Abstract
Mechanisms of telomere replication remain poorly defined. It has been suggested that G-rich telomeric strand replication by lagging mechanisms requires, in a stochastic way, the WRN protein. Here we show that this requirement is more systematic than previously thought. Our data are compatible with a situation in which, in the absence of WRN, DNA synthesis at replication forks is uncoupled, thus allowing replication to continue on the C strand, while single G strands accumulate. We also show that in cells in which both WRN and POT1 are limiting, both G- and C-rich telomeric strands shorten, suggesting a complete replication block. Under this particular condition, expression of a fragment spanning the two POT1-OB (oligonucleotide-binding) fold domains is able to restore C (but not G) strand replication, suggesting that binding of POT1 to the lagging strand allows DNA synthesis uncoupling in the absence of WRN. Furthermore, in vitro experiments indicate that purified POT1 has a higher affinity for the telomeric G-rich strand than purified RPA. We propose a model in which the relative enrichments of POT1 versus RPA on the telomeric lagging strand allows or does not allow uncoupling of DNA synthesis at the replication fork. Our study reveals an unanticipated role for hPOT1 during telomere replication.
Keywords: Telomeres, replication, POT1, WRN, lagging, leading
Replication of telomeres is typically unidirectional, with the G-rich strand being replicated by lagging mechanisms (Gilson and Geli 2007). It has been suggested that, because of the unusual base composition of this strand, lagging mechanisms require specific factors to prevent fork stalling (Crabbe et al. 2004; Karlseder 2006). This event constitutes an exceptionally threatening situation in this portion of the genome, since no converging replication forks would be available to help relieve the block. It has been shown that deficiencies in the activity of the WRN helicase are associated with an increase in the frequency of total loss of sister telomeres replicated by lagging mechanisms (Crabbe et al. 2004). On the other hand, a wealth of in vitro data indicate that WRN is indeed a 3′ > 5′ DNA helicase capable of unwinding various DNA structures formed by G-rich strands, including intrastrand G quadruplexes and t-loop-shaped molecules (Opresko et al. 2004, 2009; Sowd et al. 2008), both thought to represent potential obstacles to the progression of the replication fork at telomeres. Interestingly, WRN has been shown to interact with TRF2 and POT1 (Opresko et al. 2002, 2005), two major components of shelterin, the protein complex protecting telomeres (Liu et al. 2004; de Lange 2005). TRF2 binds the double-stranded portion of telomeres, while POT1 binds the 3′ G overhang. However, as part of shelterin, both proteins interact directly and indirectly with each other (Liu et al. 2004; Barrientos et al. 2008), suggesting that POT1 can also be present all along the telomere tract (de Lange 2005) and perhaps stimulates, together with TRF2, WRN activities during replication (Opresko et al. 2002, 2005).
Because experimentally induced WRN deficiency in human cells leads to only limited increase in the frequency of sister telomere loss (STL), it has been suggested that WRN involvement during telomere replication is rather stochastic (Multani and Chang 2007). In addition, length analyses of single telomeres using PCR-based approaches did not detect an accelerated shortening with cell division in in vitro cultures of cells from WRN patients (Baird et al. 2004), leaving open the question regarding the actual consequences of a WRN defect on telomere replication. More generally, the role of shelterin during telomere replication in vivo has just started to be explored (Sfeir et al. 2009), but many aspects of the molecular mechanisms involved remain unknown.
In this study, we used a recently described approach (Arnoult et al. 2008) that measures the efficiency of telomere replication on both lagging and leading strands to better characterize the implication of WRN and POT1 in these processes. Our results indicate that WRN is absolutely required for complete G-rich telomeric strand (lagging) replication. Our work indicates that, when WRN is absent, POT1 is required to prevent RPA binding to the accumulated G strand and to allow full C strand (leading) replication.
Results
Measuring the efficiency of lagging versus leading telomere replication by chromosome orientation fluorescent in situ hybridiszation (CO-FISH)
The quantitative telomere CO-FISH analysis is based on the degradation, on metaphasic chromosomes, of neosynthesized DNA strands during the previous S phase (Fig. 1A). After this degradation, fluorescence intensities from labeled probes specific for telomeric G- and C-rich sequences should reflect the length of the parental G and C strands, respectively, before the start of S phase. Thus, any process preventing full replication of these strands will lead to a shortening of the corresponding sister telomere. The reliability of measurements is greatly improved when telomeres are long (>10 kb), and therefore analyses are greatly facilitated when telomerase-expressing cells are used (Arnoult et al. 2008). Importantly, any telomerase-mediated elongation of telomeres during S or G2 phases will also be eliminated by the CO-FISH procedure, and will not contribute to the fluorescence intensity.
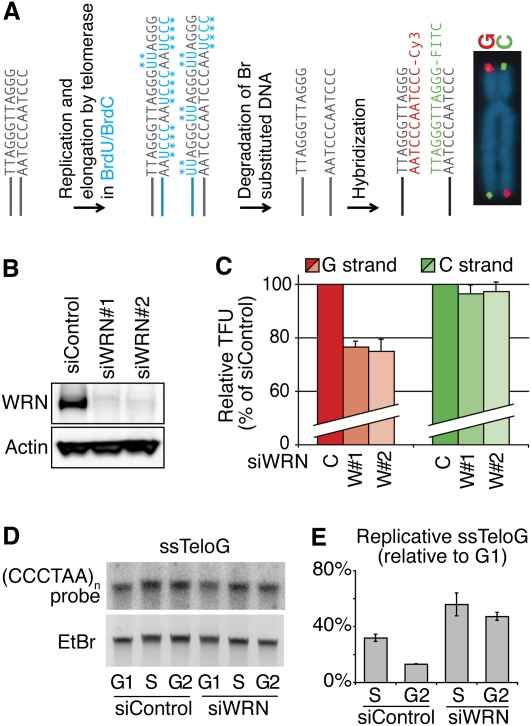
Figure 1.
Depletion of WRN leads to replication fork uncoupling at telomeres. (A) In the CO-FISH technique, BrdU and BrdC are incorporated during S phase. Br-substituted strands (i.e., neosynthesized by replication or telomerase elongation) are degraded by the CO-FISH procedure. Fluorescence intensities of hybridized probes revealing the telomeric G- and C-rich strands reflect the length of parental strands after replication. (B) Efficient knockdown (95%) of WRN in HCA2-T+ cells after transfection of siRNAs and revealed by Western blot. (C) WRN knockdown in these cells leads to G-rich strand shortening (red), while C-rich strands (green) are completely replicated. Average measurements of 30 metaphases are represented as percentages (%) of control siRNA (single intensity distributions are presented in Supplemental Fig. S1). Partial WRN knockdown (<90%) allows normal telomere replication (not shown). (D) Accumulation of replicative single G strands after WRN knockdown shown by the telomeric single-strand assay (Gomez et al. 2004). Radioactive signals are normalized with regard to total DNA input (ethidium bromide). The signal corresponding to single G strands detected in cells blocked in G1 phase is considered as the baseline. (E) Quantification of single G strand signals shows that this strand accumulates at higher levels during S phase in the absence of WRN and that it persists in G2.
