Abstract
Large-scale changes (gross chromosomal rearrangements [GCRs]) are common in genomes, and are often associated with pathological disorders. We report here that a specific pair of nearby inverted repeats in budding yeast fuse to form a dicentric chromosome intermediate, which then rearranges to form a translocation and other GCRs. We next show that fusion of nearby inverted repeats is general; we found that many nearby inverted repeats that are present in the yeast genome also fuse, as does a pair of synthetically constructed inverted repeats. Fusion occurs between inverted repeats that are separated by several kilobases of DNA and share >20 base pairs of homology. Finally, we show that fusion of inverted repeats, surprisingly, does not require genes involved in double-strand break (DSB) repair or genes involved in other repeat recombination events. We therefore propose that fusion may occur by a DSB-independent, DNA replication-based mechanism (which we term “faulty template switching”). Fusion of nearby inverted repeats to form dicentrics may be a major cause of instability in yeast and in other organisms.
Keywords: Inverted repeats, acentric and dicentric chromosomes, breakage–fusion–bridge cycle, genome instability, large palindromes, template switch
Large-scale chromosome changes (gross chromosomal rearrangements [GCRs]) occur in all genomes and can occur at a high frequency. Twin studies show frequent recombination between nearby (<1-Mb) large (>1-kb) direct repeat sequences (Bruder et al. 2008). Complex genome rearrangements cause loss or duplication of genes that affect human pathology (such as loss or gain of the PLP1 gene that affects myelination in humans) (Lee et al. 2007). Numerous large-scale changes are present in cancer cells and are thought to contribute to the cancer phenotype (Bignell et al. 2007). Bacterial genomes also suffer large-scale rearrangements (Tillier and Collins 2000). How these changes occur is largely unknown, and is of interest in understanding both pathology and genetic diversity.
The mechanisms underlying GCR formation appear complex, and may involve some combination of DNA replication, double-strand breaks (DSBs), repeat sequences, and unstable chromosome intermediates. One common view is that GCR formation is initiated by telomere or DNA replication dysfunction, leading to DSBs and subsequent fusion of two DSBs. DSB fusions may occur by any of several repair mechanisms (Wong et al. 2000; Kolodner et al. 2002; Murnane and Sabatier 2004; Weinstock et al. 2006), or the breaks may be capped by telomere addition (Chen and Kolodner 1999; Putnam et al. 2004; Sabatier et al. 2005; see Mizuno et al. 2009 for discussion). Recent studies of instability in yeast are in general consistent with the conclusion that DSBs can be intermediates in instability (Myung et al. 2001; Hackett and Greider 2003; Lemoine et al. 2005; Rattray et al. 2005; Narayanan et al. 2006). The question of whether GCRs also arise from an alternative replication-based mechanism, without a DSB intermediate, remains speculative. However, replication-based mechanisms have been inferred from complex rearrangements in human disorders (Lee et al. 2007) and rearrangements in bacteria (Bi and Liu 1996).
Repeat sequences present in genomes contribute to the formation of GCRs (whether a DSB is involved or not). Immediately adjacent repeat sequences, called palindromes, may form hairpin structures that are easily broken and precipitate further genome rearrangements (Lobachev et al. 2002). Once a damaged structure (e.g., DSB or stalled fork) is formed, repeats may provide sequence homology that directs DNA repair (Zhou et al. 2001; Dujon 2006; Schmidt et al. 2006; Voineagu et al. 2008; Pennaneach and Kolodner 2009; Putnam et al. 2009). Repeats in eukaryotes are plentiful (Richard et al. 2008), and their recombination in mammalian cells is common (Zhou et al. 2001; Lobachev et al. 2007). Pairs of nearby repeats studied in this report are of special interest, since their close proximity and extensive homology may render them highly prone to rearrangements.
A third issue concerning GCR formation is whether they involve unstable chromosome intermediates. The presence of an unstable chromosome would readily explain how one cell generates progeny with many different chromosome structures, as commonly observed in cancer cells (Weinstock et al. 2006). An apparently common unstable chromosome is a dicentric, first described by McClintock (1941); once formed, the attempted segregation of the two centromeres of the dicentric to opposite poles during mitosis leads to chromosome breakage and further rearrangements (called the breakage–fusion–bridge [BFB] cycle). Dicentrics and the BFB cycle have been invoked to explain the behavior of complex chromosome changes (Lo et al. 2002; Maser and DePinho 2002; Hackett and Greider 2003; Narayanan et al. 2006;).
In this study, we provide evidence that GCRs form when nearby inverted repeats fuse to form unstable dicentric intermediates. This study is an extension of a previous study describing a highly unstable region of a yeast chromosome (Chromosome VII [ChrVII]) that underwent multiple recombination events and formed unstable chromosomes, eventually forming a specific GCR (Fig. 1A,B; Admire et al. 2006). We reported that this highly unstable region contains inverted repeats and tRNA genes that appear to contribute to instability—instability that was further increased when DNA replication was disrupted. How the inverted repeats underwent recombination to form unstable chromosomes was unclear, and is clarified in this study. We now demonstrate that those inverted repeats first fuse to form a dicentric chromosome, which then rearranges further to form GCRs. We show that in budding yeast, fusion of nearby inverted repeats is a general phenomenon; we report that fusion occurs between naturally occurring and synthetically constructed inverted repeats present at different sites in the yeast genome. We then investigate the mechanism of fusion and find, surprisingly, that fusion does not require known DSB repair and replication fork rescue pathways, nor does it proceed by mechanisms known to mediate direct or inverted repeat recombination. We therefore suggest a replication-based mechanism of fusion of nearby inverted repeats. In an accompanying study, Mizuno et al. (2009) also present evidence for fusion of nearby inverted repeats in fission yeast, and suggest that fusion events also occur by a DSB-independent, replication-based mechanism.
Figure 1.
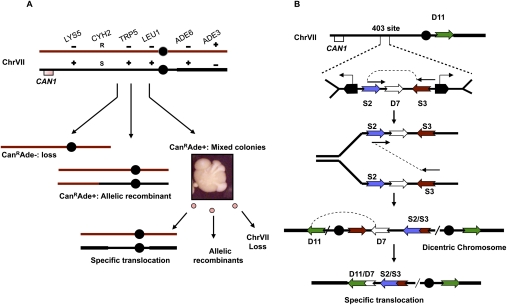
Chromosome system to detect instability. (A) Two homologs of ChrVII and mutant alleles on each allow for genetic detection of chromosome changes. The CAN1 gene has been removed from ChrV and inserted into one copy of ChrVII. Selection for the loss of the CAN1 gene allows growth of cells with any of three types of chromosome changes, including simple loss, allelic recombinants, and mixed colonies. Mixed colonies contain cells of multiple genotypes, including a specific translocation. See the Materials and Methods for details. (B) Configuration of elements in the ChrVII403 site and the geometry and order of how fusion may occur. Two tRNA genes (pentagons) transcribe toward the oncoming fork and slow replication. Fusion between the two LTR σ repeats (S2 and S3), shown diagrammatically, forms a dicentric, followed by recombination between the two LTR δ sequences (D7 and D11) to form the specific translocation.
Results
The genetic system that led us to the discovery of inverted repeat fusion is shown in Figure 1A (Carson and Hartwell 1985; Admire et al. 2006). Briefly, we constructed a haploid yeast strain that contains an extra copy of ChrVII (a disomic strain). Any changes to this extra chromosome should not compromise cell viability per se. The CAN1 gene, inserted near the left telomere of the extra chromosome, provides the means of identifying cells with chromosome changes. CAN1 encodes arginine permease that imports arginine as well as the toxic analog canavanine; only cells that lose the CAN1 gene can grow in media containing canavanine (forming CanR colonies). Using multiple genetic markers, we detected three types of chromosome changes in CanR colonies: whole-chromosome loss, normal allelic recombination, and other complex rearrangements that give rise to colonies with a sectored appearance (which we refer to as “mixed colonies”) (Fig. 1A). (Point mutations in CAN1 occur more rarely [Weinert and Hartwell 1990.]) Mixed CanR colonies contain cells of multiple genotypes, whereas normal CanR colonies contain cells of a single genotype (see the Materials and Methods). Previously, we were able to infer that a mixed colony arises from a cell with an unstable chromosome, whereas a normal colony arises from a cell with a stable chromosome (Admire et al. 2006).
The initial focus of this study is to determine the nature of the unstable chromosome that forms mixed colonies. We used rad9 mutants here merely as a tool to study unstable chromosomes, because unstable chromosomes are formed more frequently in rad9 mutants than in wild-type (Rad+) cells (Admire et al. 2006; this study). We emphasize that unstable chromosomes are formed in other mutants as well (some at an even higher frequency than in rad9 mutants) (see below).
A major clue to solving the structure of the unstable chromosome came from the structure of a specific translocation (the “Delta 7/Delta 11” [D7/D11] translocation) present only in mixed colonies (Fig. 1B). Our previous analyses suggested that this translocation probably arises by multiple deletion and fusion events centered at a site 403 kb from the left end (Fig. 1B; Admire et al. 2006). This ChrVII403 site contains long terminal repeat (LTR) sequences as well as tRNA genes that appear to contribute to instability (Admire et al. 2006; H Jones and T Weiner, unpubl.). LTRs are derived from retrotransposons (Kim et al. 1998) and are present in hundreds of copies in the yeast genome. (The Sigma 2 [S2] and Sigma 3 [S3] repeats are derived from Ty3 elements, are 278 and 245 bases long, share 82% identity, and are 3.3 kb apart, while D7 and D11 are from Ty1, are both 331 bases, share 97% identity, and are 133 kb apart.) Given the structure of the D7/D11 translocation, we suggest a relatively simple model in which one event fuses S2 and S3 inverted repeats to generate a dicentric chromosome, depicted in Figure 1B, and a second event fuses D7 and D11 direct repeats to resolve the dicentric and form the translocation. We first test the hypothesis that a dicentric intermediate is involved in ChrVII's instability.
