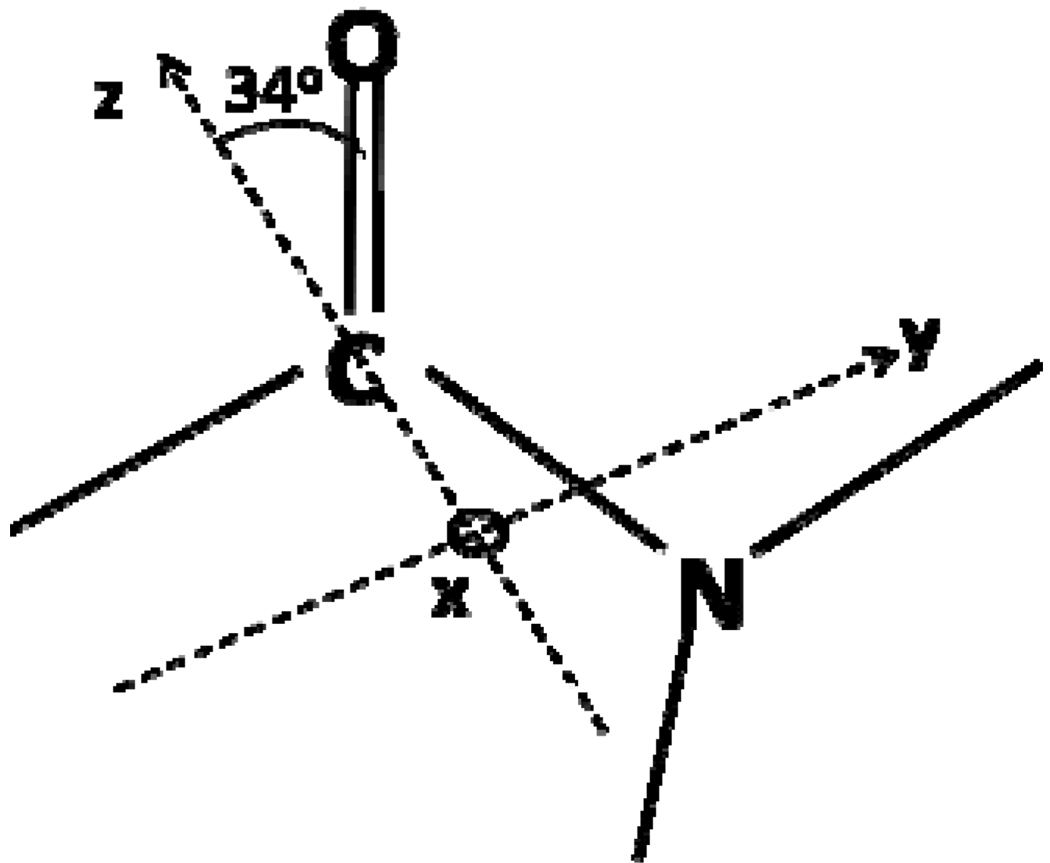
Based on the symmetry and structure of an α-helix, the Raman polarizability tensors can be obtained, as shown above. The Raman tensor components can now be quantitatively deduced from Tsuboi’s Raman tensor of aspartame,30 which can be used to represent a peptide unit in an α-helix. In order to apply Tsuboi’s Raman tensor to the α-helical structure, a transformation needs to be applied to bring Tsuboi’s Raman tensor to the first peptide unit of the α-helix in the molecular frame. Pauling proposed an accurate α-helical structure but his model was that of a left-handed helix, while most (if not all) of the α-helical structures in nature are right-handed. The left-handed and the right-handed α-helices are mirror images of each other and therefore the absolute value of the overall calculated Raman tensor is not affected. However, the right-handed version of Pauling’s helix will be used in subsequent discussions due to its relevance in nature. The Euler angles transforming Tsuboi’s Raman tensor to the first peptide unit in the right-handed version of the Pauling helix are: ϕ = 0°, θ = 133.3° and ψ = 270° using the x-convention rotation. The Euler transformation in the x-convention yields the following rotation matrix:
| (17) |
With the set of Euler angles listed above, this rotation ξ matrix becomes:
| (18) |
The rotation is applied on the coordinate system (a,b,c) which describes the tensor as:
| (19) |
Therefore,
| (20) |
Due to the uniaxial property of the α-helix, all the peptide units are assumed to be approximately identical and can thus be transformed geometrically onto each other. This makes the process of obtaining the helical total Raman tensor much simpler. The Raman polarizability tensor for each peptide unit in the α-helix can be obtained by successively performing 100 degree rotations around the helical axis to move from one peptide unit to the next. The Raman tensor of the entire α-helix can be calculated by multiplying the Raman tensor of each peptide unit with the phase factor in the vibrational mode and then summing over all of them. The A mode vibrations of all the peptide units are all in phase. Therefore, the Raman tensor for the A mode of an ideal α-helix can be calculated as:
| (21) |
The Raman polarizability tensors of the E1 modes can be calculated similarly after incorporating the phase difference between the adjacent peptide units. The deduced Raman polarizability tensors of the A and E1 modes are: For the A mode:
| (22) |
For the E1 mode:
| (23) |
These results agree very well with experimental results found in the literature. For example, αbb/αcc and αcb/αcc can be calculated to be 0.59 and 0.35, respectively. The calculated αbb/αcc value (0.59) is similar to the widely used value reported by Tsuboi et al. of 0.54.70 In general, the above calculated A and E1 mode Raman polarizability tensors of an α-helix closely match the experimentally measured values reported in the literature, which were determined with polarized Raman experiments on well-aligned α-helical samples. For the A mode, an experimental ratio of αbb/αcc within a range of 0.53 to 0.64 was reported, which is in good agreement with our value of 0.59. Many of these values are obtained using the assumption that all the α-helical structures are perfectly aligned along the z-axis (or the fiber axis when fiber samples are used) in the Raman experiments. For example, Wilser et al. measured this ratio to be 0.55 according to their polarized Raman studies on the α-helical polypeptide poly-γ-benzyl-L-glutamate.29 Using a wool fiber, Rintoul et al. measured this value to be 0.62.71 Ackermann et al. obtained a ratio of 0.62 by experiments on α-helices in intact human hair.72 Overman et al. deduced this value to be 0.58 by performing measurements on α-helical pVIII subunits in the filamentous virus Ff (fd, f1, M13).31 Tsuboi et al. studied the α-helical coat protein in filamentous bacteriophage PH75 and found the value to be 0.64.38 In the last two experiments the authors did not use the experimentally measured values as αbb/αcc, and instead, used these values and the calculated value of 0.54 to determine the orientation of the helices. Perhaps in these samples, the α-helices were actually more or less aligned along the z-axis.
The αcb/αcc value has also been measured experimentally using polarized Raman experiments. Rintoul et al. measured this value to be 0.39 by studying a wool fiber. Lee et al. investigated well-aligned poly-alanine samples and obtained a value of 0.35.37 Wilser et al. reported this ratio to be between 0.34 and 0.40 in their studies on α-helical poly-γ-benzyl-L-glutamate.29 These experimental values are all comparable to our calculated value of 0.35. Therefore we believe that this calculated value is also correct. Using these two tensors combined with the transition dipole moment calculated earlier, we can calculate the needed hyperpolarizability components for our later SFG data analysis.