WRN is required at most replication forks for normal G-rich telomeric strand (lagging) replication
To determine the impact of WRN on telomere replication, human hTERT-immortalized foreskin fibroblasts (HCA2-T+ cells) were transfected with two WRN-specific siRNAs, which led to significant down-regulation of protein expression (>90%) 48 h later (Fig. 1B). At about the same time, doubly (thymidin–aphidicolin) synchronized cells were released into S phase in the presence of BrdU/dC base substitutes. Metaphases were prepared 10 h later and chromosome spreads were analyzed by quantitative CO-FISH. As shown in Figure 1C, we detected a substantial decrease of G strand-specific fluorescence intensities with both WRN-specific siRNAs when compared with cells transfected with a siRNA control, while in both cases, C strand intensities remained largely unchanged. The distribution of parental G strand intensities (Supplemental Fig. S1) indicates that most chromosome ends had been affected to some extent by the WRN knockdown, strongly suggesting that WRN is required most of the time—perhaps always—for lagging G strand replication. Interestingly, in experiments in which the WRN knockdown was below 90%, cells showed unchanged telomere lengths after 48 h, suggesting that very little protein is sufficient to allow normal G-rich telomeric strand replication.
The absence of WRN leads to uncoupling of the replication fork
The complete replication of C strands at all extremities in spite of a global perturbation of the replication of G strands suggests that DNA polymerization on lagging and leading strands had been uncoupled in the absence of WRN. To verify that replication on the lagging G strand had been blocked, we performed experiments to determine whether single G strands accumulate during the S phase. In these experiments, genomic DNA was purified from cells at different times of the cell cycle and hybridized—in solution and under nondenaturing conditions—with radioactive probes specific for G and C strands. Probes specifically hybridizing to telomeric DNA were detected after gel migration, also under native conditions (Fig. 1D). Quantification of the radioactive signals indicated that, in the control situation, there was a transient increase, above the level detected in nonreplicating (G1) cells, of G strand-specific signals during the S phase, followed by a decrease in G2. This kinetics was identical in the parental HCA2 cells without telomerase (Supplemental Fig. S2A), clearly indicating that this accumulation of single G strands was both replication-related and telomerase-independent. As expected, very low-intensity signals were detected with probes specific for the C strand in nonreplicating cells, and no increase was detected during the S phase (Supplemental Fig. S2B). This is in agreement with the established view that synthesis on the leading strand closely follows fork progression, while synthesis on the lagging is somewhat retarded (Hamdan et al. 2009).
In the absence of WRN, the levels of single G strand signal almost doubled during S phase and, more striking, remained high in the subsequent G2 phase (Fig. 1D,E), in agreement with a situation in which synthesis on this strand has stopped, while synthesis on the opposite strand (the C strand) has continued. Interestingly, in spite of a slight delay in entering G2 by cells with depleted WRN, we could not detect any other sign of replication stress or DNA damage during S or G2 phases (Supplemental Fig. S3A,B), suggesting that, under these experimental conditions, single-strand accumulation at telomeres does not trigger a signaling pathway. Instead, uncoupling of DNA polymerizations by lagging and leading mechanisms seems to take place at every replication fork, allowing full replication of the latter strand.
The absence of WRN leads to both leading and lagging telomere shortening in cells with very long telomeres
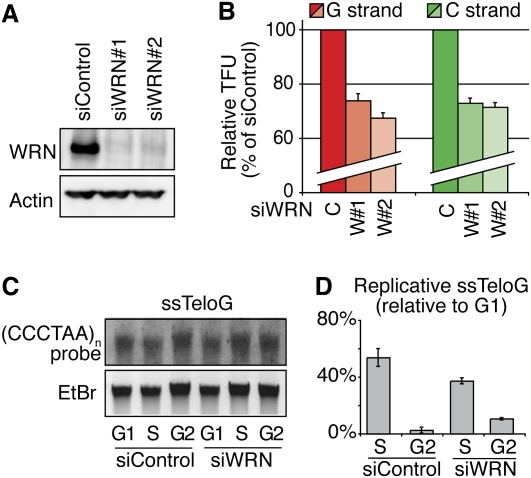
To determine whether replication fork uncoupling is a general mechanism always occurring in the absence of WRN, we tested the effect of WRN depletion in cells that carry abnormally long telomeres (>40 kb). These are HT1080 fibrosarcoma cells expressing extra copies of hTERT and hTR (the catalytic and RNA subunits of telomerase, respectively) and maintaining very long telomeres (cells are here called ST, for supertelomerase) (Cristofari and Lingner 2006). Transfection of siRNAs against WRN into ST cells achieved almost complete knockdown of the protein within 48 h (Fig. 2A). Synchronized cells were treated as above, and the CO-FISH procedure was carried out identically. As shown in Figure 2B, significant losses in fluorescence intensity were recorded for both siRNAs, this time affecting both G and C strands. Strikingly, the replicative single G strand measured by in-gel hybridization, which increased in S phase and almost disappeared in G2 in the control cells, accumulated at lower levels during S phase and further decreased in G2 in cells with depleted WRN (Fig. 2C,D). Together, these results suggest that, in ST cells, the mechanisms allowing the uncoupling of the replication fork in the absence of WRN no longer operate. Here again, all replicating telomeres were affected, indicating a systematic need of WRN for normal telomere replication (Supplemental Fig. S4). Interestingly, ST cells progressed normally throughout the cell cycle in the absence of WRN (Supplemental Fig S5A), and no signs of replication stress were detected (Supplemental Fig. S5B,C). We detected, though, a significant increase in the phosphorylation levels of ATM, specifically in G2 and in ST cells depleted for WRN (Supplemental Fig. S5C). This increase was specific to ST cells, since it was not detected in the parental HT1080 cells and depleted for WRN (Supplemental Fig. S5C). We could not apply a quantitative CO-FISH (Q-CO-FISH) on HT1080 cells (which are telomerase-positive but bear much shorter telomeres than ST cells) (Cristofari and Lingner 2006; data not shown), and therefore we do not know whether, in HT1080 cells depleted for WRN, there is replication of the C strand, while the G strand is shortened.
Figure 2.
No replication fork uncoupling after WRN knockdown in cells with very long telomeres. (A) Efficient knockdown of WRN in HT1080-ST cells revealed by Western blot. (B) In ST cells, WRN knockdown leads to both C-rich strand (green) and G-rich strand shortening (red). Average measurements of 30 metaphases, represented as percentages (%) of control siRNA (single intensity distributions are presented in Supplemental Fig. S4). (C) Accumulation of replicative single G strands after WRN knockdown shown by the telomeric single-strand assay. Radioactive signals are normalized with regard to total DNA input (ethidium bromide). The signal corresponding to single G strands detected in cells blocked in G1 phase is considered as the baseline. (D) Quantification of single G strands indicates that ST cells deficient in WRN do not accumulate more replicative single G strands during S phase and that these strands do not persist in G2.