A specific dicentric is present in cells that form mixed colonies
We employed a PCR assay to test for the presence of the specific dicentric (Fig. 2A). We reasoned that the dicentric, if formed, might be present at a low but detectable amount in a culture of cells, and the amount of dicentric might predict the frequency of CanR mixed colonies arising from that culture. We therefore grew (in rich media) each of 10 single rad9 cells (initially containing an unrearranged ChrVII) into 10 independent cultures (first to colonies on plates, then each into liquid culture). We then performed two tests on each of these 10 cultures. We isolated genomic DNA from each culture and subjected it to amplification using primers that should detect the specific dicentric chromosome (Fig. 2A). We did, in fact, detect a specific PCR fragment of the correct size and sequence if S2 and S3 inverted repeats fused to form the dicentric (see Supplemental Fig. S1 for sequence). Control PCR reactions failed to amplify the same DNA fragment from native ChrVII403 site sequences (native sequences were made by PCR-amplifying the ChrVII403 site) (data not shown). (We note that these PCR primers would amplify the same DNA fragment if it arose by an inversion event; a formal possibility that we eliminate by genetic and molecular tests [Supplemental Fig. S2; see below]).
Figure 2.
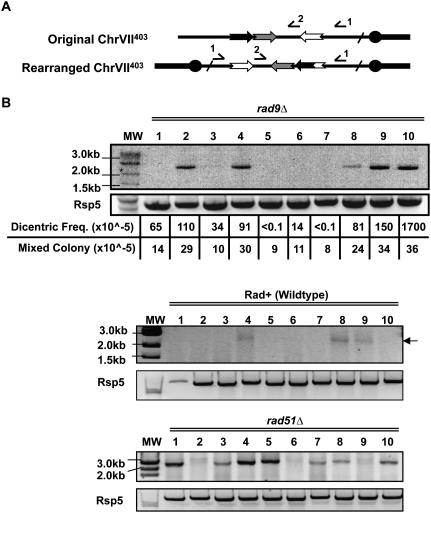
Inverted repeat fusion generates dicentrics that cause further chromosome instability. (A) Shown are the structures of normal ChrVII and the putative dicentric with the positions of PCR primers 1 and 2 used to detect the presence of the dicentric. (B) Genomic DNA from unselected cultures of Rad+, rad9, and rad51 were subjected to PCR amplification using primers 1 and 2. Gels show qualitative PCR results. The rad9 cultures were also analyzed for their frequencies of mixed colonies and quantitative PCR to determine the amount of dicentric. Spearman correlation test was performed to correlate instability and dicentrics. Spearman rank correlation coefficient (ρ) is 0.973; P-value, <0.0001. (Lanes 4,8,9) Three of the Rad+ cultures are faintly positive for the dicentric fragment. Primers to RSP5 were used as PCR controls.
We next tested whether the amount of the PCR product correlates with the frequency of mixed colonies in the 10 cultures. If so, cultures with quantitatively more PCR product should give rise to more mixed colonies. We determined the amount of dicentrics by quantitative PCR, and the frequency of mixed colonies by a quantitative genetic assay (see the Materials and Methods). We found that, in fact, cultures that had more dicentrics gave rise to a generally higher frequency of mixed colonies (correlation coefficient 0.97; P < 0.001) (Fig. 2B). The concentration of dicentrics varied between cultures, with an average of about one in 2000 rad9 cells (and one in 100,000 cells in Rad+ cells) (Supplemental Fig. S3).
We further tested the correlation between the amount of PCR product and instability by studying strains with low instability (Rad+, wild-type cells) and high instability (rad51 mutants with a defect in homologous recombination [HR]) (Fig. 2B; discussed further below). Again, we found more dicentric PCR product in rad51 mutants with high instability than in Rad+ strains with low instability (Fig. 2B). We observe this correlation in other mutants as well (see below). We conclude that the specific dicentric depicted in Figures 1B and 2A is most likely a key intermediate to instability and to formation of the specific D7/D11 translocation shown in Figure 1B.
Genetic test confirms that dicentrics cause the instability in mixed colonies
Although results from the PCR assay described above argue strongly that a specific dicentric causes instability, we cannot directly detect intact dicentric chromosomes (due to their fragility and low abundance). We therefore turned to a genetic approach to test if dicentrics are, in fact, intermediates in instability.
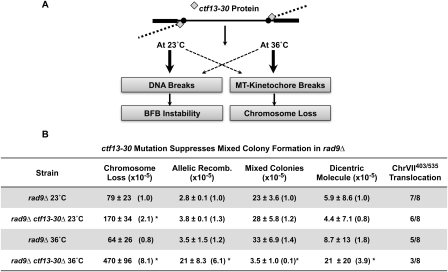
Our genetic test uses a temperature-sensitive mutant of a kinetochore protein, ctf13-30, that functions properly at 23°C but not at 36°C (Doheny et al. 1993). Ctf13 connects microtubules to the kinetochore. The idea of using this mutant is as follows: When a dicentric undergoes chromosome segregation during mitosis and Ctf13 is active, the DNA will break, but if Ctf13 is inactive, the microtubule–kinetochore connection will break instead of the DNA (Fig. 3A). (DNA breakage is required to form mixed colonies, since mixed colonies contain cells of multiple genotypes.) We therefore predicted that Ctf13 status will affect the frequency of mixed colonies if dicentrics are intermediates, whereas Ctf13 status will not affect the frequency of mixed colonies if dicentrics are not intermediates. To test this prediction, we grew rad9 and rad9 ctf13-30 cells at the permissive (23°C) or the restrictive (36°C) temperature, and determined the level of dicentrics by quantitative PCR and the level of instability from the frequency of mixed colonies (Fig. 3B). (We found that the status of Ctf13 did not drastically affect the amount of dicentrics formed, as measured by quantitative PCR; there is a fourfold increase in dicentrics in Ctf13-defective cells.) We found that Ctf13 status did, in fact, affect instability; rad9 ctf13-30 mutants had an eightfold lower frequency of mixed colonies when grown at a restrictive temperature compared with the permissive temperature, and compared with rad9 CTF13+ strains grown at either temperature. Thus, the amount of dicentric formed was largely independent of Ctf13 activity (there is actually more dicentric in Ctf13-defective cells), but the fate of the dicentrics was dependent on Ctf13 status (fewer mixed colonies in Ctf13-defective cells). This Ctf13 result is consistent with the hypothesis that the specific dicentric detected by PCR is an intermediate to instability.
Figure 3.
Instability is affected by the Ctf13 protein, suggesting that dicentrics are intermediates in instability. (A) Model of how Ctf13 function may affect the fate of a dicentric chromosome. See the text for discussion. (B) rad9 and rad9 ctf13-30 strains were grown at permissive (23°C) or restrictive (36°C) temperatures, then analyzed for instability events, translocations, and frequency of dicentrics by quantitative PCR as described in the Materials and Methods.
We note that rad9 ctf13-30 cells have a higher frequency of chromosome loss and of allelic recombination when grown at the restrictive temperature compared with the permissive temperature, as reported previously (Fig. 3B; Doheny et al. 1993). An increase in chromosome loss was expected from the model; dicentrics missegregate and are lost when the Ctf13 kinetochore connection breaks. The higher frequency of allelic recombination in ctf13-defective cells has also been reported for monocentric chromosomes in both ctf13-defective cells (Doheny et al. 1993) and for cells treated with the microtubule inhibitor benomyl (Wood and Hartwell 1982). The cause of increased allelic recombination in any of these studies is unknown, but it is unlikely that it is due to dicentric behavior (see the Discussion; Supplemental Fig. S4).
In sum, we conclude that inverted repeats in the ChrVII403 site fuse to form a dicentric that causes the instability evident in mixed colonies. We next show that fusion of nearby inverted repeats is a general phenomenon in the yeast genome.
The pairwise fusion of many nearby inverted repeats in the yeast genome
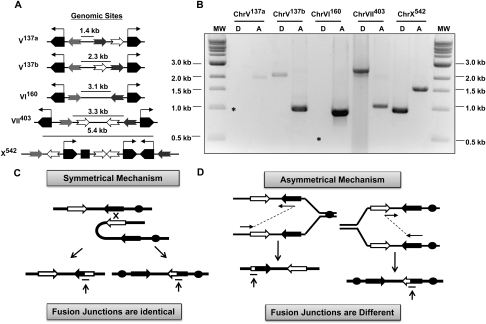
To test if inverted repeat fusion is general, we again used a PCR-based assay. By examining the yeast genome, we found that there are ∼15 sites in budding yeast that contain LTR inverted repeats flanked by tRNA genes, a geometry similar to that of the unstable ChrVII403 site (Fig. 4A). We chose five of these sites to test for their fusion of inverted repeats. We isolated genomic DNA from cells and subjected it to PCR amplification using appropriate primers (as in Fig. 2A). We developed separate sets of primers that could detect either acentrics or dicentrics, since theoretically either is equally possible (see Fig. 4D). We detected DNA fragments diagnostic of acentric and/or dicentric chromosomes at all five genomic sites tested (Fig. 4B). To get an idea of how frequently these other nearby inverted repeats fuse, we performed quantitative PCR on two additional sites (ChrVI160 and ChrX542, in addition to the ChrVII403), and found that the frequency of fusion events in Rad+ strains at all three sites was comparable (1.5 × 10−5, 4.7 × 10−5, and 8.1 × 10−5 for the three sites, respectively) (see Supplemental Fig. S5I).
