Abstract
The histone acetyltransferase complex SAGA is well characterized as a coactivator complex in yeast. In this study of Drosophila SAGA (dSAGA), we describe three novel components that include an ortholog of Spt20, a potential ortholog of Sgf73/ATXN7, and a novel histone fold protein, SAF6 (SAGA factor-like TAF6). SAF6, which binds directly to TAF9, functions analogously in dSAGA to TAF6/TAF6L in the yeast and human SAGA complexes, respectively. Moreover, TAF6 in flies is restricted to TFIID. Mutations in saf6 disrupt SAGA-regulated gene expression without disrupting acetylated or ubiquitinated histone levels. Thus, SAF6 is essential for SAGA coactivator function independent of the enzymatic activities of the complex.
Keywords: Histone acetylation, SAGA, Gcn5, PCAF, TAF, histone fold
In Drosophila melanogaster, the histone acetyltransferase (HAT) Gcn5 is the catalytic subunit of two separate high-molecular-weight complexes: ATAC and SAGA (Kusch et al. 2003; Muratoglu et al. 2003). Studies in Saccharomyces cerevisiae have shown that SAGA also has ubiquitin protease activity specific for monoubiquitinated histone H2B (ubH2B) (Henry et al. 2003). In addition to its enzymatic activities, SAGA has transcription coactivator activities mediated through its interactions with transcription activators and the TATA-binding protein (TBP) (Baker and Grant 2007). We and others have shown previously that Drosophila SAGA (dSAGA) includes the orthologs of most components of the yeast SAGA (ySAGA) complex (Supplemental Table S1; Kusch et al. 2003; Muratoglu et al. 2003; Guelman et al. 2006; Kurshakova et al. 2007; Weake et al. 2008). In addition, dSAGA contains subunits that are unique to the fly complex, such as the WD repeat-containing protein WDA (Guelman et al. 2006).
SAGA is essential for development in multicellular organisms, and Gcn5 is required for viability in both mice and Drosophila (Xu et al. 2000; Carre et al. 2005). Furthermore, mutations that disrupt the HAT activity of dSAGA, such as ada2b and wda, result in lethality in flies (Qi et al. 2004; Pankotai et al. 2005; Guelman et al. 2006). Moreover, mutations that specifically affect the ubiquitin protease activity of dSAGA are lethal, and result in defects in axon targeting in the larval eye–brain complex (Weake et al. 2008). To characterize dSAGA more fully, we sought to identify orthologs of all ySAGA components, as there are subunits present in the ySAGA and human SAGA complexes for which orthologs have not yet been identified in flies (Rodriguez-Navarro 2009). In addition, the apparent orthologs of some ySAGA subunits do not appear to function analogously in the Drosophila complex. Specifically, it has been unclear from our previous studies whether TAF6, which is a shared subunit of both the SAGA and TFIID coactivator complexes in yeast, is in fact a subunit of dSAGA (Kusch et al. 2003; Guelman et al. 2006). In the human SAGA complexes, TAF6 is replaced by TAF6L (PAF65α) (Ogryzko et al. 1998; Martinez et al. 2001; Nagy et al. 2009). However, the Drosophila homolog of TAF6L is encoded by a gene that is expressed exclusively in primary spermatocytes (Hiller et al. 2004). Thus, it is unlikely that this testis-specific TAF6L is a subunit of SAGA in the majority of Drosophila cell types.