In the absence of WRN, RPA accumulates at telomeres only in cells with very long telomeres
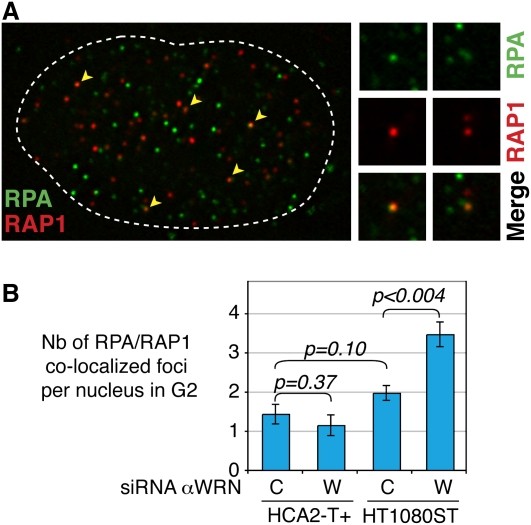
Why is uncoupling of telomere replication in the absence of WRN possible in some cells but not in others? During replication, fork stalling due to lesions that block polymerization on either the leading or lagging strands leads to accumulation of ssDNA (Dart et al. 2004). RPA, the ssDNA-binding protein that travels with the fork and that typically binds to the lagging single strand (Bae et al. 2001), also accumulates. When a threshold level is reached, a signal is transmitted to the replicative helicase, which then stops, thus preventing further accumulation of ssDNA (Nitani et al. 2008). To test whether there was any RPA accumulation at telomeres in the absence of WRN, we used an immunofluorescence (IF) approach. RPA, being very abundantly recruited at replicating chromatin during the S phase, colocalization experiments of RPA, and a telomeric protein (RAP1) were carried in G2, when RPA is still detected at telomeres by chromatin immunoprecipitation (ChIP) (Verdun and Karlseder 2006). Cells in G2 from cultures that were or were not depleted for WRN were sorted and processed for IF. Interestingly, despite the accumulation of single G strand in S and G2 phases in WRN-deficient HCA2-T+ cells (Fig. 1E), the number of RPA/RAP1 colocalization foci per nucleus in these cells was equivalent to that found in WRN-proficient cells (Fig. 3). Conversely, this number was slightly, but significantly (P < 0.004), increased in ST cells that were depleted for WRN when compared with the same cells transfected with control siRNA (Fig. 3). This experiment suggests that RPA is recruited either more abundantly or at more telomeres in the absence of WRN, but only in cells with abnormally long telomeres. Accumulation of RPA in these cells might, in turn, signal to stop fork progression, similar to what occurs when polymerization is blocked elsewhere in the genome (Dart et al. 2004; Nitani et al. 2008).
Figure 3.
Higher RPA accumulation at telomeres of ST cells upon depletion of WRN. (A) Example of colocalized RPA (green) and telomere (RAP1, red) foci revealed by deconvolution three-dimensional IF after cell sorting for cells in G2 phase. (B) Quantification of RPA/RAP1 colocalization foci. The number of RPA/RAP1 foci after WRN knockdown in G2 in normal fibroblasts (HCA2-T+) remains the same, while there is a small but highly significant increase of this number in ST cells depleted for WRN. At least 30 nuclei were analyzed.
POT1 is required for normal replication of the telomeric leading strand in the absence of WRN
What prevents RPA from binding to replicating normal-length telomeres when WRN is absent? We turned our attention to POT1, the natural telomeric single G strand-binding protein (Baumann and Cech 2001). POT1 binds to the 3′ G overhang, where it exerts well-known functions for telomere protection (Veldman et al. 2004; Hockemeyer et al. 2005; He et al. 2006; Wu et al. 2006) and for control of telomerase-dependent elongation (Loayza and de Lange 2003; Wang et al. 2007; Xin et al. 2007). It has been proposed that one of the main mechanisms by which POT1 protects telomeres is by preventing the binding of RPA to the 3′ overhang (Wu et al. 2006; Denchi and de Lange 2007). On the other hand, thanks to its interaction with other shelterin components (Barrientos et al. 2008), POT1 could also be present all along the telomeric tract and therefore near replication forks, where the G strand becomes single-stranded and a potential substrate for POT1 to bind (Gilson and Geli 2007). Thus, in the absence of WRN, binding of POT1 to the G strand at the lagging telomere may both prevent excessive RPA accumulation and allow polymerization on the C strand to continue. We surmised that if POT1 becomes limiting under conditions in which WRN is deficient, the uncoupling mechanism would be compromised. Alternatively, if the levels of POT1 are increased in cells with very long telomeres but dysfunctional WRN, RPA binding would be prevented and uncoupling of the polymerization would be restored.
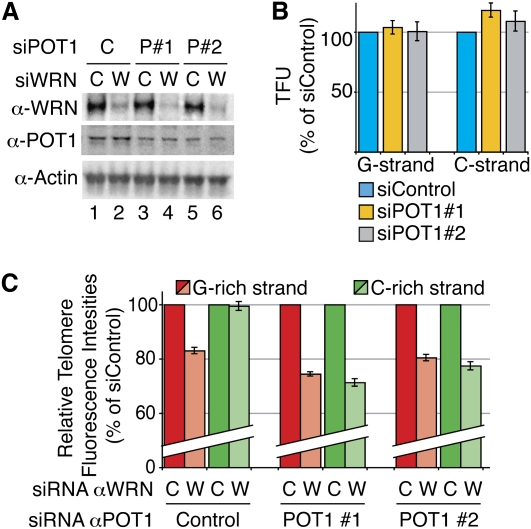
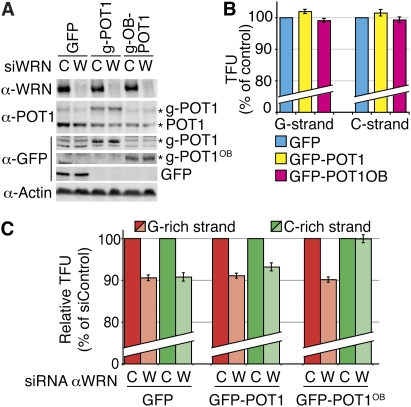
To test these hypotheses, we conducted two types of experiments. In the first experiment, we induced partial knockdown of POT1 in HCA2-T+ cells in which WRN expression had been abolished. The partial knockdown of POT1 (∼50%–60%) did not affect cell cycle progression (data not shown) or G/C strand fluorescence intensities after replication in WRN-proficient cells (Fig. 4A,B), suggesting that enough protein was still present to ensure telomere protection. On the contrary, when combined with WRN depletion, POT1 partial knockdown led to the partial loss—and to the same extent—of both G and C strands, indicating that, under these conditions, uncoupling of the replication fork had been prevented (Fig. 4C). In the second experiment, we introduced, in ST cells depleted for WRN, expression vectors carrying either the full-length (fl) POT1 or a POT1 fragment spanning the two oligonucleotide-binding (OB) fold domains, together with a nuclear localization signal. In both constructs, the POT1 protein was fused to GFP, which allowed us to control for their expression using anti-GFP antibodies by both IF and Western blotting. We could show that while GFP-POT1-OB1+2 showed a nuclear diffuse pattern, GFP-POT1(fl) was readily detected at telomeres (Supplemental Fig. S6), supporting the view that POT1 is efficiently recruited to telomeres through its interactions with other shelterin components (Barrientos et al. 2008). Interestingly, forced expression of either GFP-POT1(fl) or GFP-POT1-OB1+2 led to a decrease of endogenous levels of the protein, as has been reported previously (Loayza and de Lange 2003; Wang et al. 2007; Xin et al. 2007), and therefore the total amounts of available POT1 did not significantly exceed the levels naturally present in these cells. Next, both fusion proteins were expressed in cells in which WRN expression had, or had not, been abolished (Fig. 5A). Quantification of G and C strand fluorescence intensities revealed that neither fl POT1 nor POT1-OB1+2 affected telomere replication in a WRN+ context (Fig. 5B). On the other hand, while GFP-POT1(fl) expression did not change the efficiency of G or C strand replication in WRN-deficient ST cells, the expression of GFP-POT1-OB1+2 completely rescued the replication of the C strand, despite the fact that replication of the G strand remained defective (Fig. 5C).