Figure 4.
Inverted repeats at five sites in the yeast genome fuse to form dicentrics and/or acentrics. (A) The sites screened for fusion contain tRNA genes (pentagons), repeat sequences (gray and brown arrows are involved in the fusion reaction, whereas clear ones are not), and, in one case, an origin (square). Inverted repeat fusion forms either dicentric or acentric fragments using primers in schemes shown in C. (B) DNA fragments are detected at all five sites tested, using appropriate primers specific for each site. Primer sequences are in Supplemental Figure S9. Analysis of three sites yielded both acentric and dicentric fragments. Asterisks indicate where DNA fragments should have appeared, if they formed. The DNA sequence of all of the DNA fragments has been confirmed (Supplemental Fig. S5). (C) If the fusion junctions for acentric and dicentric chromosomes are identical, then they could have arisen by a symmetrical mechanism. This mechanism can conceptually be viewed as a crossover depicted here; one event generates both acentrics and dicentrics. (Not shown here are stalled forks as intermediates; this diagram is meant to convey the idea of a symmetrical event only.) (D) On the other hand, if the fusion junctions for acentric and dicentric chromosomes are not identical, then we infer that acentrics and dicentrics must be formed by separate events. In this case, the DNA polymerase is “jumping” between different sequences to form acentrics than it does to form dicentrics. Note that symmetric joints can also be formed by an asymmetrical mechanism, if the DNA polymerases for the acentric and dicentric “jump” between identical sequences.
By examining the DNA sequence of the fused pairs of inverted repeats, we can deduce some features of the mechanism(s) of fusion. First, we found that fusion always involves some sequence homology; fusion can occur between two sequences that share as little as 20 base pairs (bp) of homology with a 1 bp mismatch (ChrV137a site) (Supplemental Fig. S5D), or as much as 148 bases of exact homology (ChrVII403 dicentric) (summarized in Supplemental Fig. S5H). Genetic tests described below are consistent with the involvement of only limited homology in the fusion reactions.
Second, we found that fusion of inverted repeats can happen between repeats that are as far as 5.4 kb away from each other (ChrX542 site) (Supplemental Fig. S5H). Since this site had the largest distance between the two repeats, and still readily formed both dicentric and acentric chromosomes, it is likely that the maximum distance for fusion is larger (Supplemental Fig. S5I). Distance requirements for fusion will be the focus of a future study.
Third, we found evidence that bears on the issue of whether one event generates both an acentric and dicentric chromosome (Fig. 4C), or whether one event generates either an acentric or a dicentric chromosome but not both (Fig. 4D). We term these two options “symmetric” and “asymmetric” events, respectively. The evidence that bears on this question comes from the sequence of the fusion junctions for sites where both acentric and dicentric chromosomes are formed. Surprisingly, we found that, for two of them (ChrVII403 and ChrV137b), the fusion junction in the acentric product was different from the dicentric product (using different bases within the repeats as depicted in Fig. 4D; actual sequences in Supplemental Figs. S1, S5). For the ChrVII403 site, we found that 16 of 16 acentrics used the same sequence (within 8 bases of homology between S2 and S3), whereas 13 of 13 dicentrics analyzed used a different sequence (within 150 bases of homology between S2 and S3). For the third site (ChrX542), the fusion to form acentrics and dicentrics does occur in the same sequence (as in Fig. 4C), so there is a formal possibility that, for this site, both acentrics and dicentrics are generated by a single event. Formation of acentric and dicentric chromosomes by an asymmetric event has mechanistic implications that are addressed in the Discussion.
Synthetic inverted repeats also fuse to form dicentrics associated with instability
To further test the generality of the fusion reaction, we generated a synthetic inverted repeat consisting of non-LTR sequences (all of the events analyzed thus far contain LTR sequences). This synthetic inverted repeat also does not contain any tRNA genes that likely contribute to instability. We divided the URA3 gene into three fragments, with each pair of consecutive fragments sharing ∼200 bp of sequence homology (designated as “UR,” “RA,” and “A3”) (Fig. 5A; Supplemental Fig. S6). We first inserted the URA3 modules into the ChrVII sites that form the dicentric and D7/D11 translocation (the 403 and 535 sites, respectively) (see Figs. 1B, 5A). We allowed instability of these genetically modified cells to occur, identified mixed colonies by selection for CanR, and analyzed cells in the mixed colonies for URA3-mediated events. Overall, we found that the URA3-mediated events were virtually identical to the LTR-mediated events. First, we found that the URA3 sequence-containing inverted repeats formed dicentric chromosomes, just as the LTR sequences S2 and S3 had (Fig. 5B). Second, the URA3 dicentric intermediate was frequently resolved to form a translocation (Supplemental Fig. S7), just as with the LTR-containing dicentric. As discussed further below, we also found that the genetic requirements for fusion of the URA3-based inverted repeats is the same as for fusion of the S2 and S3 repeats (neither reaction requires the RAD52 or RAD59 genes) (Fig. 5B; see below).
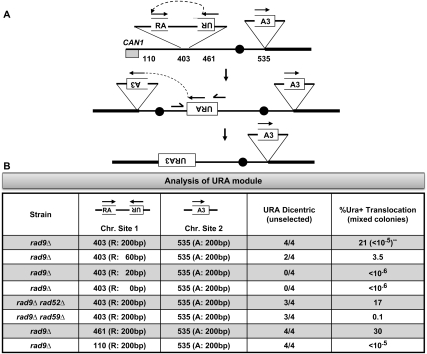
Figure 5.
Synthetic inverted repeats fuse to form dicentrics, and then recombine to form translocations. (A) Three segments of the URA3 gene were cloned into two modules (see Supplemental Fig. S6 for details of module construction and chromosome insertion). The two modules were then inserted into the specific sites in ChrVII as indicated. Inverted repeat recombination joins UR to RA modules via recombination of shared “R” sequences to form a dicentric. A second recombination event joins “A” to “A3” to form an intact URA3 gene, rendering the cells Ura+. (B) Each rad9 strain contains a UR or RA module in one of three sites on the left arm of ChrVII, and all strains contain the A3 module at one site on the right arm. The UR modules differ in the amount of sequence homology shared with the RA module (size of “R” is 200, 60, 20, or 0 bp of homology, as indicated). Each modified rad9 strain was analyzed for the presence of the specific PCR fragment diagnostic of a dicentric chromosome, in four independent cultures, using dicentric URA primers (Supplemental Fig. S9). CanR mixed colonies were generated from each rad9 strain, and the frequency of Ura+ cells in cells from mixed colonies was determined (see the Materials and Methods). (**) The frequency of Ura+ colonies in Cans cells is shown in Supplemental Figure S8.
We also used the synthetic inverted repeat module to test whether fusion is for some reason constrained to occur in the ChrVII403 site. We inserted the synthetic inverted repeat module into two additional ChrVII sites (the ChrVII110 and ChrVII461 sites) that do not contain LTR fragments, do not contain tRNA genes, and are in completely different regions of the chromosome (one 110 kb from the telomere, and the other 37 kb from the centromere). We were able to identify URA3-containing dicentrics formed at both sites (Fig. 5B), confirming that nearby inverted repeat fusion is a general phenomenon. These studies show that fusion of nearby inverted repeats is not restricted to types of DNA sequences, to sequences near tRNA genes, or to specific regions of a chromosome. (We also identified Ura+ translocations, formed by resolution of the dicentric, from Ura modules inserted at the ChrVII461 site but not the ChrVII110 site, a result for which we have no explanation).
Finally, we used the synthetic inverted repeat module to re-examine how much sequence homology is required to form dicentrics. We found that 60 bases of homology, but not 20 or 0, allowed dicentric formation (Fig. 5B). In sum, the lengths of homology involved in LTR and URA3 inverted repeat fusion are consistent with homologies of 60 bases and greater and, in some cases, as few as 20 bp undergoing fusion.