2.4. SFG data analysis for α-helices based on the calculated IR transition dipole moment and Raman polarizability
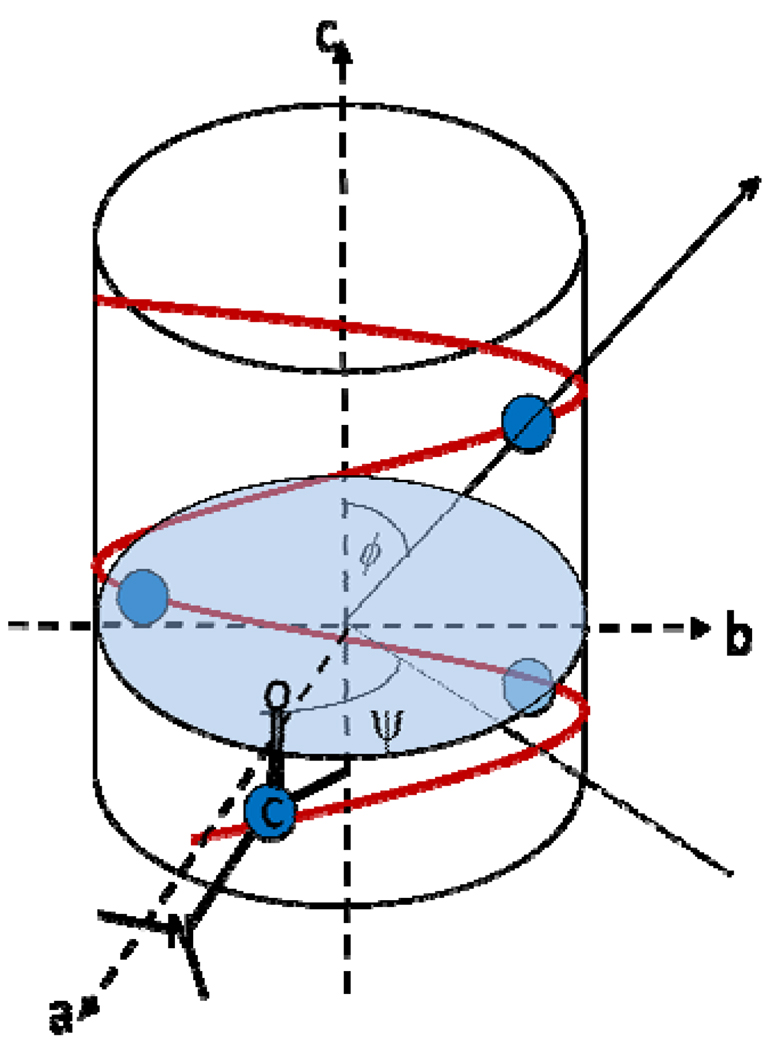
The SFG hyperpolarizability tensor is a third-rank tensor with 27 elements. It is a tensor product of the Raman polarizability tensor and the IR transition dipole moment (Eq. (1)). The theoretical background of SFG has been discussed in great details in many publications39–61,73–76 and will not be repeated in this paper. The discussion will now be steered toward the application of SFG in the orientation analysis of α-helical structures. An α-helical peptide’s orientation can be measured by analyzing SFG amide I spectra collected under the polarization combinations ssp (s-polarized SFG signal beam, s-polarized visible input beam, and p-polarized IR input beam) and ppp. The SFG susceptibility tensor element χijk (i, j, k=x,y,z) is related to the SFG molecular hyperpolarizability tensor element βlmn (l, m, n = a, b, c) by a Euler angle projection:59,75,77,78
| (24) |
where Ns is the surface density of α-helical repeating units, and the notation “< >” indicates the average value.
For vibrational modes of different symmetries, these relations can be quite different, and have been discussed in detail in the literature.75,78 According to the symmetry of the α-helix, the following relationships can be expressed:58–60 For the A mode:
| (25) |
| (26) |
| (27) |
where For the E1 mode:
| (28) |
| (29) |
| (30) |
where the notation “< >” indicates the average value, and θ is the angle between the principal helical axis and the surface normal. When assuming that θ has a delta-distribution, <cosθ>= cosθ. The orientation distribution can be more complicated than a delta-distribution, which will be discussed in more detail later.
Experimentally, the SFG signals from the A and E1 modes cannot be resolved due to the resolution of our SFG system. The amide I signal can therefore be considered as arising from a contribution of both modes:
| (31) |
| (32) |
| (33) |
From the above expressions, if βaac/βccc and βaca/βccc are known and the combined χzzz/χyyz or χzzz/χyzy are measured, the orientation angle θ should be able to be deduced. Since the SFG hyperpolarizability is a product of the Raman polarizability and the IR transition dipole moment, Eqs. (1), (8), (9) and (18) can be combined to give:
| (34) |
| (35) |
Here, the complex conjugate of the Raman polarizability tensor is used since the Raman process involved in SFG is an anti-Stokes Raman process.
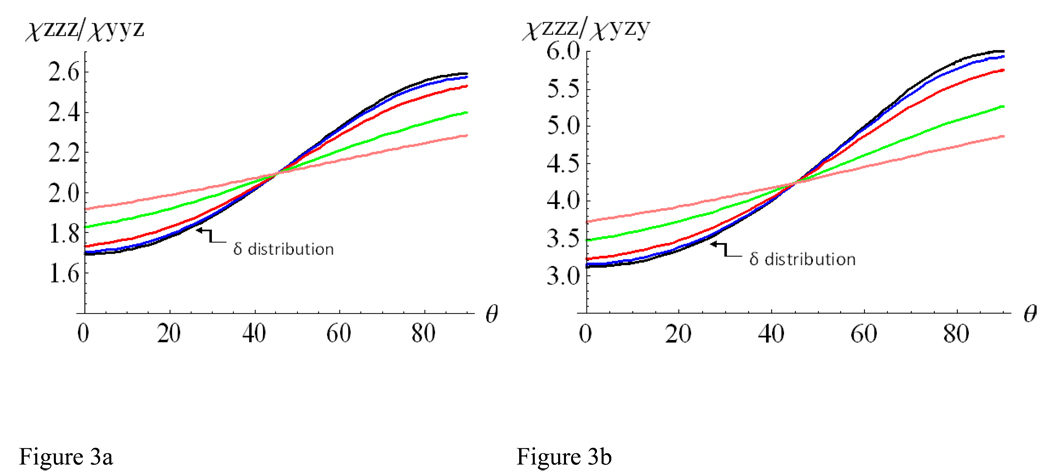
Assuming the orientation angle θ to be a delta-distribution, meaning that all of the α-helices adopt an identical orientation, the relationships between each of the ratios, χzzz/χyyz and χzzz/χyzy, and θ can be described by the curves shown in Figures 3a and 3b. However, it can be the case that not all of the helices adopt the exact same orientation, and that instead a distribution of orientations is present. In this scenario, the orientation can be assumed as a Gaussian (normal) distribution with a standard deviation σ:
| (36) |
| (37) |
| (38) |
Figure 3.