Figure 4.
POT1 partial knockdown prevents replication fork uncoupling at telomeres of WRN-depleted HCA2-T+ cells. (A) Efficient knockdown of WRN (>90%) (lanes 2,4,6) combined to partial knockdown of POT1 (50%–60% compared with control) (lanes 1,2) with two different POT1 siRNAs (P#1, lanes 3,4; P#2, lanes 5,6) in HCA2-T+, revealed by Western blot. (B) POT1 partial knockdowns have no effect on telomere replication. The histogram represents mean CO-FISH signal intensities after POT1 knockdown, represented as percentages (%) of control siRNA. (C) POT1 knockdown leads to shortening of both C-rich (green) and G-rich (red) strands in HCA2-T+ cells depleted for WRN. Quantification results are represented in percentages of control siRNA for WRN.
Figure 5.
Forced expression of GFP-POT1-OB1+2 in ST cells depleted for WRN rescues replication fork uncoupling at telomeres. (A) Efficient knockdown of WRN (>90%) combined with forced expression of GFP-POT1(fl) or GFP-POT1-OB1+2. POT1 antibodies reveal both endogenous POT1 and GFP-POT1(fl) (g-POT1). GFP antibodies reveal proteins expressed from all three transient transfection experiments: control (GFP), GFP- POT1(fl) (g-POT1), and GFP-POT1-OB1+2 (g-POT1OB). (B) Neither GFP-POT1(fl) nor GFP-POT1-OB1+2 expressions by themselves affect parental telomere length after replication. CO-FISH signal quantifications after POT1 knockdown are represented in percentages (%) of control transfected with the GFP plasmid. (C) Expression of GFP-POT1-OB1+2 in ST cells depleted for WRN is able to completely rescue C strand (but not G strand) replication, while forced expression of GFP-POT1(fl) has no impact. Quantification results are represented in percentages of control siRNA for WRN.
hPOT1 displays higher affinity than RPA for the G-rich telomeric strand in vitro
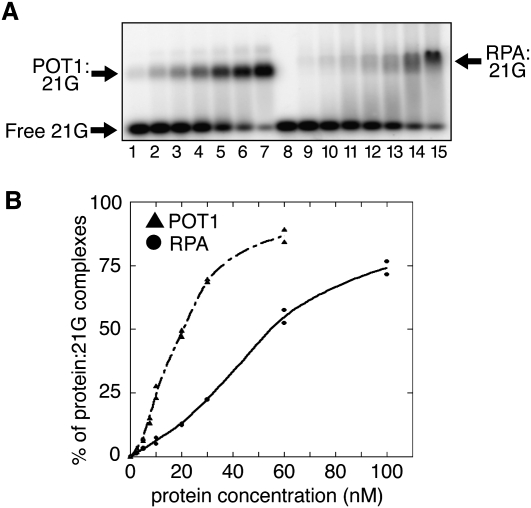
As noted above, POT1 specifically binds to the telomeric G-rich overhang (Lei et al. 2004), where it may prevent RPA from binding (Wu et al. 2006; Denchi and de Lange 2007). Our observations strongly suggest that, in the absence of WRN, POT1 also binds to the G-rich single strand that accumulates during telomere replication, thus preventing RPA from doing so, and, in this particular case, from triggering full replication fork stalling. During replication, however, the conditions for this competition are probably more demanding, since RPA is a very abundant protein, is part of the normal replication apparatus (Wold 1997), and readily accumulates along stretches of ssDNA produced by stalled replication forks (Raderschall et al. 1999; Robison et al. 2004). To gain further insight into the way POT1 may facilitate C strand replication, we carried out in vitro experiments using both purified POT1 and RPA proteins and measured their relative binding affinities toward a synthetic telomere G-rich oligonucleotide (21G). As indicated in Figure 6, both proteins were able to efficiently bind 21G in a concentration-dependent manner, as evidenced by the shift in the gel (Fig. 6A). Quantification of several independent experiments also indicated that POT1 presents a threefold higher affinity for this substrate than RPA (Fig. 6B). These experiments lend support to the hypothesis that POT1 can efficiently compete with RPA for the binding of the G-rich single strand accumulated during replication in the absence of WRN.
Figure 6.
POT1 binds to telomeric G-rich single strands with higher affinity than RPA. (A) Titration of 21G as a function of hPOT1 or hRPA concentration. 32P-21G (20 nM) (lane 8: without protein) was incubated with different amounts of POT1 (lane 1: 2.5 nM; lane 2: 5 nM; lane 3: 7.5 nM; lane 4: 10 nM; lane 5: 20 nM; lane 6: 30 nM; lane 7: 60 nM) or RPA (lane 9: 2.5 nM; lane 10: 5 nM; lane 11: 10 nM; lane 12: 20 nM; lane 13: 30 nM; lane 14: 60 nM; lane 15: 100 nM) and separated on a 1% agarose gel. (POT1:21G) Noncovalent complexes between 21G and POT1; (RPA:21G) noncovalent complexes between 21G and RPA. (B) Quantitation of POT1 and RPA titrations. Each band was analyzed and quantified with a PhosphorImager Storm 860 instrument (Molecular Dynamics). The data from two independent experiments are shown. POT1 binding is represented by triangles connected by a dashed line, and RPA binding is represented by circles connected by a solid line.