Major DNA repair and replication fork pathways and the mechanism of inverted repeat fusion
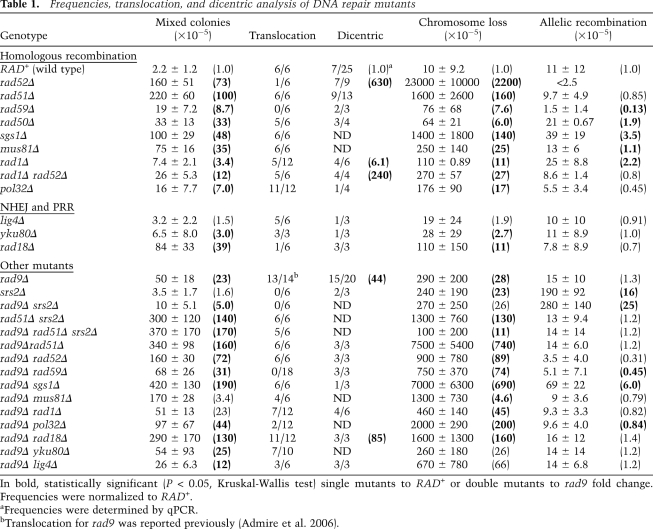
We next used genetic analyses to address questions of mechanism: How does the cell normally prevent such a deleterious fusion reaction, and, when not prevented, how does the fusion reaction then occur? We first considered the possibility that fusion of inverted repeats occurs by the same mechanisms by which other types of repeats recombine. The mechanisms of recombination between direct repeats (Klein 1988; Thomas and Rothstein 1989), between inverted repeats (Bai and Symington 1996; Jinks-Robertson et al. 1993; Fasullo et al. 2001) and between sister chromosomes (Paulovich et al. 1999, Fasullo et al. 2001) has been thoroughly studied in budding yeast. The key observations in those studies are that recombination is decreased >100-fold by mutations in either RAD52 or RAD52 and RAD1. Rad52 is a single-strand annealing (SSA) protein required for most HR reactions (Krogh and Symington 2004), and Rad1 is a nuclease with roles in HR and nucleotide excision repair. We therefore determined the frequency of dicentrics and instability at the ChrVII403 site in rad52, rad1, and rad52 rad1 mutants. Strikingly, we found that inverted repeat fusions were actually increased (>10-fold) in rad52 and rad52 rad1 double mutants compared with wild-type (Rad+) cells (determined from dicentric PCR or mixed colonies) (Table 1). (Some of the fusion events that occurred in rad52 mutants may be Rad1-dependent, since the level of dicentric in rad52 rad1 double mutants was twofold lower by quantitative PCR [Table 1]; this does not change our overall conclusion that the inverted repeat fusion occurs by a RAD52, RAD1-independent mechanism. We also found that rad52 and rad52 rad1 also gave rise to the specific D7/D11 translocation, a product formed when the dicentric intermediate is resolved). Other mutants shown in other studies to decrease inverted repeat inversions (e.g., rad51 and rad59) (Bai and Symington 1996) also lead in our study to an increase in inverted repeat fusion at the ChrVII403 site (Table 1). We also analyzed the roles of RAD52 and RAD1 in fusion at two other sites—ChrVI160 and ChrX542—by using quantitative PCR. We found that fusion between inverted repeats occurred efficiently in rad52 and rad52 rad1 mutants; fusion products are present at nearly the levels formed in wild-type cells (Supplemental Fig. S5I). We do not know why the ChrVII403 site is more unstable in a rad52 mutant than the other two sites. The ChrVII403 site is at a replication terminus, and that might contribute to its instability and different genetic regulation (Admire et al. 2006). We therefore conclude that inverted repeat fusion occurs by a mechanism that is very different from “conventional” recombination between inverted and direct repeats (forming inversions or deletions, respectively).
Table 1.
Frequencies, translocation, and dicentric analysis of DNA repair mutants
In bold, statistically significant (P < 0.05, Kruskal-Wallis test) single mutants to RAD+ or double mutants to rad9 fold change. Frequencies were normalized to RAD+.
aFrequencies were determined by qPCR.
bTranslocation for rad9 was reported previously (Admire et al. 2006).
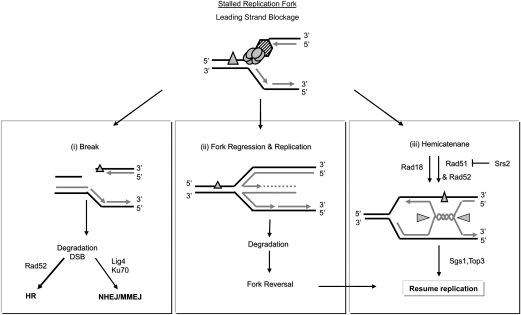
To further address the mechanisms that underlie inverted repeat fusion, we focused on the observation that fusions are increased when DNA replication forks stall (either by lowering dNTPs, by defects in a DNA helicase, or by tRNA gene transcription) (Admire et al. 2006; H Jones and T Weiner, unpubl.). We therefore suggest that inverted repeats fuse by a mechanism related to stalled replication forks and perhaps to how they recover. To guide our studies, we provided a working model of replication fork biology (Fig. 6; Atkinson and McGlynn 2009; Branzei and Foiani 2009). A stalled replication fork (Fig. 6, top) may resume replication (data not shown), undergo a DSB (Fig. 6, left), or undergo sister strand annealing reactions to form a regressed fork (Fig. 6, center) or a hemicatenane (Fig. 6, right). Some outcomes from these structures are shown, and others are possible as well (e.g., the regressed fork may be cleaved by a Holliday junction resolvase to form a DSB) (Atkinson and McGlynn 2009).
Figure 6.
A model of the pathways that may act on stalled replication forks. DNA polymerase stalls at a lesion (triangle). The stalled fork may undergo a DSB (i), or sister strand annealing, template switch events forming a regressed fork (ii) or hemicatenane (iii). DSBs may undergo DNA rejoining by NHEJ, MMEJ, HR, SSA, or telomere addition (not shown). Some genes tested in this study that may regulate each pathway are shown.
To begin to test if any of these pathways are relevant to the fusion of inverted repeats to form dicentrics, we measured the frequency of mixed colony formation in a number of mutants thought to have roles in replication fork recovery. The frequency of mixed colonies is a suitable quantitative surrogate for the frequency of dicentric formation (see Fig. 2), and for some mutants we also performed quantitative dicentric PCR (Table 1). Our genetic and molecular analyses uncovered several relationships between replication fork recovery pathways and nearby inverted repeat fusion. First, as noted above, we found that the Rad52 protein was not required for dicentric formation; fusion of inverted repeats is Rad52-independent. Second, we found that defects in other end-joining pathways—such as nonhomologous end-joining (NHEJ), microhomology-mediated end-joining (MMEJ), and SSA—had little effect on instability as single mutants (Ku70, Lig4, and Rad1; NHEJ/MMEJ, SSA) (Paques and Haber 1999). In strains where events occur more frequently (i.e., rad9 mutants), these events do not require any of these genes (i.e., rad9 and rad9 lig4 mutants have similar phenotypes). Inverted repeat fusion also does not require either Rad50, a component of the MRX complex that is required for both NHEJ and MMEJ, or Mus81, a prominent nuclease. Indeed, fusion events increase ∼30-fold in both of these mutants, implying that they have a prominent role in the prevention of fusion events. These results suggest that fusion of inverted repeats coming from stalled forks has little to do with NHEJ, MMEJ, or SSA repair pathways.
We next tested the possible role of hemicatenane in inverted repeat fusion (see Fig. 6). These structures are proposed to involve Rad51 and Rad18, among other genes. Currently, it is believed that Rad18 and Rad51 promote a template switch event to facilitate formation of a hemicatenane that is resolved by Sgs1 and Top3 (Liberi et al. 2005; Branzei et al. 2006, 2008). We found that mutations in RAD18, RAD51, RAD52, and SGS1 significantly increased instability. These results suggest two things: First, these error-free recovery pathways limit nearby inverted repeat fusion, and second, these genes are not required to fuse inverted repeats.
We also tested the possible role of Srs2, a protein that removes and therefore inhibits Rad51 from acting on ssDNA (Krejci et al. 2003; Veaute et al. 2003). We found that a mutation in SRS2 suppressed instability in a rad9 background (rad9 srs2 has lower instability than rad9). We wondered whether this suppression is due to increased Rad51 activity in a srs2 mutant. We therefore compared the instability of rad9, rad9srs2, rad51, rad9rad51, rad51srs2, and rad9 srs2 rad51 triple mutant. In a srs2 mutant that is Rad51 plus, instability was low, while in the srs2 mutant where rad51 was mutant, instability was as high as the rad51 single mutant; rad51 is epistatic to srs2. In sum, regulation of the activity of Rad51 by Srs2 is important in preventing nearby inverted repeat fusion.
Rad59
In addition to Rad51, the SSA protein Rad59 is thought to define a second distinct Rad52-dependent HR pathway that mediates annealing between sequences with shorter regions of homology (Spell and Jinks-Robertson 2003; Pannunzio et al. 2008). We found that strains lacking Rad59 showed an increase in mixed colony formation, and readily formed both dicentric and acentric chromosomes (Table 1; Supplemental Fig. S8); therefore, Rad59 acts like Rad52 and Rad51 to normally prevent inverted repeat fusion. We did make one surprising observation studying Rad59 that bears on how the dicentric is resolved. We found that Rad59 is required for resolution of the dicentric (involving recombination between D7 and D11 to from the D7/D11 translocation); neither rad59 nor rad9 rad59 double mutants generated this translocation (Table 1), while both mutants generated dicentric chromosomes. Events associated with the URA3 synthetic module also obey the same genetics as LTR recombination in all respects: The “URA”-containing inverted repeats fuse to form the dicentric in the absence of RAD59, but the resolution of the dicentric to form a URA3-containing translocation is reduced 200-fold in rad9 rad59 mutants (Fig. 5B, cf. rad9 and rad9 rad59).
Pol32 and break-induced replication (BIR)-mediated events
There is increasing evidence that links replication stress to microhomology-mediated fusion events (for review, see Hastings et al. 2009a). A mechanism termed microhomology-mediated BIR (MMBIR) has been proposed to resolve stalled forks by mediating fusion events (for review, see Hastings et al. 2009b). Both homology-driven and microhomology-driven BIR require a nonessential subunit of DNA polymerase δ, Pol32 (Lydeard et al. 2007; Payen et al. 2008). We thus tested if Pol32 was required for inverted repeat fusion, and for resolution of the dicentric to form the translocation. We found that instability is actually increased in pol32 mutants, and in rad9 pol32 double mutants when compared with Rad+ and rad9 strains. We therefore infer that neither Pol32 nor BIR events are involved in either fusion of inverted repeats to form the dicentric or in resolution of the dicentric to form the D7/D11 translocation.
Conclusion of genetic analysis of inverted repeat fusion
In conclusion, our genetic studies first indicate that inverted repeat fusion occurs by a mechanism distinct from repeat recombination events reported previously in budding yeast. Second, fusion is prevented by genes proposed to act in replication fork recovery to form hemicatenanes (RAD51, RAD52, and RAD18). Third, fusion does not require genes that are involved in DSB repair, BIR, or hemicatenane formation (RAD1, RAD50, RAD51, RAD59, RAD52, yKU80, LIG4, POL32, and RAD18). These genetic results rule out several models, and lead us to speculate on a replication-based model addressed in the Discussion.