To further characterize the subunit composition and function of dSAGA, we used affinity purification and MudPIT (multidimensional protein identification technology) analysis. These efforts revealed proteins in SAGA, including the products of genes CG17689, CG9866, and CG3883. Sequence analysis shows that CG17689 encodes an ortholog of Spt20/p38IP, and CG9866 encodes a potential ortholog of Sgf73. The third subunit, encoded by an uncharacterized gene, CG3883, is a novel histone fold domain (HFD)-containing protein that we named SAF6 (SAGA factor-like TAF6). We show in this study that SAF6 is homologous but not orthologous to TAF6, and can functionally substitute for TAF6 within the majority of dSAGA complexes, binding directly to the HFD of TAF9. In contrast, TAF6 in flies is restricted to TFIID. Hence, SAF6 provides a means by which the functions of the TAF octamer in SAGA can be examined, independent of TFIID. We show that TAF function within SAGA, as examined through SAF6, is critical for Drosophila development, as saf6 mutant animals die as second instar larva. However, loss of SAF6 did not affect global levels of acetylated or ubiquitinated histones, and is therefore unlikely to affect the integrity or enzymatic activities of the dSAGA complex. Instead, we show that SAF6 is essential for the coactivator function of SAGA in regulating gene expression, independent of the catalytic activities of the complex.
Results and Discussion
Drosophila SAGA contains three previously unidentified polypeptides
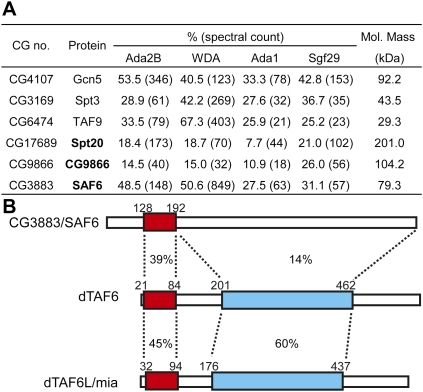
Orthologs of all subunits identified in the well-characterized ySAGA complex have not yet been identified in Drosophila. We sought to identify additional components of dSAGA, and to characterize subunits involved in formation of the TAF octamer complex within SAGA (Selleck et al. 2001). To identify candidates for dSAGA subunits, we isolated SAGA using tandem Flag-HA affinity purification from S2 cell nuclear extracts with the SAGA-specific subunits Ada2B, WDA, and Ada1, and the shared ATAC/SAGA subunit Sgf29 as bait proteins. The composition of affinity-purified SAGA was determined by MudPIT (Florens and Washburn 2006). Peptides from three novel proteins were consistently identified in affinity-purified SAGA: CG17689, CG9866, and CG3883 (Fig. 1A). These three polypeptides were present at levels similar to those of the SAGA subunits Gcn5, Spt3, and TAF9 (Figs. 1A, 2A), and were not found in control purifications from cells expressing a nonspecific tagged bait protein, or in samples from cells lacking a tagged protein (Supplemental Table S2). Moreover, none of these polypeptides were identified in purifications of the Gcn5-containing ATAC complex (Suganuma et al. 2008).
Figure 1.
(A) Peptides from CG17689, CG9866, and CG3883 are identified in SAGA purifications from S2 cells using Ada2B, WDA, Ada1, and Sgf29 as bait proteins. Sequence coverage (percentage) and number of peptides (spectral count) are shown for each polypeptide, relative to the characterized SAGA subunits Gcn5, Spt3, and TAF9. (B) Bar diagram comparison of Drosophila CG3883/SAF6, TAF6, and TAF6L. The percentage of consensus residues conserved between the HFDs (red box) and TAF6 domains (blue box) of TAF6L and SAF6 relative to TAF6 are shown between the dotted lines. The amino acid positions of the domains are indicated above the boxes.
Figure 2.