Discussion
The original experiments suggesting a role for WRN in telomere lagging replication were carried out in primary telomerase-negative human cells or in tumor cells in which telomerase activity had been abolished (Crabbe et al. 2004). In those experiments, WRN dysfunction led to a limited increase in STL (defined by a qualitative criterion, the presence or absence of a probe signal) exclusively affecting the sister chromatid replicated by lagging mechanisms. This defect was fully corrected in telomerase-expressing cells (Crabbe et al. 2004). In the present study, the Q-CO-FISH technique allowed us to study WRN deficiency in telomerase-positive cells, this time using a quantitative criterion: the length of parental telomeric G and C strands after replication. Our data conclusively show that, in the absence of WRN, most, if not all, replicating G strands are shortened. While our experiments confirm that C strand replication is unaffected in the absence of WRN, they clearly indicate that the involvement of this protein during G strand replication is more systematic than thought previously. On the other hand, very little of WRN may be required to carry out this task, as suggested by the observation (data not shown) that cells with partial knockdown of the WRN gene allowing an expression level of at least 10% of the protein display normal telomere replication.
Outstandingly, our results indicate that, in the absence of WRN, the shelterin protein hPOT1 plays a totally unanticipated role, facilitating the progression of the replicative helicases and allowing DNA polymerization on the leading strand (Fig. 7A,B). The POT1 forced expression experiments showed that the two OB fold domains of POT1 are sufficient to ensure this role (Fig. 7C,D). Since the domains that mediate protein–protein interactions with other shelterin components have been removed from this peptide, this experiment strongly suggests that binding to the telomeric single G strands is involved. In agreement with this hypothesis, our in vitro experiments using purified proteins indicate for the first time that POT1 has a higher affinity for its specific substrate than RPA, whose ssDNA-binding activity is nonspecific (Alani et al. 1992). This in vitro system, in which other shelterin components are absent, reflects the activity of the two OB fold domains included in the construction that we used in cells depleted for WRN and shown to rescue C strand replication. However, it is possible that high affinity for its substrate is not sufficient to render POT1 proficient for this particular role, as suggested by the failure to restore leading G strand replication upon forced expression of fl POT1—a failure that may also be explained by down-regulation of the endogenous POT1. In fact, POT1 may need to be enriched at the telomeric replication fork, perhaps through its interaction with TRF2 (Barrientos et al. 2008).
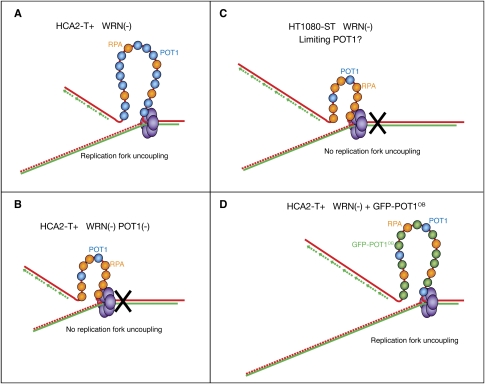
Figure 7.
Model for replication fork uncoupling upon WRN loss. (A) In normal fibroblasts (HCA2-T+), the single G strands that accumulate during replication are bound by POT1, thus preventing accumulation of RPA and fork stalling. This accumulation therefore enables polymerization to continue on the leading strands up to the end of the chromosome. (B) When combined with POT1 deficiency, WRN depletion in HCA2-T+ cells leads to RPA accumulation on single G strands, thus inducing replication fork stall. (C) In ST cells, levels of POT1 are limiting, but enough to ensure normal telomere replication. After WRN knockdown, there is a deficit of POT1 molecules so that RPA accumulates on single G strands, thus preventing replication fork uncoupling, leading to both G-rich and C-rich strand shortening. (D) Forced expression of the OB fold domains of POT1 prevents RPA from binding to single G strands, thereby restoring the replication fork uncoupling mechanism, which allows complete C strand replication.
Our experiments also indicate that single G strands transiently accumulate during normal (i.e., in the presence of WRN) S phase, while single C strands are barely detectable. Whether or not POT1 binds to these replicative single G strands remains to be determined. In fact, our in vitro experiments using purified hPOT1 and RPA leave open the possibility that POT1, together or in competition with RPA, binds to this strand during normal replication—perhaps, as suggested above, through its interactions with TRF2 (Barrientos et al. 2008). Importantly, WRN also interacts with TRF2 (Opresko et al. 2002), and thus both WRN and POT1 may be together near the replication fork. This colocalization may have functional implications, since it has been shown both that POT1 and WRN interact in vitro, and that the presence of POT1 stimulates WRN unwinding activity on substrates believed to form on the lagging G strand or believed to be present at the base of the T-loop (Opresko et al. 2005; Sowd et al. 2008). Therefore, binding of POT1 to the replicative single G strand might facilitate WRN's role, even during normal telomere replication.
Our observation that WRN is required in a systematic way for telomere replication begs the question about the type of lesion encountered by the fork and for which WRN's activities are so essential. Importantly, the experiments in which GFP-POT1-OB1+2 forced expression totally rescued replication of the leading but not the lagging G strand strongly suggest that the obstacle encountered by the replication fork in cells with very long telomeres and depleted WRN is of a similar nature to the one arising in cells with standard telomere length. One possibility, already suggested (Crabbe et al. 2004), is the formation of G quadruplexes by the lagging G strand, although such structures so far have been detected only in lower eukaryotes (Paeschke et al. 2005). However, both POT1 and RPA have been shown to prevent G quadruplex formation in vitro (Zaug et al. 2005; Salas et al. 2006), and perhaps these activities are present during telomere replication in vivo whenever WRN is absent. On the other hand, formation of G quadruplex structures by the lagging G strand may also be counteracted by other helicases such as FANCJ (Wu et al. 2008), RTEL1 (Ding et al. 2004; Sfeir et al. 2009), or BLM (Li et al. 2001; Mohaghegh et al. 2001). Interestingly, BLM has also been shown to interact with TRF2, although its biological role at telomeres in normal cells needs to be explored further (Opresko et al. 2002; Lillard-Wetherell et al. 2004). Therefore, other lesions or secondary structures, different from G quadruplexes, may be responsible for blocking G strand polymerization. Among these structures, the D-loop at the base of the t-loop could prevent progression of the replication fork in the absence of WRN. However, given the current model for t-loop formation in which the G overhang displaces the G strand to pair with the C strand (de Lange 2004), the persistence of such structures would be expected to completely block the progression of the fork and not just the polymerization on the lagging strand.
Finally, in cells with very long telomeres and depleted for WRN, we detected a significant increase in the phosphorylation of ATM in G2. This is compatible with the onset of a local DNA damage response perhaps triggered by the persistence of unreplicated chromosome ends that must be processed. It may also correspond to an exaggeration of the normal damage response detected during G2 at telomeres and believed to participate in the remodeling and protection of just replicated telomeres (Verdun and Karlseder 2006, 2007). Whatever the case, we were unable to detect any sign of replication stress in spite of a robust block of telomere replication. It is possible that replication stress signals arising from single telomeres (which, in humans, replicate at specific times all along the S phase) (N Arnoult, C Schluth, A Letessier, I Draskovic, R Bouarich, J Campisi, S-h Kim, B Dutrillaux, A Ottaviani, F Magdinier, et al., in prep.) are both transient and of low intensity and therefore difficult to detect. Another possibility is that cells, in the absence of converging replication forks at telomeres able to rescue eventual blocks, have developed mechanisms to dampen any strong signal taking place at damaged telomeric forks. As long as telomere-capping functions are not compromised, such a strategy would guarantee unperturbed S-phase progression. In the case of WRN deficiency, it is patent that uncoupling of polymerization on lagging and leading G strands is beneficial, since at least one sister chromatid will have its telomere completely replicated.