Discussion
In this study, we report and discuss below two major findings. First, we found that nearby inverted repeats fuse to form either dicentric or acentric chromosomes, and that dicentric formation is linked to chromosome instability. This fusion is general; it happens at multiple sites in the yeast genome, and at a synthetic site placed at three different locations on a single chromosome. Second, fusion of nearby inverted repeats is suppressed by many DNA repair pathways, and when it does occur, fusion is surprisingly independent of DSB repair and other major repair pathways. These and other results lead to a replication-based model of how fusions occur.
Inverted repeat instability in budding yeast
Inverted repeats represent potential sites of instability in both prokaryotic and eukaryotic genomes. Here, we found that nearby inverted repeats, consisting of either LTRs or synthetic inverted repeats, fuse to form acentric or dicentric chromosomes. (This reaction is mechanistically very different from an inversion between inverted repeats, so we used the phrase “inverted repeat fusion.”) A search of the literature indicates that nearby inverted repeat fusion is an event that occurs in bacteria and in fission yeast as well (see below). The extent to which three nearby inverted repeats destabilize the budding yeast genome is not yet clear, and will depend in part on defining sequence parameters required for fusion—parameters that may involve chromosome topology as well. We did find that the fusion events in budding yeast occur between inverted repeats with as few as ∼20 bases of homology and separated by ∼1.3–5.5 kb of DNA (Supplemental Fig. S5I). The frequency of inverted repeat fusion is about one in 100,000 wild-type cells at the ChrVII403 site to eight in 100,000 at the ChrX542 site. The inverted repeats in ChrVII403 do appear to cause most of that chromosome's instability (H Jones and T Weinert, unpubl.); other inverted repeats on ChrVII probably also cause instability, though they have not yet been tested. Sites of inverted repeats also appear to be sites of translocation events that led to speciation of Saccharomyces species (Admire et al. 2006; AL Paek and T Weinert, unpubl.).
Inverted repeat fusion and instability in other genomes
After we identified inverted repeat fusion in budding yeast, we examined the literature seeking similar observations in other organisms. Inverted repeat fusions similar to that described in this study have been reported in two studies in bacteria and two in fission yeast. In particular, in one bacterial study, nearby inverted repeats present on a plasmid were found to undergo a complex rearrangement that did not involve RecA (Rad51 ortholog) (Bi and Liu 1996), and in a second study, DNA intermediates were identified that also appeared to be undergoing inverted repeat fusion (Ahmed and Podemski 1998). The geometry and RecA independence of these reactions suggest to us that they may occur by a similar mechanism as the events in budding yeast (see below for discussion of mechanism). Finally a third study of instability in fission yeast also appears due to nearby inverted repeat fusion. Albrecht et al. (2000) selected for the amplification of Sod2 that confers resistance to lithium chloride, and isolated strains that contained high copies of acentric chromosomes. Examination of the acentric suggests it was formed by fusion of two nearby inverted repeats. Finally, an accompanying study by Mizuno et al. (2009) also shows that nearby inverted repeats can form acentric and dicentric chromosomes in fission yeast.
Similarities and differences in instability associated with palindromes and nearby inverted repeats
Palindromes are a class of inverted repeat sequences separated by very few base pairs of DNA. Nearby inverted repeats can be separated by many kilobases of DNA. Both of these classes of repeat sequences lead to formation of dicentric and acentric chromosomes, but how they do so appears to differ. In budding yeast, Lobachev and colleagues (Narayanan et al. 2006; Voineagu et al. 2008) found that palindromes formed a hairpin or cruciform structure that rearranges to form acentric and dicentric chromosomes. Similarly, Lemoine et al. (2005) also found that palindromes cause instability, presumably also by formation of a cruciform. In Schizosaccharomyces pombe, 80-bp perfect inverted repeats form hairpin and cruciform structures and confer mitotic instability (Farah et al. 2002). Due to their unstable nature, palindromes are underrepresented in the human genome (Lobachev et al. 2000). Although the nearby inverted repeat fusion events reported here also lead to the formation of dicentric and acentric chromosomes, they must do so by a different mechanism, since the repeats are separated by many kilobases of DNA and hence are unlikely to form hairpin or cruciform structures (Lobachev et al. 2000).
A replication-based mechanism for fusion of nearby inverted repeats
The mechanism of inverted repeat fusion remains speculative. What we do know is that many repair and checkpoint control pathways normally suppress fusion of nearby inverted repeats (Fig. 6; Table 1; Admire et al. 2006). It is also clear that fusions occur by a mechanism very different from those mediating recombination between inverted and direct repeats—events that can be stimulated by DSBs (Sugawara and Haber 1992; Bai and Symington 1996; Fasullo et al. 2001). Whatever the mechanism of inverted repeat fusion is, it should account for (1) the geometry of the fusion reaction, (2) the lack of involvement of Rad52 and other repair proteins, (3) the increased fusion of inverted repeats upon disruption of DNA replication, and (4) an asymmetric mechanism of fusion.
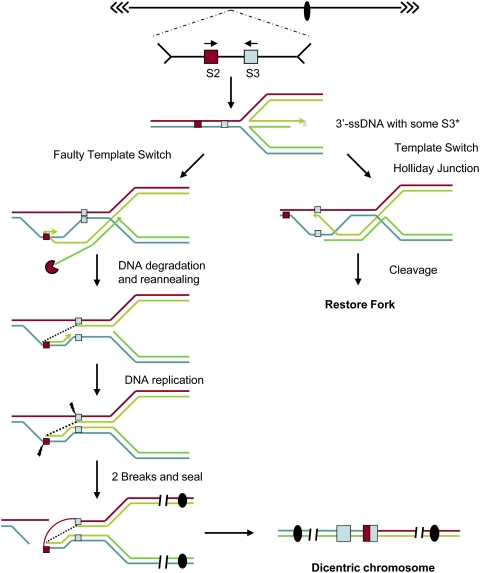
To account for these observations, we suggest a replication-based mechanism we call “faulty template switching” (Fig. 7). This mechanism bears a similarity to mechanisms proposed from studies in bacteria (Goldfless et al. 2006; Long and Kreuzer 2008; Atkinson and McGlynn 2009). In this model, a stalled fork fails to be rescued by error-free pathways (the Rad51, Rad18-dependent hemicatenane pathway, for example) (see Fig. 6), and undergoes a different template switch event (fork regression, replication, and reversal) (Fig. 6) to form a so-called “chicken foot” structure. We suggest that, during fork restart, the nascent DNA strand may either undergo fork reversal (data not shown), pair with the correct sequence and be resolved by an error-free mechanism (as summarized by Atkinson and McGlynn 2009), or pair with an incorrect, nearby sequence (faulty template switch). This chicken foot-based faulty template switch reaction appears to be Rad18-independent. (Rad18 has been proposed to be involved in hemicatenane-based template switch [Fig. 6; Branzei et al. 2008].) How the pairing reaction occurs is unclear, since strand annealing proteins (i.e., Rad52) are not required. A speculative strand annealing reaction without Rad52 has been discussed recently (Hastings et al. 2009b). Once strand annealing has occurred, completion of DNA replication leads fairly directly to formation of a dicentric (or acentric) chromosome.
Figure 7.
DNA replication-based mechanism of inverted repeat fusion. The top shows a chromosome region that contains inverted repeat sequences (red box, S2; blue box, S3), separated by several kilobases of DNA. The right fork stalls (not shown), then regresses to form the “chicken foot” structure with some S3 sequence at the 3′ end of the regressed strand. On the right, the regressed fork normally reinvades at the appropriate S3 sequence, and forms a Holliday structure that then is cleaved and resolved to restore a fork (after Atkinson and McGlynn 2009). On the left, the regressed fork reinvades at the wrong S2 sequence. Then, the unpaired regressed strand (dark green) is degraded (pacman), and the invaded strand begins to replicate using the blue strand as a template (displacing the blue strand from the red strand and allowing annealing of light green to red, forming the S2–S3 joint; dotted line). Upon completion of replication, ssDNA breaks must separate the left red and blue strands and the gap is filled (thin red arc, details not shown), forming the dicentric with hybrid S2–S3 repeat and one intact S2 repeat. The dicentric may then undergo further instability.
Although speculative, this model illustrates how a relatively simple error in fork restart (selection of the wrong sequence) can lead to a chromosome change. The model makes specific testable predictions; we hypothesize that faulty template switching requires a 5′–3′ exonuclease for degradation of the regressed fork, and an endonuclease to resolve the structure after annealing to a nonallelic sequence. Further work linking gene function to DNA structure will allow us to further test how inverted repeats fuse. Models having a DSB as an intermediate are also plausible.
Other examples of possible template switching
Models involving some form of template switching have been proposed previously. In bacteria, a faulty template switch model was proposed to explain rearrangements between direct repeats (Goldfless et al. 2006). In that study, rearrangements were independent of RecA (a Rad51 ortholog), as inverted repeat fusion events reported here were. (In the bacterial study, rearrangements required a gene [DnaK] known to interact with DNA polymerases.) Fusion of nearby inverted repeats in bacteria (mentioned above) also suggest a template switch mechanism (Bi and Liu 1996; Ahmed and Podemski 1998). In yeast, template switching can occur during BIR, and is thought to be due to the unstable nature of replication forks during BIR (Lydeard et al. 2007; Smith et al. 2007, 2009). Smith et al. (2007, 2009) showed switching between allelic and nonallelic sequences, though those events require proteins involved in HR. Finally, a template switch reaction has been suggested to explain complex rearrangements in a human disorder (Lee et al. 2007). The template switch mechanism Lee et al. (2007) propose involves fusion between sequences sharing only very short stretches of sequence homology, seemingly an event that would not involve strand annealing activities (Hastings et al. 2009b). The extent to which a faulty template switch model between nonallelic sequences might explain rearrangements in budding yeast remains to be determined (Myung et al. 2001; Payen et al. 2008).