Relationships between the (a) χzzz/χyyz or (b) χzzz/χyzy ratio and θ for an α-helix in terms of different Gaussian distribution widths σ. Black: σ = 0, blue: σ = 5°, red: σ = 10°, green: σ =20°, and pink: σ = 30°. When σ is zero, the distribution is a delta-distribution.
The above mean values 〈cos (θ)〉 and 〈cos3(θ)〉 can now be used in the Eqs. (25) to (30) to describe the relationship between θ and the SFG susceptibility component χijk. The variation of the relationship between the ratios, χzzz/χyyz and χzzz/χyzy, and θ in terms of different Gaussian distribution widths σ is displayed in Figures 3a and 3b.
2.5. The effect of varying the number of peptide units in an α-helical structure on the SFG data analysis
The theoretical framework discussed so far has been for ideal α-helical structures, either a unit cell with eighteen peptide units (for five turns) or an infinitely long α-helix. One may pose the question of whether the above SFG data analysis method is still valid if the α-helical species under study does not possess a perfect α-helical structure that has a multiple of repeated helical units, e.g., 18, 36, or 54 amino acids. In nature, many α-helical structures do not have a multiple of repeated helical units; this concern will be addressed in this section.
If we assume that each peptide unit in an ideal α-helical unit (defined as 18 residues) is a “normal” peptide unit, while any peptide units beyond a multiple number of repeated helical units are “extra” units, when the α-helix is very long, the number of “extra” units is much less than that of “normal” units. In that case, it seems that these “extra” units should not make the SFG data analysis deviate too much from that for an ideal α-helix. If every 3.6 units (one turn) is considered as a repeating unit for an α-helix, then for any α-helix longer than 18 peptide units, the “extra” units are much less than the normal units. For example, for an α-helix with 23 peptide units, 21.6 are normal units, and only 1.4 are extra units. In this case, the deviation from the α-helical symmetry should be minimal, and the SFG data analysis for a perfect α-helix can be approximately applied. If the method discussed above to calculate the hyperpolarizability component ratios αbb/αcc and αcb/αcc was used for α-helices longer than 18 peptide units, their values should be reasonably similar. Hence, the SFG measured susceptibility and orientation angle relation should also be similar.
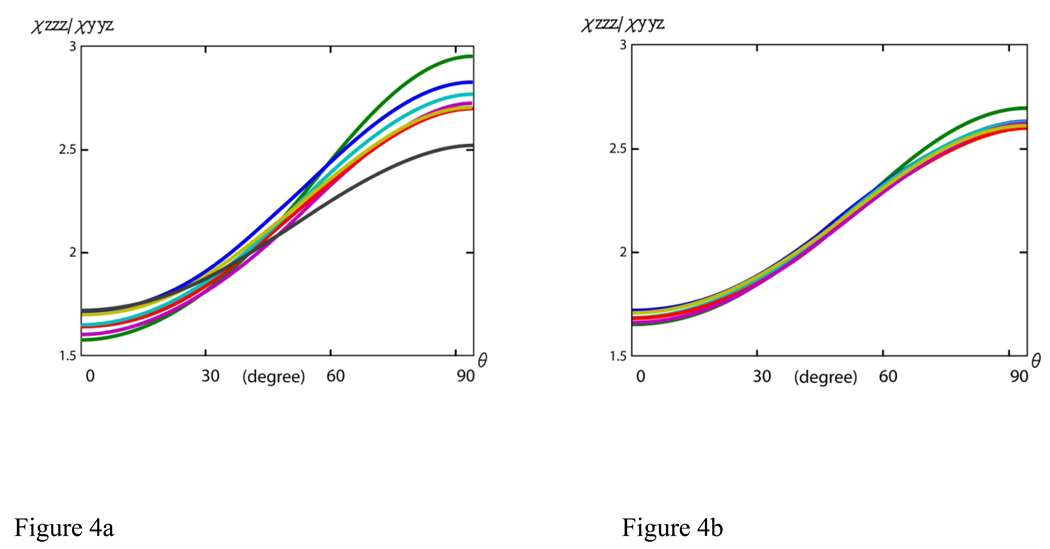
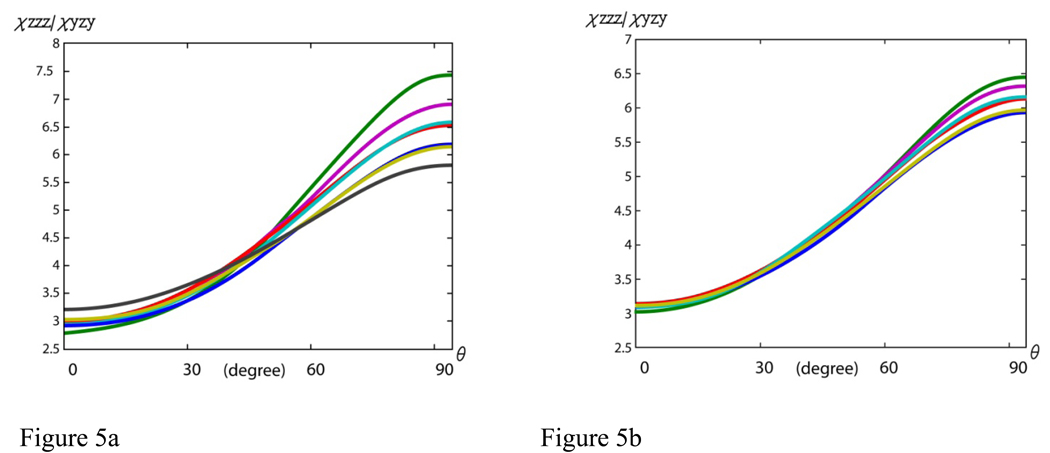
The above reasoning may not apply, however, for relatively short α-helical structures (e.g., less than 10 residues long). In this case, the data analysis for any α-helical peptides that have their number of peptide units not close to 3.6 or 7.2 (e.g., containing 2, 5, or 6 peptide units) may be influenced by a breaking of the symmetry. However, the occurrence of these short α-helical structures in nature is rare and does not necessitate a discussion here. The focus, instead, will be placed on α-helical structures that have more than ten but less than eighteen peptide units. For these α-helices, the magnitude of the effect of breaking the symmetry of a perfect α-helix on the hyperpolarizability ratios will be discussed. The hyperpolarizability ratios βaac/βccc and βaca/βccc of α-helical structures that have 10, 12, 13, 15, 16 and 17 peptide units were calculated. The dependence of the α-helix’s orientation curve on the helix length is deduced below in order to understand how the SFG data analysis method is affected by changing the peptide unit number in an α-helix away from eighteen. For comparison, the calculations were also done for α-helical structures with a number of peptide units ranging from 28 to 35. The comparison between these two sets of orientation curves should provide an idea of how the length of the structures affects the deviation from the symmetry of an α-helix unit. This comparison can be found in Figure 4a, 4b, Figure 5a and 5b. It is clear from the two sets of curves in both the χzzz/χyyz and χzzz/χyzy plots that as the number of amino acid residues of the structure becomes larger, the result is less dependent on the number of amino acid residues, leading to less deviations in the relationship between the ratio, χzzz/χyyz or χzzz/χyzy, and θ from the ideal case. Our findings show that for shorter α-helices, it is important to calculate the molecular hyperpolarizability ratios βaac/βccc and βaca/βccc using the bond additivity model with the number of amino acid residues in the particular α-helix under study. Then the relationship between χzzz/χyyz or χzzz/ χyzy and θ can be deduced for SFG data analysis. By taking into account the effect of the peptide chain lengths to establish a specific orientation curve for each α-helical structure, it should provide a more accurate result than using the curve for an ideal helix. For longer α-helices, it is a valid approximation to use the SFG data analysis method developed for an α-helix unit.