In addition, a recent study in the fission yeast suggests that uncoupling of DNA synthesis at telomeres also occurs under normal conditions (Moser et al. 2009). In that report, kinetics studies of polymerase recruitment at telomeres using ChIP showed that polymerization on the leading strand largely precedes polymerization on the lagging strand, with accumulation of RPA on the latter before POT1 is recruited. Interestingly, recruitment of RPA leads to recruitment of checkpoint proteins RAD3–RAD26 without perturbing S-phase progression, indicating again that signaling at telomeres is somehow diminished. Clearly, more studies are needed to unveil all the idiosyncrasies of replication mechanisms at work in these particular regions of the genome.
Materials and methods
Cell culture and synchronization
HCA2-T+ cells were human foreskin fibroblast HCA2-immortalized by transduction of a pBABE-derived retrovirus carrying hTERT. Both parental and derived cells were obtained from J. Campisi (Lawrence Berkeley Laboratories). The HT1080 human lung sarcoma cell line and its hTERT/hTERC derivative were a generous gift from G. Cristofari and J. Lingner (Cristofari and Lingner 2006). HCA2-T+ and HT1080ST were grown in IMDM (Gibco) supplemented with, respectively, 20% or 10% of heat-inactivated fetal calf serum, at 37°C with 5% CO2. All cell lines were synchronized by a double thymidin/aphidicholin block. Details are provided in the Supplemental Material.
siRNA and plasmid transfections
Cells were transfected with siRNAs and plasmids using Lipofectamine 2000 (Invitrogen) according to the instructions of the manufacturer. Details and siRNA sequences are provided in the Supplemental Material.
The plasmids used were pEGFP-C1 (Clontech) for GFP control plasmid. The pEGFP-C1∷POT1 plasmid has been described in Gomez et al. (2006). The construction of pEGFP-C1∷OB1+2 POT1 + NLS of SV40 is described in the Supplemental Material.
Metaphase preparations and CO-FISH
After release from the aphidicolin block, cells were incubated with 10 μM BrdU and 3.3 μM BrdC and, 9 h after release from the aphidicolin block, cells were arrested in mitosis with 1.5-h incubation in colcemid (0.1 μg/mL) before 40 min of hypotonic shock in 0.8 g/L sodium citrate at 37°C, and were fixed in ethanol/acetic acid. Metaphase spreads were obtained by dropping suspensions of fixed cells onto clean glass slides and were rapidly used for hybridization.
The CO-FISH procedure was carried out as described previously (Londoño-Vallejo et al. 2004; Arnoult et al. 2008) with a few modifications (see Supplemental Material). The quantification of telomeric signals after hybridization was carried out essentially as described before (Londoño-Vallejo et al. 2001; Arnoult et al. 2008). The data are represented either as a mean intensity of all metaphases or the distribution of at least 2000 intensity values. Statistical analyses were done using the Wilcoxon rank-sum test.
Telomeric single-strand assay
The nondenaturing hybridization assay to detect the replicative single G strand was carried out as described previously with some modifications (Gomez et al. 2004). Transfected and synchronized cells were collected in G1, S, and G2, according to cell cycle analysis. DNA was extracted using Nucleospin Tissue kit (Macherey Nagel) and was quantified using nanodrop. Three micrograms of DNA were incubated overnight at 50°C with 0.5 pmol of radiolabeled telomeric C-rich [(CCCTAA)6] or G-rich [(TTAGGG)6] probe in hybridization buffer (50 mM NaCl, 10 mM Tris HCl at pH 8, 1 mM EDTA at pH 8.0). The free probe was separated from bulk DNA by electrophoresis on a 0.8% agarose gel containing 0.01% ethidium bromide for 2 h at 80 V. The gel was dried at 50°C, washed in water, dried again a few minutes, and exposed. Radioactive signals were detected in a PhosphorImager apparatus, quantified, and normalized with regard to total DNA in that sample given by the ethidium bromide signals.
POT1/RPA-binding assays
Purified recombinant hPOT1, prepared after baculovirus expression, was a generous gift from Dr. D. Gomez (Institut de Pharmacologie et de Biologie Structurale, Toulouse, France). Recombinant hRPA was provided by Dr. Lavrik (Institute of Chemical Biology and Fundamental Medicine, Russia) and purified, as a trimer, as described previously (Salas et al. 2006).
The oligonucleotide used in the study was 21G (5′-GGGTTAGGGTTAGGGTTAGGG-3′) and was labeled with [γ-32P]ATP using T4 polynucleotide kinase. 32P-labeled 21G was purified using denaturing 15% polyacrylamide gel electrophoresis (PAGE). hRPA and hPOT1 were diluted and preincubated (10 min at 4°C) in buffer containing 50 mM Tris-HCl (pH 7.5), 100 mM KCl, 1 mM DTT, 10% glycerol, 0.2 mg/mL BSA, and 0.1 mM EDTA for hRPA, and 20% glycerol for hPOT1. The radioactively labeled 21G (20 nM) was mixed with various amounts of protein in 10 μL of reaction buffer (50 mM Hepes at pH 7.9, 0.1 mg/mL BSA, 2% glycerol, 100 mM KCl, 4% sucrose, 0.02% Bromophenol Blue, 0.1 mM EDTA). Protein:21G-binding reactions were conducted for 10 min at 20°C. Individual reaction mixtures were loaded on 1% agarose gel in 0.5× TBE and migrated for 45 min at room temperature at 7 V/cm. The gels were dried and analyzed with a PhosphorImager Storm 860 instrument (Molecular Dynamics). Data were analyzed using ImageQuant software (Amersham), and results were expressed as the fraction of bound oligonucleotide relative to total oligonucleotide as a function of the protein concentration. Values correspond to the mean value of two independent experiments.
Acknowledgments
We thank Gaël Cristofari and Joachim Lingner for providing ST cells, Judy Campisi for providing HCA2 cells, Dennis Gomez for providing purified hPOT1, and Olga Lavrik for purified hRPA. We gratefully acknowledge Zofia Maciorowski and Annick Viguier for help with cell sorting, the Nikon Imaging Center of Institut Curie for help with deconvolution microscopy, Céline Douarre for technical advice on single-strand assay, Irena Draskovic for help with image treatment, and Silvia Bacchetti for comments on the manuscript. We thank all the members of the Telomere and Cancer Laboratory for helpful discussions. This work was supported by the ANR (grant NT05-4_41705) and ARC (to A.L.-V.). N.A. is the recipient of a PhD fellowship from MENRST and ARC.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.544009.