Clearly, understanding the mechanism of template switching is a technical challenge, in part given how infrequently these events occur. Recently, Mizuno et al. (2009) described a system that may permit dissection of replication fork stalling and template switching. Their results bear similarities to those reported here in two aspects: They found that replication forks stall in nearby inverted repeats, and that stalling causes the fusion of those inverted repeats to form acentric and dicentric chromosomes. In addition, Mizuno et al. (2009) failed to detect any DSBs as intermediates to fusion, and they suggested a template switch model with some similarities to that shown in Figure 7. However, the genetic requirements of those events in fission yeast are similar in some aspects (events are independent of Lig4 and Ku70) but differ in others (85% of the events require genes involved in HR); thus, the mechanistic similarities are as yet unclear. The role of genes in HR lead Mizuno et al. (2009) to propose that, following a template switch, a Holliday structure is formed and resolved to generate symmetric products. We do not have an explanation for the differences, though it is possible that initiation of faulty template switch events is similar, but resolution of intermediates may differ.
The role of Rad59
We also report here the surprising finding that Rad59 has a role clearly independent of Rad52. Specifically, we found that resolution of the dicentric to the D7/D11 translocation required Rad59, but not Rad52 or Rad51 (see Fig. 1B; Table 1). This result was surprising, because previous genetic analyses suggested that Rad52 is required for Rad59 function (Bai and Symington 1996; Davis and Symington 2001). However, recently, Pannunzio et al. (2008) found that chromosomal rearrangements in rad52 mutants were partially dependent on Rad59. Several other studies have shown that dicentric chromosomes are often resolved by translocations involving repetitive elements (Haber and Thorburn 1984; Surosky and Tye 1985; Pennaneach and Kolodner 2009). It is not clear whether these translocations require Rad59. The Rad59-dependent resolution of the dicentric chromosome will be the focus of a further study.
Broader implications for genome stability in mammalian genomes
It is clear that many eukaryotic genomes harbor nearby inverted repeats that might destabilize their chromosomes. The human genome contains a high frequency of large inverted repeats with high sequence homology (Warburton et al. 2004). Wang and Leung (2009) estimated the frequency of inverted repeats of >30 bp in length to be around one in 10,000 bp in the human genome. The extent to which inverted repeats fuse or cause instability in mammalian genomes is unknown. A recent study by Lange et al. (2009) found that fusion of large inverted repeats (0.3- to 2.9-Mb arms separated by 2.1–169 kb) on the human Y chromosome leads to the formation of dicentric chromosomes. Patients that harbored dicentric chromosomes exhibited spermatogenic failure, and in some cases Turner syndrome, a sex disorder that is characterized by “XO” mosaicism. Lange et al. (2009) hypothesize that the “XO” mosaicism seen in at least some of these patients is due to an unstable dicentric Y chromosome. Interestingly it has been shown recently that individuals with Turner syndrome have a higher incidence of cancer than the general population (Schoemaker et al. 2008). It is not yet known whether the increased cancer risk is in fact due to the existence of an unstable dicentric chromosome. Also, the extent to which these fusion events are caused by a faulty template switching mechanism, as opposed to a crossover event between sister chromatids, is unclear.
Materials and methods
Yeast strains
Yeast strains are derivatives of the A364a strain described in previous studies and references therein (Weinert and Hartwell 1990), and are derived from TY200 or TY206 by transformation. TY200 is a ChrVII disome generated by T. Formosa, M. Carson, and L. Hartwell that is MATα +/hxk2∷CAN1 lys5/+ cyhr/CYHs trp5/+ leu1/+ centromere ade6/+ +/ade3, ura3-52. TY206 harbors a rad9-null mutation, rad9∷ura3, described by Weinert and Hartwell (1990). Derivatives were made by transformation (Gietz and Woods 2001) of appropriate DNA fragments, and genotypes were verified by Southern analysis, PCR, and/or phenotype. Mutant DNA fragments were generated by PCR amplification of KanMX4-marked mutations from the Euroscarf strain using primers that flank each gene. Candidate mutants were analyzed by PCR using outside primers, and restriction digests were performed in cases where mutant and wild-type alleles had similar-sized fragments. Genotyping for all strains was unambiguous. In some cases, we introduced a URA3 allele to replace the KanMX4 allele. The rad1 mutation was generated by PCR-amplifying the hygromycin gene from pAG32 with primers that included 45 bp of homology with DNA flanking the rad1 ORF. rad9 ctf13-30 alleles were generated by a “pop-in–pop-out” strategy. TY206 was transformed with HindIII-linearized pBFS218 ctf13-30 (gift of F. Spencer) under selection for Ura+. We then isolated 5-Fluoroorotic acid-resistant derivatives (“pop-outs”) that were screened for temperature sensitivity at 37°C. Further details of strain constructions are available upon request.
The URA3 inverted repeat module was constructed as shown in Supplemental Figure S5. Briefly, fragments of the URA3 gene were joined to either the Nat1 gene (RA:NatR:RU cassette) or to KanMX4 (A3:KanMX4). These cassettes were inserted into plasmids that contain ∼500 bp of sequence-flanking sites 110, 403, 461, or 535. Each module was inserted into ChrVII by restriction digestion and transformed into cells, and inserts were selected by growth on drug-containing plates. Candidate insertions were confirmed by genetic analyses. For example, the rad9 430:RA:NatR:RU cassette is linked to the CAN1 gene, and resides genetically between TRP5 and CYH2, mapped by analysis of allelic recombinants. Canavanine (Can; 60 μg/mL), geneticin (100 or 500 μg/mL), hygromycin B (300 μg/mL), and nourseothricin (Nat; 50 μg/mL) were used at the concentrations given.
Chromosome instability assays
Genetic analysis of ChrVII instability was performed as described in Admire et al. (2006). Briefly, we grew cells on synthetic media lacking adenine (Ade) to ensure selection for both ChrVII homologs. We then plated cells on rich media (YEPD, 2% dextrose) and, after 2–3 d of growth at 30°C, identified single colonies that contained 1 × 106 to 3 × 106 cells. Synthetic media were as described (Sherman et al. 1986). To identify the types of chromosome changes that occurred as cells grew on rich media, we resuspended individual colonies in water, counted, and plated on selective media consisting of either synthetic media supplemented with 60 μg/mL Can and all essential amino acids except arginine and serine, or the same selective media also lacking Ade. Cells plated on selective media containing Ade identify chromosome loss events as colonies that prove to be CanR Ade− after replica plating on media lacking Ade (see Fig. 1A). Cells plated on selective media without Ade identified allelic recombinants and mixed colonies (both CanR Ade+). We scored the morphology of colonies as either “round,” which generally proved to be stable allelic recombinants, or “sectored/mixed,” which generally proved to contain cells of many different genotypes. We performed “lineage assays” to confirm that cells in allelic recombinants contained the same genotype (typically >95% of the cells had the same genotype), and that cells in mixed colonies contained multiple genotypes (see Admire et al. 2006), including chromosome loss and allelic recombinants in multiple genetic intervals shown in Figure 1A. In all strains, we verified that round and mixed colonies contained single or multiple genotypes, and corrected the frequencies for the rare strains where this was not the case. Frequencies were determined from analysis of at least six colonies; the average and standard deviations are shown. Statistical tests were performed using the Kruskal-Wallis method (Kruskal and Wallis 1952).
Molecular analysis of altered chromosomes.
We analyzed CanR Ade+ allelic and mixed colonies for the presence of the specific translocation. CanR Ade+ cells were grown in 2–3 mL of rich media, and genomic DNA was prepared and subjected to PCR amplification using ChrVII403-535 primers (Supplemental Fig. S11 for coordinates). We sequenced the 1.1-kb fragment generated by the translocation to confirm it had the recombination junction shown in Figure 1B (Admire et al. 2006). We typically analyzed four independent cultures by PCR to judge if the translocation was generated. For rad59 mutants that failed to make the translocation, we analyzed >30 mixed colonies. We also used a PCR assay to detect dicentric or acentric chromosomes formed at natural inverted repeats or in the URA module. Again, genomic DNA was isolated from cells grown in rich media, and was subjected to PCR amplification using the appropriate PCR primers (Supplemental Fig. S11). We confirmed that the URA module generates Ura+ translocations, using both a PCR assay of the recombination junction (data not shown) and pulse field gels probed with a ChrVII-specific probe (YGL250) (Supplemental Fig. S6). For pulse-field gels, chromosomes were separated (Iadonato and Gnirke 1996) using conditions that optimize for separation of 1100-kb (native) and 1200-kb (translocation) chromosomes.
Real-time PCR quantification
The level of dicentric chromosome was analyzed on a LightCycler 1.0 (Roche Diagnostics). A 20-μL amplification mixture contained 2 μL of template DNA, 3 mM MgCl2, 5 μM primers (Supplemental Fig. S11), and LightCycler FastStart Master SYBR Green I Mix (Roche Diagnostics). The thermal cycling conditions consisted of 10 min at 95°C, and 40 cycles of 15 sec at 95°C, 10 sec at 58°C, and 90 sec at 72°C. For quantification, standard curves were prepared by serial 15-fold dilutions of PCR-purified ChrVII403dicentric DNA fragment. We detected between five and 106 dicentric molecules. Crossing points, representing the PCR cycles at which products were detectable, were determined for each sample using the second derivative maximum method (LightCycler 3.5.3 software, Roche Diagnostics). Quality control was ensured by a nontemplate control and determination of the melting point of each PCR product. Only samples in exact concordance with the melting point of a sequence-proofed amplicon were included. In each LightCycler run, the calibrator and unknown sample were measured in duplicate. Variability of the real-time PCRs was 6.3%.