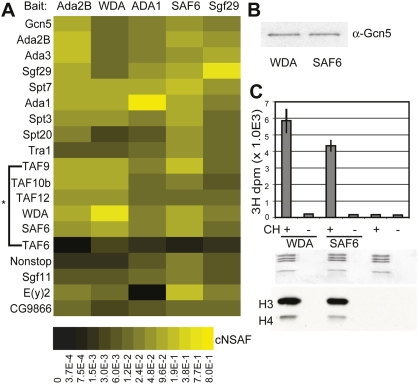
(A) Heat map showing the relative spectral abundance of SAGA subunits expressed as cNSAF. These calculations include the values for TAF6 in purifications using Ada2B, WDA, ADA1, SAF6, or Sgf29 as bait proteins. The cNSAF scale is shown at the bottom of A, with the highest abundance subunits represented in yellow, and absent or underrepresented subunits in black. cNSAF values used to generate the heat map are provided in Supplemental Table S2. (*) P-value = 0.018 as calculated by Student's t-test. (B) Western blot of SAF6- or WDA-purified SAGA probed with anti-Gcn5. (C) The HAT activity of Flag-purified WDA and SAF6 SAGA complexes was assayed using core histones as substrate. Incorporation of 3H-acetyl CoA was assayed by scintillation counting (top panel) and fluorography (bottom panel), and the migrations of histones H3 and H4 were determined by Coomassie staining (middle panel).
We then asked whether these polypeptides had similarity to any subunits of the ySAGA complex. The uncharacterized CG17689 gene (FBgn0036374) encodes a 1873-amino-acid polypeptide with a predicted molecular mass of 201 kDa. Psi-BLAST searches revealed that CG17689 shares specific sequence similarity with SPT20 (p38IP) in mammals, which has been identified recently as a subunit of mammalian SAGA (Nagy et al. 2009). Thus, CG17689 will be referred to hereafter as Spt20. The uncharacterized CG9866 (FBgn0031420) gene encodes a 971-amino-acid polypeptide with a predicted molecular mass of 104.2 kDa. Iterative Psi-BLAST searching identified weak similarity between the N-terminal region of CG9866 and the first nonglobular Sgf11-related domain of S. cerevisiae Sgf73 and human ATXN7. However, in contrast to Sgf73 and ATAXN7, CG9866 does not contain a recognizable SCA7 domain. Although it shares one homologous domain with Sgf73/ATXN7, it is unclear whether CG9866 also regulates the histone deubiquitination activity of SAGA-like yeast Sgf73 (Kohler et al. 2008; Lee et al. 2009). For this reason, we refrained from naming CG9866 until functional similarity between this protein and Sgf73 can be established.
The third novel polypeptide identified in affinity-purified SAGA, CG3883 (FBgn0031281), has a predicted molecular mass of 79.3 kDa, and shares no discernable overall similarity with any of the known ySAGA subunits. However, closer examination of CG3883 using sequence similarity searches revealed the presence of a HFD with significant similarity to the H4-like HFDs that are confined to the N termini of TAF6 and TAF6L (Fig. 1B; Supplemental Fig. S1). However, there is no significant sequence conservation between CG3883 and TAF6 or TAF6L outside of this region (Fig. 1B). Due to the similarity between the HFDs of CG3883 and TAF6, we named CG3883 SAF6.
SAF6 is a subunit of dSAGA
To confirm that SAF6 is a bona fide subunit of dSAGA, we purified tagged SAF6 from S2 cell nuclear extract by tandem Flag-HA affinity chromatography. MudPIT analysis of affinity-purified SAF6 revealed the presence of all known subunits of SAGA (Fig. 2A; Supplemental Table S2). In contrast, no peptides for ATAC-specific subunits were identified. We then asked whether SAF6-purified SAGA had comparable HAT activity to SAGA purified through other SAGA-specific subunits, such as WDA (Guelman et al. 2006). HAT assays were performed using Gcn5-normalized SAF6- and WDA-purified SAGA complexes (Fig. 2B) using HeLa core histones as substrate. SAF6-purified SAGA exhibited a similar level of HAT activity and histone preference to WDA-purified SAGA (Fig. 2C; Guelman et al. 2006). While these studies were conducted in S2 cells, the broad temporal expression of SAF6 suggests that it is a primary component of SAGA in flies (Supplemental Fig. S2). We concluded from these results that SAF6 is indeed a bona fide subunit of dSAGA.