Supplemental material is available at http://www.genesdev.org.
References
- Alani E, Thresher R, Griffith JD, Kolodner RD. Characterization of DNA-binding and strand-exchange stimulation properties of y-RPA, a yeast single-strand-DNA-binding protein. J Mol Biol. 1992;227:54–71. doi: 10.1016/0022-2836(92)90681-9. [DOI] [PubMed] [Google Scholar]
- Arnoult N, Shin-Ya K, Londoño-Vallejo JA. Studying telomere replication by Q-CO-FISH: The effect of telomestatin, a potent G-quadruplex ligand. Cytogenet Genome Res. 2008;122:229–236. doi: 10.1159/000167808. [DOI] [PubMed] [Google Scholar]
- Bae SH, Bae KH, Kim JA, Seo YS. RPA governs endonuclease switching during processing of Okazaki fragments in eukaryotes. Nature. 2001;412:456–461. doi: 10.1038/35086609. [DOI] [PubMed] [Google Scholar]
- Baird DM, Davis T, Rowson J, Jones CJ, Kipling D. Normal telomere erosion rates at the single cell level in Werner syndrome fibroblast cells. Hum Mol Genet. 2004;13:1515–1524. doi: 10.1093/hmg/ddh159. [DOI] [PubMed] [Google Scholar]
- Barrientos KS, Kendellen MF, Freibaum BD, Armbruster BN, Etheridge KT, Counter CM. Distinct functions of POT1 at telomeres. Mol Cell Biol. 2008;28:5251–5264. doi: 10.1128/MCB.00048-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann P, Cech TR. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science. 2001;292:1171–1175. doi: 10.1126/science.1060036. [DOI] [PubMed] [Google Scholar]
- Crabbe L, Verdun RE, Haggblom CI, Karlseder J. Defective telomere lagging strand synthesis in cells lacking WRN helicase activity. Science. 2004;306:1951–1953. doi: 10.1126/science.1103619. [DOI] [PubMed] [Google Scholar]
- Cristofari G, Lingner J. Telomere length homeostasis requires that telomerase levels are limiting. EMBO J. 2006;25:565–574. doi: 10.1038/sj.emboj.7600952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dart DA, Adams KE, Akerman I, Lakin ND. Recruitment of the cell cycle checkpoint kinase ATR to chromatin during S phase. J Biol Chem. 2004;279:16433–16440. doi: 10.1074/jbc.M314212200. [DOI] [PubMed] [Google Scholar]
- de Lange T. T-loops and the origin of telomeres. Nat Rev Mol Cell Biol. 2004;5:323–329. doi: 10.1038/nrm1359. [DOI] [PubMed] [Google Scholar]
- de Lange T. Shelterin: The protein complex that shapes and safeguards human telomeres. Genes & Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- Denchi EL, de Lange T. Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature. 2007;448:1068–1071. doi: 10.1038/nature06065. [DOI] [PubMed] [Google Scholar]
- Ding H, Schertzer M, Wu X, Gertsenstein M, Selig S, Kammori M, Pourvali R, Poon S, Vulto I, Chavez E, et al. Regulation of murine telomere length by Rtel: An essential gene encoding a helicase-like protein. Cell. 2004;117:873–886. doi: 10.1016/j.cell.2004.05.026. [DOI] [PubMed] [Google Scholar]
- Gilson E, Geli V. How telomeres are replicated. Nat Rev Mol Cell Biol. 2007;8:825–838. doi: 10.1038/nrm2259. [DOI] [PubMed] [Google Scholar]
- Gomez D, Paterski R, Lemarteleur T, Shin-Ya K, Mergny JL, Riou JF. Interaction of telomestatin with the telomeric single-strand overhang. J Biol Chem. 2004;279:41487–41494. doi: 10.1074/jbc.M406123200. [DOI] [PubMed] [Google Scholar]
- Gomez D, O'Donohue MF, Wenner T, Douarre C, Macadre J, Koebel P, Giraud-Panis MJ, Kaplan H, Kolkes A, Shin-ya K, et al. The G-quadruplex ligand telomestatin inhibits POT1 binding to telomeric sequences in vitro and induces GFP-POT1 dissociation from telomeres in human cells. Cancer Res. 2006;66:6908–6912. doi: 10.1158/0008-5472.CAN-06-1581. [DOI] [PubMed] [Google Scholar]
- Hamdan SM, Loparo JJ, Takahashi M, Richardson CC, van Oijen AM. Dynamics of DNA replication loops reveal temporal control of lagging-strand synthesis. Nature. 2009;457:336–339. doi: 10.1038/nature07512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H, Multani AS, Cosme-Blanco W, Tahara H, Ma J, Pathak S, Deng Y, Chang S. POT1b protects telomeres from end-to-end chromosomal fusions and aberrant homologous recombination. EMBO J. 2006;25:5180–5190. doi: 10.1038/sj.emboj.7601294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D, Sfeir AJ, Shay JW, Wright WE, de Lange T. POT1 protects telomeres from a transient DNA damage response and determines how human chromosomes end. EMBO J. 2005;24:2667–2678. doi: 10.1038/sj.emboj.7600733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlseder J. Telomeric proteins: Clearing the way for the replication fork. Nat Struct Mol Biol. 2006;13:386–387. doi: 10.1038/nsmb0506-386. [DOI] [PubMed] [Google Scholar]
- Lei M, Podell ER, Cech TR. Structure of human POT1 bound to telomeric single-stranded DNA provides a model for chromosome end-protection. Nat Struct Mol Biol. 2004;11:1223–1229. doi: 10.1038/nsmb867. [DOI] [PubMed] [Google Scholar]
- Li JL, Harrison RJ, Reszka AP, Brosh RM, Jr, Bohr VA, Neidle S, Hickson ID. Inhibition of the Bloom's and Werner's syndrome helicases by G-quadruplex interacting ligands. Biochemistry. 2001;40:15194–15202. doi: 10.1021/bi011067h. [DOI] [PubMed] [Google Scholar]
- Lillard-Wetherell K, Machwe A, Langland GT, Combs KA, Behbehani GK, Schonberg SA, German J, Turchi JJ, Orren DK, Groden J. Association and regulation of the BLM helicase by the telomere proteins TRF1 and TRF2. Hum Mol Genet. 2004;13:1919–1932. doi: 10.1093/hmg/ddh193. [DOI] [PubMed] [Google Scholar]
- Liu D, O'Connor MS, Qin J, Songyang Z. Telosome, a mammalian telomere-associated complex formed by multiple telomeric proteins. J Biol Chem. 2004;279:51338–51342. doi: 10.1074/jbc.M409293200. [DOI] [PubMed] [Google Scholar]
- Loayza D, de Lange T. POT1 as a terminal transducer of TRF1 telomere length control. Nature. 2003;423:1013–1018. doi: 10.1038/nature01688. [DOI] [PubMed] [Google Scholar]