Acknowledgments
We thank F. Spencer for plasmids, and Sue Lovett for bringing to our attention the bacterial literature on template switching. We also thank JMST for useful discussions. T.W was funded by an NACRP/NCI grant and NIH 1R01 GM076186, S.K. was supported by GM008659-11A1, and A.P. was funded by NSF-IGERT grant DGE-0114420. H.J. was supported by award number F31GM087120 from the National Institute of General Medical Sciences.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1862709.
Supplemental material is available at http://www.genesdev.org.
References
- Admire A, Shanks L, Danzl N, Wang M, Weier U, Stevens W, Hunt E, Weinert T. Cycles of chromosome instability are associated with a fragile site and are increased by defects in DNA replication and checkpoint controls in yeast. Genes & Dev. 2006;20:159–173. doi: 10.1101/gad.1392506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed A, Podemski L. Observations on template switching during DNA replication through long inverted repeats. Gene. 1998;223:187–194. doi: 10.1016/s0378-1119(98)00159-0. [DOI] [PubMed] [Google Scholar]
- Albrecht EB, Hunyady AB, Stark GR, Patterson TE. Mechanisms of sod2 gene amplification in Schizosaccharomyces pombe. Mol Biol Cell. 2000;11:873–886. doi: 10.1091/mbc.11.3.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson J, McGlynn P. Replication fork reversal and the maintenance of genome stability. Nucleic Acids Res. 2009;37:3475–3492. doi: 10.1093/nar/gkp244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y, Symington LS. A Rad52 homolog is required for RAD51-independent mitotic recombination in Saccharomyces cerevisiae. Genes & Dev. 1996;10:2025–2037. doi: 10.1101/gad.10.16.2025. [DOI] [PubMed] [Google Scholar]
- Bi X, Liu LF. DNA rearrangement mediated by inverted repeats. Proc Natl Acad Sci. 1996;93:819–823. doi: 10.1073/pnas.93.2.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bignell GR, Santarius T, Pole JCM, Butler AP, Perry J, Plesance E, Greenman C, Menzies A, Taylor S, Edkins S, et al. Architectures of somatic genomic rearrangement in human cancer amplicons at sequence-level resolution. Genome Res. 2007;17:1296–1303. doi: 10.1101/gr.6522707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branzei D, Foiani M. The checkpoint response to replication stress. DNA Repair (Amst) 2009;8:1036–1046. doi: 10.1016/j.dnarep.2009.04.014. [DOI] [PubMed] [Google Scholar]
- Branzei D, Sollier J, Liberi G, Zhao X, Maeda D, Seki M, Enomoto T, Ohta K, Foiani M. Ubc9- and MMS21-mediated sumoylation counteracts recombinogenic events at damaged replication forks. Cell. 2006;127:509–522. doi: 10.1016/j.cell.2006.08.050. [DOI] [PubMed] [Google Scholar]
- Branzei D, Vanoli F, Foiani M. SUMOylation regulates Rad18-mediated template switch. Nature. 2008;456:915–920. doi: 10.1038/nature07587. [DOI] [PubMed] [Google Scholar]
- Bruder CE, Piotrowski A, Gijsbers AACJ, Andersson R, Erickson S, Diaz de Ståhl T, Menzel U, Sandgren J, von Tell D, Poplawski A, et al. Phenotypically concordant and discordant monozygotic twins display different DNA copy-number-variation profiles. Am J Hum Genet. 2008;82:763–771. doi: 10.1016/j.ajhg.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson MJ, Hartwell L. CDC17: An essential gene that prevents telomere elongation in yeast. Cell. 1985;42:249–257. doi: 10.1016/s0092-8674(85)80120-3. [DOI] [PubMed] [Google Scholar]
- Chen C, Kolodner RD. Gross chromosomal rearrangements in Saccharomyces cerevisiae replication and recombination defective mutants. Nat Genet. 1999;23:81–85. doi: 10.1038/12687. [DOI] [PubMed] [Google Scholar]
- Davis AP, Symington LS. The yeast recombinational repair protein Rad59 interacts with Rad52 and stimulates single-strand annealing. Genetics. 2001;159:515–525. doi: 10.1093/genetics/159.2.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doheny KF, Sorger PK, Hyman AA, Tugendreich S, Spencer F, Hieter P. Identification of essential components of the S. cerevisiae kinetochore. Cell. 1993;73:761–774. doi: 10.1016/0092-8674(93)90255-O. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dujon B. Yeasts illustrate the molecular mechanisms of eukaryotic genome evolution. Trends Genet. 2006;22:375–387. doi: 10.1016/j.tig.2006.05.007. [DOI] [PubMed] [Google Scholar]
- Farah JA, Hartsuiker E, Mizuno K, Ohta K, Smith GR. A 160-bp palindrome is a Rad50.Rad32-dependent mitotic recombination hotspot in Schizosaccharomyces pombe. Genetics. 2002;161:461–468. doi: 10.1093/genetics/161.1.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasullo M, Giallanza P, Dong Z, Cera C, Bennett T. Saccharomyces cerevisiae rad51 mutants are defective in DNA damage-associated sister chromatid exchanges but exhibit increased rates of homology-directed translocations. Genetics. 2001;158:959–972. doi: 10.1093/genetics/158.3.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Woods RA. Genetic transformation of yeast. BioTechniques. 2001;30:816–831. doi: 10.2144/01304rv02. [DOI] [PubMed] [Google Scholar]
- Goldfless SJ, Morag AS, Belisle KA, Sutera VA, Jr, Lovett ST. DNA repeat rearrangements mediated by DnaK-dependent replication fork repair. Mol Cell. 2006;21:595–604. doi: 10.1016/j.molcel.2006.01.025. [DOI] [PubMed] [Google Scholar]
- Haber JE, Thorburn PC. Healing of broken linear dicentric chromosomes in yeast. Genetics. 1984;106:207–226. doi: 10.1093/genetics/106.2.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett JA, Greider CW. End resection initiates genomic instability in the absence of telomerase. Mol Cell Biol. 2003;23:8450–8461. doi: 10.1128/MCB.23.23.8450-8461.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings PJ, Lupski JR, Rosenberg SM, Ira G. Mechanisms of change in gene copy number. Nat Rev Genet. 2009a;10:551–564. doi: 10.1038/nrg2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings PJ, Ira G, Lupski JR. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet. 2009b;5:e1000327. doi: 10.1371/journal.pgen.1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iadonato SP, Gnirke A. RARE-cleavage analysis of YACs. Methods Mol Biol. 1996;54:75–85. doi: 10.1385/0-89603-313-9:75. [DOI] [PubMed] [Google Scholar]
- Jinks-Robertson S, Michelitch M, Ramcharan S. Substrate length requirements for efficient mitotic recombination in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:3937–3950. doi: 10.1128/mcb.13.7.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JM, Vanguri S, Boeke JD, Gabriel A, Voytas DF. Transposable elements and genome organization: A comprehensive survey of retrotransposons revealed by the complete Saccharomyces cerevisiae genome sequence. Genome Res. 1998;8:464–478. doi: 10.1101/gr.8.5.464. [DOI] [PubMed] [Google Scholar]
- Klein HL. Different types of recombination events are controlled by the RAD1 and RAD52 genes of Saccharomyces cerevisiae. Genetics. 1988;120:367–377. doi: 10.1093/genetics/120.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodner RD, Putnam CD, Myung K. Maintenance of genome stability in Saccharomyces cerevisiae. Science. 2002;297:552–557. doi: 10.1126/science.1075277. [DOI] [PubMed] [Google Scholar]
- Krejci L, Van Komen S, Li Y, Villemain J, Reddy MS, Klein H, Ellenberger T, Sung P. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature. 2003;423:305–309. doi: 10.1038/nature01577. [DOI] [PubMed] [Google Scholar]
- Krogh BO, Symington LS. Recombination proteins in yeast. Annu Rev Genet. 2004;38:233–271. doi: 10.1146/annurev.genet.38.072902.091500. [DOI] [PubMed] [Google Scholar]
- Kruskal WH, Wallis WA. Use of ranks in one-criterion variance analysis. J Am Stat Assoc. 1952;47:583–621. [Google Scholar]
- Lange J, Skaletsky H, van Daalen SK, Embry SL, Korver CM, Brown LG, Oates RD, Silber S, Repping S, Page DC. Isodicentric Y chromosomes and sex disorders as byproducts of homologous recombination that maintains palindromes. Cell. 2009;138:855–869. doi: 10.1016/j.cell.2009.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JA, Carvalho CM, Lupski JR. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell. 2007;131:1235–1247. doi: 10.1016/j.cell.2007.11.037. [DOI] [PubMed] [Google Scholar]
- Lemoine FJ, Degtyareva NP, Lobachev K, Petes TD. Chromosomal translocations in yeast induced by low levels of DNA polymerase a model for chromosome fragile sites. Cell. 2005;120:587–598. doi: 10.1016/j.cell.2004.12.039. [DOI] [PubMed] [Google Scholar]
- Liberi G, Maffioletti G, Lucca C, Chiolo I, Baryshnikova A, Cotta-Ramusino C, Lopes M, Pellicioli A, Haber JE, Foiani M. Rad51-dependent DNA structures accumulate at damaged replication forks in sgs1 mutants defective in the yeast ortholog of BLM RecQ helicase. Genes & Dev. 2005;19:339–350. doi: 10.1101/gad.322605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo AW, Sabatier L, Fouladi B, Pottier G, Ricoul M, Murnane JP. DNA amplification by breakage/fusion/bridge cycles initiated by spontaneous telomere loss in a human cancer cell line. Neoplasia. 2002;4:531–538. doi: 10.1038/sj.neo.7900267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobachev KS, Stenger JE, Kozyreva OG, Jurka J, Gordenin DA, Resnick MA. Inverted Alu repeats unstable in yeast are excluded from the human genome. EMBO J. 2000;19:3822–3830. doi: 10.1093/emboj/19.14.3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobachev KS, Gordenin DA, Resnick MA. The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell. 2002;108:183–193. doi: 10.1016/s0092-8674(02)00614-1. [DOI] [PubMed] [Google Scholar]