Figure 4.
Relationship between the χzzz/ χyyz ratio and θ for α-helices with different chain lengths: (a) blue: 10, green: 12, red: 13, cyan: 15, purple: 16, yellow: 17, black: 18 residues; (b) blue: 28, green: 30, red: 31, cyan: 33, purple: 33, and yellow: 35 residues.
Figure 5.
Relationship between the χzzz/ χyzy ratio and θ for α-helices with different chain lengths: (a) blue: 10, green: 12, red: 13, cyan: 15, purple: 16, yellow: 17, and black: 18 residues; (b) blue: 28, green: 30, red: 31, cyan: 33, purple: 33, and yellow: 35 residues.
2.6. Combination of measurements using different vibrational spectroscopic techniques
In section 2.4 the possibility that all α-helices in a sample may not adopt the exact same orientation was discussed, as well as the prospect of using a Gaussian distribution to describe the orientation distribution. To do this, the average orientation and orientation distribution width need to be simultaneously deduced. Also in the same section it was shown that SFG can measure two orientational parameters: <cosθ> and <cos3θ>. The two macroscopic hyperpolarizability ratios χzzz/ χyyz and χzzz/ χyzy have been extensively discussed in the orientation analysis above. These two measurements are not independent and thus can only be used to cross-check the accuracy of the data analysis method and the reliability of the experimental measurements. To measure <cosθ> or <cos3θ> independently, the absolute intensity of SFG signal needs to be obtained, as discussed in detail in our previous publications.60,79 With two independent measurements, the Gaussian distribution can be deduced. However, the orientation distribution may sometimes be more complicated than a Gaussian distribution.
The same α-helices may adopt two different orientations with separate orientation angles θ1 and θ2. If N is the fraction of α-helical molecules tilting at θ1 from the surface normal, and (1-N) is the fraction of molecules tilting at θ2 from the surface normal:
| (39) |
| (40) |
There are now three unknowns: θ1, θ2 and N. As discussed previously, SFG can only measure two parameters for the orientation angle (for α-helices). Solely using SFG to deduce all the three unknowns is therefore impossible. As demonstrated in earlier work, it is possible to combine SFG and ATR-FTIR measurements to deduce the three unknowns.60 ATR-FTIR has been extensively used in the studies of proteins/peptides at interfaces.14,15,23,28,33 These studies involve the investigations of the adsorption amount, secondary structures, as well as the orientations of proteins and peptides at interfaces. Excellent reviews on this technique in such studies are available;15,28 here, a brief review of some of the most relevant aspects of this technique, and its complementarity to SFG in the studies of the orientation of helical structures at interfaces, will be discussed.
In ATR-FTIR studies, the tilt angle of an α-helix can be calculated from the order parameter (Sθ), which is defined as:
| (41) |
with θ being the tilt angle between the helix’s principal axis and the surface normal. The bracket denotes the time and ensemble average. Theoretically, <cos2θ> can be determined from the measured intensity ratio in ATR-FTIR using p- and s-polarized IR light.28 If θ is assumed to have the simplest delta-distribution, the orientation of the helix can be determined from this measured intensity ratio. The advantages and disadvantages of ATR-FTIR have been mentioned in our previous discussions28,56,74 and will not be reiterated in this paper. Here, the use of <cos2θ> as a third measured parameter obtained by ATR-FTIR, in addition to the two measured SFG parameters, will be shown in solving the two delta-distributions case discussed above. For ATR-FTIR:
| (42) |
By combining SFG and ATR-FTIR measurements, it is possible to measure θ1, θ2 and N simultaneously. The orientation of α-helices may be even more complicated, thus requiring additional measured parameters to deduce these complex orientations. In these cases, other vibrational spectroscopic techniques, such as Raman and four-wave mixing (FWM), can be utilized.80 Also, a maximum entropy distribution function can be used as a trial function for determining the orientation distribution.59,79 Mathematically, this function has the minimum bias with a certain number of measured parameters available. If the orientation distribution is still difficult to deduce after using combined vibrational spectroscopic studies, isotope labeled proteins can be used, similar to those in NMR studies.
2.7 Discussion on the measurement of χzzz with the near total reflection geometry
The SFG susceptibility components, χzzz/χyyz and χzzz/χyzy, can be experimentally probed using SFG spectra collected with different polarization combinations of the input laser beams and output signal beam:
| (43) |
| (44) |
| (45) |
where Lii(ω) is a Fresnel coefficient and local field correction factor and β, β1 and β2 are angles of the signal, visible and IR beams with respect to the surface normal, respectively. For an α-helix on an isotropic surface, χxzx = χzxx. Also, in a near total reflection SFG experimental geometry,81
| (46) |
Thus these two terms cancel each other out in Eq. (43), leading to:
| (47) |
If the input or output beam angle is close to the critical angle of the total internal reflection, Lxx (ω) is close to zero. Therefore, for the near total reflection geometry,81
| (48) |
This analysis indicates that the ppp signal is the result of destructive interference between the χxxz and χzzz components. When a near total reflection geometry is adopted in the SFG experiment, the ppp signal probes χzzz.
3. Orientation determination of a 3–10 helix
Although not the most common helical structure in nature, the 3–10 helical structure was proposed almost ten years earlier than the abundant α-helical structure.82 There have been studies on whether the 3–10 helical structure is actually more common as it may be involved as an intermediate step in the protein folding process.83,84,84–89 A 3–10 helix is characterized by the hydrogen bonds formed between the ith C=O group to the (i+3)th H-N group. The hydrogen bonds in 3–10 helices are stronger than those in α-helices, causing a shorter distance between the oxygen and the hydrogen atoms in the hydrogen bond.33,90 In this helix, the angular distance between two adjacent amino acid residues is 120° with an axial translation of 1.95 Å. The pitch is then 5.94 Å, and there are three residues per turn. For 3–10 helices, only A and E1 modes are both IR and Raman active, which makes them observable by SFG.33,90 Below, a similar methodology as that applied for α-helical structures will be applied to the orientation analysis of stable 3–10 helical structures at interfaces. The bond additivity model is applied to calculate the SFG molecular hyperpolarizability ratios βaac/βccc and βaca/βccc which will be used to construct the relationship between the macroscopic SFG susceptibility component ratio, χzzz/χyyz or χzzz/χyzy, and the tilt angle θ of the helix.