- Londoño-Vallejo JA, DerSarkissian H, Cazes L, Thomas G. Differences in telomere length between homologous chromosomes in humans. Nucleic Acids Res. 2001;29:3164–3171. doi: 10.1093/nar/29.15.3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Londoño-Vallejo JA, Der-Sarkissian H, Cazes L, Bacchetti S, Reddel R. Alternative lengthening of telomeres is characterized by high rates of inter-telomeric exchange. Cancer Res. 2004;64:2324–2327. doi: 10.1158/0008-5472.can-03-4035. [DOI] [PubMed] [Google Scholar]
- Mohaghegh P, Karow JK, Brosh Jr RM, Jr, Bohr VA, Hickson ID. The Bloom's and Werner's syndrome proteins are DNA structure-specific helicases. Nucleic Acids Res. 2001;29:2843–2849. doi: 10.1093/nar/29.13.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser BA, Subramanian L, Chang YT, Noguchi C, Noguchi E, Nakamura TM. Differential arrival of leading and lagging strand DNA polymerases at fission yeast telomeres. EMBO J. 2009;28:810–820. doi: 10.1038/emboj.2009.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Multani AS, Chang S. WRN at telomeres: Implications for aging and cancer. J Cell Sci. 2007;120:713–721. doi: 10.1242/jcs.03397. [DOI] [PubMed] [Google Scholar]
- Nitani N, Yadani C, Yabuuchi H, Masukata H, Nakagawa T. Mcm4 C-terminal domain of MCM helicase prevents excessive formation of single-stranded DNA at stalled replication forks. Proc Natl Acad Sci. 2008;105:12973–12978. doi: 10.1073/pnas.0805307105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opresko PL, von Kobbe C, Laine JP, Harrigan J, Hickson ID, Bohr VA. Telomere-binding protein TRF2 binds to and stimulates the Werner and Bloom syndrome helicases. J Biol Chem. 2002;277:41110–41119. doi: 10.1074/jbc.M205396200. [DOI] [PubMed] [Google Scholar]
- Opresko PL, Otterlei M, Graakjaer J, Bruheim P, Dawut L, Kolvraa S, May A, Seidman MM, Bohr VA. The Werner Syndrome helicase and exonuclease cooperate to resolve telomeric D loops in a manner regulated by TRF1 and TRF2. Mol Cell. 2004;14:763–774. doi: 10.1016/j.molcel.2004.05.023. [DOI] [PubMed] [Google Scholar]
- Opresko PL, Mason PA, Podell ER, Lei M, Hickson ID, Cech TR, Bohr VA. POT1 stimulates RecQ helicases WRN and BLM to unwind telomeric DNA substrates. J Biol Chem. 2005;280:32069–32080. doi: 10.1074/jbc.M505211200. [DOI] [PubMed] [Google Scholar]
- Opresko PL, Sowd G, Wang H. The Werner syndrome helicase/exonuclease processes mobile D-loops through branch migration and degradation. PLoS One. 2009;4:e4825. doi: 10.1371/journal.pone.0004825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paeschke K, Simonsson T, Postberg J, Rhodes D, Lipps HJ. Telomere end-binding proteins control the formation of G-quadruplex DNA structures in vivo. Nat Struct Mol Biol. 2005;12:847–854. doi: 10.1038/nsmb982. [DOI] [PubMed] [Google Scholar]
- Raderschall E, Golub EI, Haaf T. Nuclear foci of mammalian recombination proteins are located at single-stranded DNA regions formed after DNA damage. Proc Natl Acad Sci. 1999;96:1921–1926. doi: 10.1073/pnas.96.5.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robison JG, Elliott J, Dixon K, Oakley GG. Replication protein A and the Mre11.Rad50.Nbs1 complex co-localize and interact at sites of stalled replication forks. J Biol Chem. 2004;279:34802–34810. doi: 10.1074/jbc.M404750200. [DOI] [PubMed] [Google Scholar]
- Salas TR, Petruseva I, Lavrik O, Bourdoncle A, Mergny JL, Favre A, Saintome C. Human replication protein A unfolds telomeric G-quadruplexes. Nucleic Acids Res. 2006;34:4857–4865. doi: 10.1093/nar/gkl564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sfeir A, Kosiyatrakul ST, Hockemeyer D, MacRae SL, Karlseder J, Schildkraut CL, de Lange T. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009;138:90–103. doi: 10.1016/j.cell.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowd G, Lei M, Opresko PL. Mechanism and substrate specificity of telomeric protein POT1 stimulation of the Werner syndrome helicase. Nucleic Acids Res. 2008;36:4242–4256. doi: 10.1093/nar/gkn385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldman T, Etheridge KT, Counter CM. Loss of hPot1 function leads to telomere instability and a cut-like phenotype. Curr Biol. 2004;14:2264–2270. doi: 10.1016/j.cub.2004.12.031. [DOI] [PubMed] [Google Scholar]
- Verdun RE, Karlseder J. The DNA damage machinery and homologous recombination pathway act consecutively to protect human telomeres. Cell. 2006;127:709–720. doi: 10.1016/j.cell.2006.09.034. [DOI] [PubMed] [Google Scholar]
- Verdun RE, Karlseder J. Replication and protection of telomeres. Nature. 2007;447:924–931. doi: 10.1038/nature05976. [DOI] [PubMed] [Google Scholar]
- Wang F, Podell ER, Zaug AJ, Yang Y, Baciu P, Cech TR, Lei M. The POT1–TPP1 telomere complex is a telomerase processivity factor. Nature. 2007;445:506–510. doi: 10.1038/nature05454. [DOI] [PubMed] [Google Scholar]
- Wold MS. Replication protein A: A heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu Rev Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- Wu L, Multani AS, He H, Cosme-Blanco W, Deng Y, Deng JM, Bachilo O, Pathak S, Tahara H, Bailey SM, et al. Pot1 deficiency initiates DNA damage checkpoint activation and aberrant homologous recombination at telomeres. Cell. 2006;126:49–62. doi: 10.1016/j.cell.2006.05.037. [DOI] [PubMed] [Google Scholar]
- Wu Y, Shin-ya K, Brosh RM., Jr FANCJ helicase defective in Fanconia anemia and breast cancer unwinds G-quadruplex DNA to defend genomic stability. Mol Cell Biol. 2008;28:4116–4128. doi: 10.1128/MCB.02210-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin H, Liu D, Wan M, Safari A, Kim H, Sun W, O'Connor MS, Songyang Z. TPP1 is a homologue of ciliate TEBP-β and interacts with POT1 to recruit telomerase. Nature. 2007;445:559–562. doi: 10.1038/nature05469. [DOI] [PubMed] [Google Scholar]
- Zaug AJ, Podell ER, Cech TR. Human POT1 disrupts telomeric G-quadruplexes allowing telomerase extension in vitro. Proc Natl Acad Sci. 2005;102:10864–10869. doi: 10.1073/pnas.0504744102. [DOI] [PMC free article] [PubMed] [Google Scholar]