- Lobachev KS, Rattray A, Narayanan V. Hairpin- and cruciform-mediated chromosome breakage: Causes and consequences in eukaryotic cells. Front Biosci. 2007;12:4208–4220. doi: 10.2741/2381. [DOI] [PubMed] [Google Scholar]
- Long DT, Kreuzer KN. Regression supports two mechanisms of fork processing in phage T4. Proc Natl Acad Sci. 2008;105:6852–6857. doi: 10.1073/pnas.0711999105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lydeard JR, Jain S, Yamaguchi M, Haber JE. Break-induced replication and telomerase-independent telomere maintenance require Pol32. Nature. 2007;448:820–823. doi: 10.1038/nature06047. [DOI] [PubMed] [Google Scholar]
- Maser RS, DePinho RA. Connecting chromosome, crisis, and cancer. Science. 2002;297:565–569. doi: 10.1126/science.297.5581.565. [DOI] [PubMed] [Google Scholar]
- McClintock B. The stability of broken ends of chromosomes in zea mays. Genetics. 1941;26:234–282. doi: 10.1093/genetics/26.2.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno K, Lambert S, Baldacci G, Murray JM, Carr AM. Nearby inverted repeats fuse to generate acentric and dicentric palindromic chromosomes by a replication template exchange mechanism. Genes & Dev. 2009 doi: 10.1101/gad.1863009. (this issue). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murnane JP, Sabatier L. Chromosome rearrangements resulting from telomere dysfunction and their role in cancer. Bioessays. 2004;26:1164–1174. doi: 10.1002/bies.20125. [DOI] [PubMed] [Google Scholar]
- Myung K, Chen C, Kolodner RD. Multiple pathways cooperate in the suppression of genome instability in Saccharomyces cerevisiae. Nature. 2001;411:1073–1076. doi: 10.1038/35082608. [DOI] [PubMed] [Google Scholar]
- Narayanan V, Mieczkowski PA, Kim HM, Petes TD, Lobachev KS. The pattern of gene amplification is determined by the chromosomal location of hairpin-capped breaks. Cell. 2006;125:1283–1296. doi: 10.1016/j.cell.2006.04.042. [DOI] [PubMed] [Google Scholar]
- Pannunzio NR, Manthey GM, Bailis AM. RAD59 is required for efficient repair of simultaneous double-strand breaks resulting in translocations in Saccharomyces cerevisiae. DNA Repair (Amst) 2008;7:788–800. doi: 10.1016/j.dnarep.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulovich AG, Armour CD, Hartwell LH. The Saccharomyces cerevisiae RAD9, RAD17, RAD24 and MEC3 genes are required for tolerating irreparable, ultraviolet-induced DNA damage. Genetics. 1999;150:75–93. doi: 10.1093/genetics/150.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payen C, Koszul R, Dujon B, Fischer G. Segmental duplications arise from Pol32-dependent repair of broken forks through two alternative replication-based mechanisms. PLoS Genet. 2008;4:e1000175. doi: 10.1371/journal/pgen.1000175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennaneach V, Kolodner RD. Stabilization of dicentric translocations through secondary rearrangements mediated by multiple mechanisms in S. cerevisiae. PLoS One. 2009;4:6389. doi: 10.1371/journal.pone.0006389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam CD, Pennaneach V, Kolodner RD. Chromosome healing through terminal deletions generated by de novo telomere additions in Saccharomyces cerevisiae. Proc Natl Acad Sci. 2004;101:13262–13267. doi: 10.1073/pnas.0405443101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam CD, Hayes TK, Kolodner RD. Specific pathways prevent duplication-mediated genome rearrangements. Nature. 2009;460:984–989. doi: 10.1038/nature08217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattray AJ, Shafer BK, Neelam B, Strathern JN. A mechanism of palindromic gene amplification in Saccharomyces cerevisiae. Genes & Dev. 2005;19:1390–1399. doi: 10.1101/gad.1315805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard GF, Kerrest A, Dujon B. Comparative genomics and molecular dynamics of DNA repeats in eukaryotes. Microbiol Mol Biol Rev. 2008;72:686–727. doi: 10.1128/MMBR.00011-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatier L, Ricoul M, Pottier G, Murnane JP. The loss of a single telomere can result in instability of multiple chromosomes in a human tumor cell line. Mol Cancer Res. 2005;3:139–150. doi: 10.1158/1541-7786.MCR-04-0194. [DOI] [PubMed] [Google Scholar]
- Schmidt KH, Wu J, Kolodner RD. Control of translocations between highly diverged genes by Sgs1, the Saccharomyces cerevisiae homolog of the Bloom's syndrome protein. Mol Cell Biol. 2006;26:5406–5420. doi: 10.1128/MCB.00161-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F, Fink GR, Hicks JB. Methods in yeast genetics. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1986. [Google Scholar]
- Schoemaker MJ, Swerdlow AJ, Higgins CD, Wright AF, Jacobs PA. Cancer incidence in women with Turner syndrome in Great Britain: A national cohort study. Lancet Oncol. 2008;9:239–246. doi: 10.1016/S1470-2045(08)70033-0. [DOI] [PubMed] [Google Scholar]
- Smith CE, Llorente B, Symington LS. Template switching during break-induced replication. Nature. 2007;447:102–105. doi: 10.1038/nature05723. [DOI] [PubMed] [Google Scholar]
- Smith CE, Lam AF, Symington LS. Aberrant double-strand break repair resulting in half crossovers in mutants defective for Rad51 or the DNA polymerase δ complex. Mol Cell Biol. 2009;29:1432–1441. doi: 10.1128/MCB.01469-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spell RM, Jinks-Robertson S. Role of mismatch repair in the fidelity of RAD51- and RAD59-dependent recombination in Saccharomyces cerevisiae. Genetics. 2003;165:1733–1744. doi: 10.1093/genetics/165.4.1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara N, Haber JE. Characterization of double-strand break-induced recombination: Homology requirements and single-stranded DNA formation. Mol Cell Biol. 1992;12:563–575. doi: 10.1128/mcb.12.2.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surosky RT, Tye BK. Resolution of dicentric chromosomes by Ty-mediated recombination in yeast. Genetics. 1985;110:397–419. doi: 10.1093/genetics/110.3.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas BJ, Rothstein R. The genetic control of direct-repeat recombination in Saccharomyces: The effect of rad52 and rad1 on mitotic recombination at GAL10, a transcriptionally regulated gene. Genetics. 1989;123:725–738. doi: 10.1093/genetics/123.4.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillier ERM, Collins RA. Genome rearrangement by replication-directed translocation. Nat Genet. 2000;26:195–197. doi: 10.1038/79918. [DOI] [PubMed] [Google Scholar]
- Veaute X, Jeusset J, Soustelle C, Kowalczykowski SC, Le Cam E, Fabre F. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature. 2003;423:309–312. doi: 10.1038/nature01585. [DOI] [PubMed] [Google Scholar]
- Voineagu I, Narayanan V, Lobachev KS, Mirkin SM. Replication stalling at unstable inverted repeats: Interplay between DNA hairpins and fork stabilizing proteins. Proc Natl Acad Sci. 2008;105:9936–9941. doi: 10.1073/pnas.0804510105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Leung FC. A study on genomic distribution and sequence features of human long inverted repeats reveals species-specific intronic inverted repeats. FEBS J. 2009;276:1986–1998. doi: 10.1111/j.1742-4658.2009.06930.x. [DOI] [PubMed] [Google Scholar]
- Warburton PE, Giordano J, Cheung F, Gelfand Y, Benson G. Inverted repeat structure of the human genome: The X-chromosome contains a preponderance of large, highly homologous inverted repeats that contain testes genes. Genome Res. 2004;14:1861–1869. doi: 10.1101/gr.2542904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert TA, Hartwell LH. Characterization of RAD9 of Saccharomyces cerevisiae and evidence that its function acts posttranslationally in cell cycle arrest after DNA damage. Mol Cell Biol. 1990;10:6554–6564. doi: 10.1128/mcb.10.12.6554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstock DM, Elliott B, Jasin M. A model of oncogenic rearrangements: Differences between chromosomal translocation mechanisms and simple double-strand break repair. Blood. 2006;107:777–780. doi: 10.1182/blood-2005-06-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong KK, Chang S, Weiler SR, Ganesan S, Chaudhuri J, Zhu C, Artandi SE, Rudolph KL, Gottlieb GJ, Chin L, et al. Telomere dysfunction impairs DNA repair and enhances sensitivity to ionizing radiation. Nat Genet. 2000;26:85–88. doi: 10.1038/79232. [DOI] [PubMed] [Google Scholar]
- Wood JS, Hartwell LH. A dependent pathway of gene functions leading to chromosome segregation in Saccharomyces cerevisiae. J Cell Biol. 1982;94:718–726. doi: 10.1083/jcb.94.3.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou ZH, Akgun E, Jasin M. Repeat expansion by homologous recombination in the mouse germ line at palindromic sequences. Proc Natl Acad Sci. 2001;98:8326–8333. doi: 10.1073/pnas.151008498. [DOI] [PMC free article] [PubMed] [Google Scholar]