3.1 IR transition dipole moment of a 3–10 helix amide I mode
For A mode (the parallel mode):
| (49) |
For E1 mode (the perpendicular mode):
| (50) |
According to Eqs. (49) and (50), the ratio M(y+x)/Mx is calculated to be 0.71. In this calculation, the angle between the dipole moment and the helical axis is assumed to be 45°, which was back-calculated from Choi’s calculated values of the perpendicular and parallel modes of the transition dipole moment.69 A value of 45.6° would make our calculation match with Choi’s value perfectly. However, upon studies of poly(α-aminoisobutyric acid) using electron diffraction, Malcolm and Walkinshaw approximately set an upper limit of this angle to be about 45°.90 The value of 45° is therefore believed to be reasonable to use in the calculation.
3.2 Raman polarizability tensor of a 3–10 helix amide I mode
To start the process of analyzing the total polarizability of a 3–10 helix, its crystal structure is first required. Malcolm and Walkinshaw have successfully proposed the crystal structure of poly(α-aminoisobutyric acid) using the average values from the crystal structure determinations of 17 independent residues.91 The coordinates of this crystal structure are shown in Table 1. According to this crystal structure, the peptide unit is approximately planar with the C=O bond almost parallel to the y-axis. The Euler angles that transform Tsuboi’s Raman tensor to the first link of the 3–10 helix were calculated to be ϕ=0°, θ=301.7° and ψ= 270° using the x-convention rotation. The rotation matrix takes the following form:
| (51) |
Therefore,
| (52) |
Table 1.
Cartesian coordinates of the first peptide link in a 3–10 helix.
| x (Å) | y (Å) | z (Å) | |
|---|---|---|---|
| C' | 1.12 | 0.54 | 1.189 |
| O | 1.64 | 0.51 | 2.304 |
| N | −0.07 | 1.12 | 0.957 |
| H | −0.45 | 1.21 | 0.018 |
In the case of the 3–10 helix, the uniaxial property is not strictly satisfied, which is different from the α-helix case. However, the C=O bonds only orient slightly (a few degrees) away from the helical axis. Therefore, the uniaxial property can still be considered so that all the peptide units are assumed to be approximately identical and can be transformed geometrically into each other. The Raman tensor of a 3–10 helix can then be calculated by successively performing the rotation of the Raman tensor of the peptide unit around the helical axis 120° from one unit to the next and summing over them. For an ideal 3–10 helix, the Raman tensor can be calculated as:
| (53) |
The calculated polarizability of the A and E1 amide I modes are: For the A mode:
| (54) |
For the E1 mode:
| (55) |
3.3 SFG data analysis for 3–10 helices
Combining the calculated transition dipole moment and the Raman polarizability above, with the assumption that the 3–10 helical structure adopts a delta-distribution, we can calculate the two ratios βaac/ βccc and βaca/ βccc, which are used in the orientation analysis:
| (56) |
| (57) |
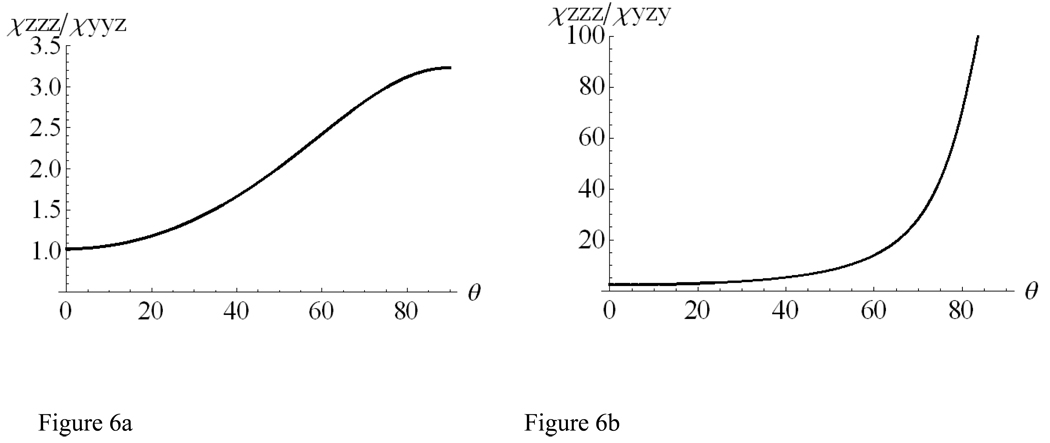
The relationship between each of the ratios, χzzz/χyyz or χzzz/χyzy, and the orientation angle θ for 3–10 helices can be deduced using the same methodology as what was adopted to treat the α-helical structures discussed in the previous sections. These relationships are plotted in Figures 6a and 6b. Figure 6b indicates that it is difficult to experimentally determine the orientation angle θ using the relationship between χzzz/χyzy and θ because very weak sps signal is expected.
Figure 6.
Relationship between the (a) χzzz/ χyyz or (b) χzzz/ χyzy ratio and θ for a 3–10 helix with a delta-distribution.
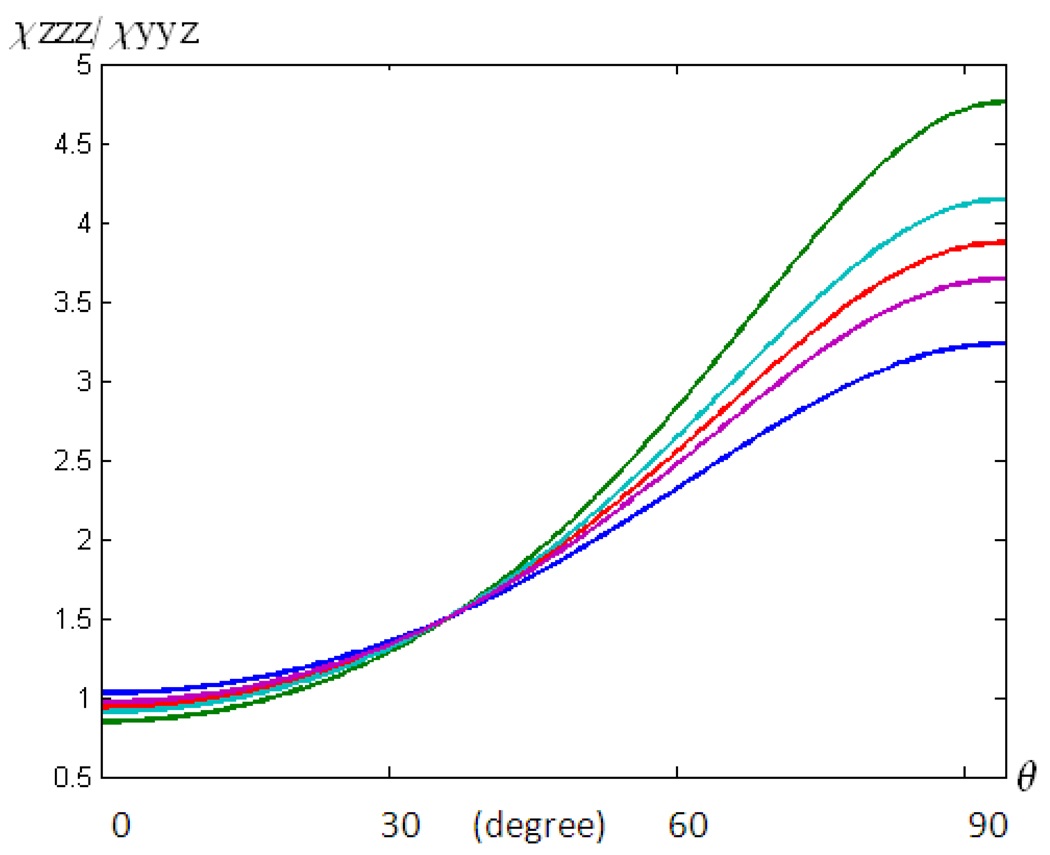
Similar to α-helices, for 3–10 helices, the above relationships between the SFG susceptibility component ratio and the helix orientation angle can also be determined when the orientation distribution is assumed to be a Gaussian distribution instead of a delta-distribution. Also, the dependency of the relationship between χzzz/χyyz and θ on the number of peptide units in the 3–10 helical structure can be investigated, as was done for α-helices above. This dependency (illustrated in Figure 7) suggests that the relationship between χzzz/χyyz and θ is varied when θ is larger than sixty degrees (meaning that the helix orients nearly parallel the surface). When θ is not large, such a variation is not substantial.
Figure 7.
Relationship between the χzzz/ χyyz ratio and θ for 3–10 helices with different chain lengths: blue: 3, green: 4, red: 5, cyan: 7, and purple: 8 residues.
4. Conclusion
In this paper, a methodology to measure the orientation of helical structures, including α-helices and 3–10 helices, using polarized SFG measurements, was systematically presented. By adopting the bond additivity model, certain SFG hyperpolarizability component ratios of a helix were computed by calculating the IR transition dipole moment and Raman polarizability tensor of the helix. The calculated values for the transition dipole moment and the Raman polarizability tensor matched experimental IR and Raman measurements reported in the literature quite well. How the number of peptide units in a helix influences the SFG orientation determination was examined, and a methodology to determine the orientation of any helix that is not ideal or perfect regarding the number of peptide units was developed. This method has been recently applied to determine the membrane orientations of a variety of α-helical peptides such as magainin 2, MSI-78 and pardaxin, the orientation of α-helical cecropin chemically immobilized on polymer surfaces, and the membrane orientation of the 3–10 helical alamethicin. These studies further validate the method presented in this article and will be reported in forthcoming articles. This method is likely general and can probably be applied to investigate all other helical structures (e.g., π-helices and DNA helical structures) in the future.
Acknowledgement
This research is supported by National Institute of Health (1R01GM081655-01A2) and Office of Naval Research (N00014-02-1-0832 and N00014-08-1-1211). SVLC acknowledges the Molecular Biophysics Training Grant from the University of Michigan and the NSF Graduate Research Fellowship.
Biography

Khoi Nguyen received his first bachelor’s degree in electrical engineering from the Institute of Telecommunications of Saigon in 2002. He then received two bachelor’s degrees in mathematics and chemistry from the New Mexico Institute of Mining and Technology in 2006. Currently he is a chemistry graduate student at the University of Michigan. His research interests include the orientation analysis of secondary structures of proteins in model cell membranes using Sum Frequency Generation and other linear vibrational spectroscopic techniques.

Stephanie Le Clair received her B.S. in Chemistry from Saint Mary’s College of California in 2006. She took a year off before graduate school and worked as an intern at Roche Palo Alto LLC and then Argonne National Laboratory. She is currently working towards obtaining her Ph.D. in Analytical Chemistry from the University of Michigan. As an NSF graduate research fellow, she is conducting research on amyloid peptides and their interactions with model cell membranes using Sum Frequency Generation and solid-state NMR.

Shuji Ye received his B.S. degree from the University of Science and Technology of China in 1997 and earned his Ph.D. degree in Chemical System Engineering from the University of Tokyo in 2004. His present research includes the study of molecular interfacial structures of anti-biofouling polymers and membrane-associated peptides/proteins using nonlinear vibrational spectroscopy. Dr. Ye is currently a Research Fellow in the group of Professor Zhan Chen at the University of Michigan. He was previously a postdoctoral associate in the group of Professor Andrea Markelz at the State University New York, Buffalo.

Zhan Chen received his B.S. degree in chemistry from Peking University in 1988. He then received his M.S. degree in physics from Institute of Physics, Chinese Academy of Sciences in 1991. After he worked in Institute of Physics for two years as a research scientist, he went to the Department of Chemistry at the University of California at Berkeley in 1993. He received his Ph.D. degree in 1998, advised by Prof. Herbert Strauss. He then worked as a postdoctoral research fellow in Prof. Gabor Somorjai’s group at Berkeley. He started his own research group in 2000 at the University of Michigan. Currently he is a professor of Chemistry there. His research is to understand molecular structures of polymers and biological molecules at interfaces.
References
- 1.Pauling L, Corey RB, Brandon HR. Proc Natl Acad Sci U S A. 1951;37:205–211. doi: 10.1073/pnas.37.4.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matsuzaki K. Biochim. Biophys. Acta. 1999;1462:1–10. doi: 10.1016/s0005-2736(99)00197-2. [DOI] [PubMed] [Google Scholar]
- 3.Sitaram N, Nagaraj R. Biochim. Biophys. Acta. 1999;1462:29–54. doi: 10.1016/s0005-2736(99)00199-6. [DOI] [PubMed] [Google Scholar]
- 4.Brogden KA. Nat. Rev. Microbiol. 2005;3:238–250. doi: 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]
- 5.Epand RM, Vogel HJ. Biochim. Biophys. Acta. 1999;1462:11–28. doi: 10.1016/s0005-2736(99)00198-4. [DOI] [PubMed] [Google Scholar]
- 6.Hancock RE, Diamond G. Trends Microbiol. 2000;8:402–410. doi: 10.1016/s0966-842x(00)01823-0. [DOI] [PubMed] [Google Scholar]
- 7.Hamasaki KM, editor. Membrane Proteins: Structure, Function, and Expression Control: International Symposium; Kyushu University Press; Japan. 1997. [Google Scholar]
- 8.Rydström J, editor. Membrane Proteins: Structure, Function, Assembly. Cambridge: Cambridge University Press; 1987. [Google Scholar]
- 9.Azzi A, Masotti L, Vecli A, editors. Membrane Proteins: Isolation and Characterization. New York: Springer-Verlag; 1986. [Google Scholar]
- 10.Capaldi RA, editor. Membrane Proteins and Their Interactions with Lipids. New York: Marcel Dekker; 1977. [Google Scholar]
- 11.Eisenberg D. Proc. Natl. Acad. Sci. 2003;100:11207–11210. doi: 10.1073/pnas.2034522100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pauling L, Corey R. Proc. Natl. Acad. Sci. 1951;37:3235–3240. doi: 10.1073/pnas.37.5.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Todd LA. The Legacy of Sir Lawrence Bragg. Northwood: Science Reviews Limited; 1990. [Google Scholar]
- 14.Axelsen PH, Kaufman BK, McElhaney RN, Lewis RN. Biophys. J. 1995;69:2770–2781. doi: 10.1016/S0006-3495(95)80150-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Axelsen PH, Citra MJ. Prog. Biophys. molec. Biol. 1996;66:227–253. doi: 10.1016/s0079-6107(97)00007-2. [DOI] [PubMed] [Google Scholar]
- 16.Bechinger B, Ruysschaert J-M, Goormaghtigh E. Biophys. J. 1999;76:552–563. doi: 10.1016/S0006-3495(99)77223-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bradbury EM, Brown L, Downie AR, Elliot A, Fraser RDB, Hanby WE. J. Mol. Biol. 1962;5:230–247. doi: 10.1016/s0022-2836(62)80086-2. [DOI] [PubMed] [Google Scholar]
- 18.Fraser RDB. J. Chem. Phys. 1953;21:1511–1515. [Google Scholar]
- 19.Higgs PW. Proc. R. Soc. Lond. A. 1953;220:472–485. [Google Scholar]
- 20.Lee S-H, Krimm S. Biopolymers. 1998;46:283–317. [Google Scholar]
- 21.Marsh D. Methods Enzymol. 1999;294:59–92. doi: 10.1016/s0076-6879(99)94007-7. [DOI] [PubMed] [Google Scholar]
- 22.Marsh D. Biophys. J. 1998;75:354–358. doi: 10.1016/S0006-3495(98)77519-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marsh D, Müller M, Schmitt F-J. Biophys J. 2000;78:2499–2510. doi: 10.1016/S0006-3495(00)76795-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miyazawa T, Blout ER. J. Am. Chem. Soc. 1961;83:712–719. [Google Scholar]
- 25.Reisdorf W, Jr, Krimm S. Biophys. J. 1995;69:271–273. doi: 10.1016/S0006-3495(95)79898-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suzuki E. Spectrochim. Acta. A. 1967;23:2303–2308. [Google Scholar]
- 27.Tsuboi M. J. Polymer Sci. 1962;59:139–153. [Google Scholar]
- 28.Tamm LK, Tatulian SA. Q. ReV. Biophys. 1997;30:365–429. doi: 10.1017/s0033583597003375. [DOI] [PubMed] [Google Scholar]
- 29.Wilser WT, Fitchen DB. J. Chem. Phys. 1975;62:720–724. [Google Scholar]
- 30.Tsuboi M, Ikeda T, Ueda T. J. Raman Spectrosc. 1991;22:619–626. [Google Scholar]
- 31.Overman AS, Tsuboi M, Thomas GJ., Jr J. Mol. Biol. 1996;259:331–336. doi: 10.1006/jmbi.1996.0323. [DOI] [PubMed] [Google Scholar]
- 32.Chen LX, Strauss HL, Snyder RG. Biophys. J. 1993;64:1533–1541. doi: 10.1016/S0006-3495(93)81522-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dwivedi AM, Krimm S, Malcolm BR. Biopolymers. 1984;23:2025–2065. doi: 10.1002/bip.360230509. [DOI] [PubMed] [Google Scholar]
- 34.Fanconi B, Tomlinson B, Nafie LA, Small W, Peticolas WL. J. Chem. Phys. 1969;51:3993–4005. doi: 10.1063/1.1672620. [DOI] [PubMed] [Google Scholar]
- 35.Fanconi B. Biopolymers. 1973;12:2759–2776. doi: 10.1002/bip.1973.360121210. [DOI] [PubMed] [Google Scholar]
- 36.Koenig JL, Sutton PL. Biopolymers. 1970;9:1229–1237. doi: 10.1002/bip.1970.360091003. [DOI] [PubMed] [Google Scholar]
- 37.Lee S-H, Krimm S. J. Raman Spectrosc. 1998;29:73–80. [Google Scholar]
- 38.Tsuboi M, Benevides JM, Bondre P, Thomas GJ., Jr Biochemistry. 2005;44:3091–3100. [Google Scholar]
- 39.Ma G, Allen HC. Langmuir. 2006;22:5341–5349. doi: 10.1021/la0535227. [DOI] [PubMed] [Google Scholar]
- 40.Moore FG, Richmond GL. Acc. Chem. Res. 2008;41:739–748. doi: 10.1021/ar7002732. [DOI] [PubMed] [Google Scholar]
- 41.Opdahl A, Koffas TS, Amitay-Sadovsky E, Kim J, Somorjai GA. J. Phys.: Condens. Matter. 2004;16:659–677. [Google Scholar]
- 42.Richmond GL. Chem. Rev. 2002;102:2693–2724. doi: 10.1021/cr0006876. [DOI] [PubMed] [Google Scholar]
- 43.Rupprechter G, Weilach C. J. Phys.: Condens. Matter. 2008;20:184019–184036. [Google Scholar]
- 44.Shen YR, Ostroverkhov V. Chem. Rev. 2006;106:1140–1154. doi: 10.1021/cr040377d. [DOI] [PubMed] [Google Scholar]
- 45.Shultz MJ, Baldelli S, Schnitzer C, Simonelli D. J. Phys. Chem. B. 2002;106:5313–5324. doi: 10.1021/jp014466v. [DOI] [PubMed] [Google Scholar]
- 46.Stiopkin IV, Jayathilake HD, Bordenyuk AN, Benderskii AV. J. Am. Chem. Soc. 2008;130:2271–2275. doi: 10.1021/ja076708w. [DOI] [PubMed] [Google Scholar]
- 47.Voges AB, Stokes GY, Gibbs-Davis JM, Lettan RB, Bertin PA, Pike RC, Nguyen ST, Scheidt KA, Geiger FM. J. Phys. Chem. C. 2007;11:1567–1578. [Google Scholar]
- 48.Baldelli S. Acc. Chem. Res. 2008;41:421–431. doi: 10.1021/ar700185h. [DOI] [PubMed] [Google Scholar]
- 49.Santos C, Baldelli S. J. Phys. Chem. C. 2008;112:11459–11467. [Google Scholar]
- 50.Anglin TC, Conboy JC. Biophys. J. 2008;95:186–193. doi: 10.1529/biophysj.107.118976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dreesen L, Sartenaer Y, Humbert C, Mani AA, Methivier C, Pradier CM, Thiry PA, Peremans A. Chem. Phys. Chem. 2004;5:1719–1725. doi: 10.1002/cphc.200400213. [DOI] [PubMed] [Google Scholar]
- 52.Gautam KS, Dhinojwala A. Phys. Rev. Lett. 2002;88:145501.1–145501.4. doi: 10.1103/PhysRevLett.88.145501. [DOI] [PubMed] [Google Scholar]
- 53.Gracias DH, Chen Z, Shen YR, Somorjai GA. Acc. Chem. Res. 1999;32:930–940. [Google Scholar]
- 54.Holman J, Ye S, Neivandt DJ, Davies PB. J. Am. Chem. Soc. 2004;126:14322–14323. doi: 10.1021/ja046954x. [DOI] [PubMed] [Google Scholar]
- 55.Iwahashi T, Miyamae T, Kanai K, Seki K, Kim D, Ouchi Y. J. Phys. Chem. B. 2008;112:11936–11941. doi: 10.1021/jp8021908. [DOI] [PubMed] [Google Scholar]
- 56.Ye S, Nguyen KT, Le Clair SV, Chen Z. J. Struct. Biol. 2009 doi: 10.1016/j.jsb.2009.03.006. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen X, Wang J, Sniadecki JJ, Even MA, Chen Z. Langmuir. 2005;21:2262–2264. doi: 10.1021/la050048w. [DOI] [PubMed] [Google Scholar]
- 58.Lee S-H, Wang J, Krimm S, Chen Z. J. Phys. Chem. A. 2006;110:7035–7044. doi: 10.1021/jp057200n. [DOI] [PubMed] [Google Scholar]
- 59.Wang J, Lee S-H, Chen Z. J. Phys. Chem. B. 2008;112:2281–2290. doi: 10.1021/jp077556u. [DOI] [PubMed] [Google Scholar]
- 60.Chen X, Wang J, Boughton AP, Kristalyn CB, Chen Z. J. Am. Chem. Soc. 2007;129:1420. doi: 10.1021/ja067446l. [DOI] [PubMed] [Google Scholar]
- 61.Chen X, Boughton AP, Tesmer JJG, Chen Z. J. Am. Chem. Soc. 2007;129:12658–12659. doi: 10.1021/ja075542w. [DOI] [PubMed] [Google Scholar]
- 62.Elliott A. Proc. R. Soc. 1954;226:408–409. [Google Scholar]
- 63.Rabolt JF, Mo WH, Krimm S. Macromolecules. 1977;10:1065–1074. doi: 10.1021/ma60059a034. [DOI] [PubMed] [Google Scholar]
- 64.Itoh K, Nakahara T, Shimanouchi T, Oya M, Uno K, Iwakura Y. Biopolymers. 1968;6:1759. doi: 10.1002/bip.1968.360061211. [DOI] [PubMed] [Google Scholar]
- 65.Itoh K, Shimanouchi T, Oya M. Biopolymers. 1969;7:649–658. [Google Scholar]
- 66.Masuda Y, Fukushima K, Fujii T, Miyazawa T. Biopolymers. 1969;8:91–99. [Google Scholar]
- 67.Itoh K, Shimanouchi T. Biopolymers. 1970;9:383–399. doi: 10.1002/bip.1970.360090402. [DOI] [PubMed] [Google Scholar]
- 68.Koenig JL, Sutton PL. Biopolymers. 1969;8:167–171. [Google Scholar]
- 69.Choi J-H, Hahn S, Cho M. Biopolymers. 2006;83:519–536. doi: 10.1002/bip.20583. [DOI] [PubMed] [Google Scholar]
- 70.Yokote Y, Kubo Y, Takahashi R, Ikeda T, Akahane K, Tsuboi M. Bull. Chem. Soc. Jpn. 2007;80:1148–1156. [Google Scholar]
- 71.Rintoul L, Carter EA, Stewart SD, Fredericks PM. Biopolymers. 2000;57:19–28. doi: 10.1002/(SICI)1097-0282(2000)57:1<19::AID-BIP4>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 72.Ackermann KR, Koster J, Schlucker S. J. Biophotonics. 2008;1:419–424. doi: 10.1002/jbio.200810015. [DOI] [PubMed] [Google Scholar]
- 73.Lambert AG, Davies PB, Neivandt DJ. App. Spec. Rev. 2005;40 [Google Scholar]
- 74.Chen X, Clarke ML, Wang J, Chen Z. Intern. J. Mod. Phys. B. 2005;19:691–713. [Google Scholar]
- 75.Moad JA, Simpson G. J. Phys. Chem. B. 2004;108:3548–3562. [Google Scholar]
- 76.Li Q, Hua R, Cheah IJ, Chou KC. J. Phys. Chem. B. 2008;112:694–697. doi: 10.1021/jp072147j. [DOI] [PubMed] [Google Scholar]
- 77.Simpson G, Perry JM, Ashmore-Good CL. Phys. Rev. B. 2002;66:165437. [Google Scholar]
- 78.Hirose C, Akamatsu N, Domen K. J. Chem. Phys. 1992;96:997–1004. [Google Scholar]
- 79.Wang J, Paszti Z, Even MA, Chen Z. J. Am. Chem. Soc. 2002;124:13302–13305. doi: 10.1021/ja026881m. [DOI] [PubMed] [Google Scholar]
- 80.Wang J, Paszti Z, Clarke ML, Chen X, Chen Z. J. Phys. Chem. B. 2007;111:6088–6095. doi: 10.1021/jp070383o. [DOI] [PubMed] [Google Scholar]
- 81.Wang J, Even MA, Chen X, Schmaier AH, Waite JH, Chen Z. J. Am. Chem. Soc. 2003;125:9914–9915. doi: 10.1021/ja036373s. [DOI] [PubMed] [Google Scholar]
- 82.Huggins M. Chem. Rev. 1943;32:195–218. [Google Scholar]
- 83.Millhauser GL. Acc. Chem. Res. 1999;32:1027–1033. [Google Scholar]
- 84.Tirado-Rives J, Jorgensen WL. Biochemistry. 1991;30:3864–3871. doi: 10.1021/bi00230a009. [DOI] [PubMed] [Google Scholar]
- 85.Tobias DJ, Brooks CL. Biochemistry. 1991;30:6059–6070. doi: 10.1021/bi00238a033. [DOI] [PubMed] [Google Scholar]
- 86.Tobias DJ, Mertz JE, Brooks CL. Biochemistry. 1991;30:6054–6058. doi: 10.1021/bi00238a032. [DOI] [PubMed] [Google Scholar]
- 87.Brooks CL. J. Phys. Chem. 1996;100:2546–2549. [Google Scholar]
- 88.Sheinerman FB, Brooks CL. J. Am. Chem. Soc. 1995;117:10098–10103. [Google Scholar]
- 89.Brooks CLI, Case DA. Chem. Rev. 1993;93:2487–2502. [Google Scholar]
- 90.Malcolm BR, Walkinshaw MD. Biopolymers. 1986;25:607–625. [Google Scholar]
- 91.Paterson Y, Rumsey SM, Benedetti E, Nemethy G, Sheraga HA. J. Am. Chem. Soc. 1981:2947–2955. [Google Scholar]