SAF6 replaces TAF6 in dSAGA
Due to the similarity between the HFDs of SAF6 and TAF6, we wondered whether SAF6 might be present instead of TAF6 within dSAGA complexes. Studies in S. cerevisiae have shown that TAF6, TAF9, TAF12, and Ada1 within ySAGA combine to form a TAF octamer complex with marked similarities to the histone octamer (Selleck et al. 2001; Tora 2002). Peptides corresponding to orthologs of TAF9, TAF12, and Ada1 were consistently identified with a high percent of sequence coverage in purifications of dSAGA (Fig. 2A; Supplemental Table S2). However, TAF6 is consistently underrepresented or absent from these same purifications. When comparing cNSAF (complex-specific spectral abundance factor) (Florens et al. 2006) values for TAF6 and its presumed binding partner, TAF9, across five different SAGA purifications, TAF6 detection is significantly lower than TAF9 (P-value = 0.018). TAF6 cNSAF values are also significantly lower than the mean SAGA subunit cNSAF value (P-value = 0.017).
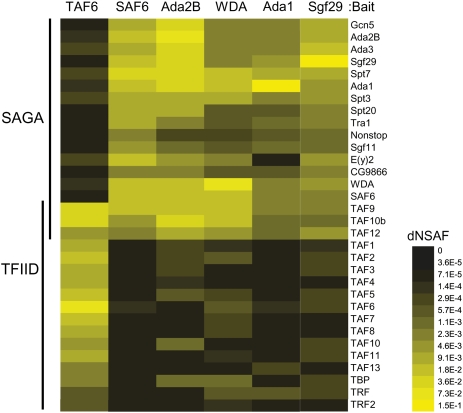
Only one SAGA-specific subunit, WDA, consistently copurifies more than a few peptides from TAF6 (Supplemental Table S2). The higher coverage of TAF6 peptides in WDA-purified SAGA raised the possibility that a subset of SAGA complexes might contain TAF6 in addition to, or instead of, SAF6. Thus, to determine whether TAF6 is a bona fide subunit in any fraction of dSAGA complexes, we purified tagged TAF6 from S2 cell nuclear extract by tandem Flag-HA affinity chromatography and analyzed the resulting complex. TAF6 is a known subunit of TFIID and, as expected, high numbers of peptide spectra corresponding to the other TAF components of TFIID were identified in the TAF6 purification (Fig. 3). However, very low levels of SAGA-specific subunits were identified in the TAF6 purification (Fig. 3; Supplemental Table S2). A comparison of the dNSAF (distributive normalized SAF) (Mosley et al. 2009) values from SAGA-specific purifications with values from the TAF6 purification reveals a low level of TFIID that copurifies with SAGA (Fig. 3). No nucleases are used in our purifications; therefore, it is possible that the limited amount of TFIID in the SAGA purifications results from co-occupancy of these coactivator complexes at some genomic loci (Zhang et al. 2008). Alternatively, there may be a weak interaction between these complexes. Notably, TAF6 is not present at levels higher than any of the other non-SAGA TAFs in SAGA-specific purifications, indicating that its presence is likely to result from being a component of TFIID. We then asked whether a paralog of TAF6, TAF6L, is present in dSAGA instead of TAF6, as it replaces TAF6 in the human SAGA complexes (Ogryzko et al. 1998; Martinez et al. 2001; Nagy et al. 2009). Drosophila TAF6L is encoded by the testis-specific gene meiosis I arrest (mia) (Hiller et al. 2004). Consistent with the spermatocyte-specific expression of TAF6L in flies, no peptides for TAF6L were detected in SAGA isolated from S2 cells. Thus, neither TAF6 nor TAF6L are stable subunits of the majority of dSAGA complexes in S2 cells; therefore, we conclude that the majority of dSAGA complexes contain SAF6 instead of TAF6.
Figure 3.
Heat map showing the relative spectral abundance of SAGA and TFIID subunits in purifications using TAF6, SAF6, Ada2B, WDA, ADA1, or Sgf29 as bait proteins expressed as dNSAF. The dNSAF scale shows the highest-abundance subunits represented in yellow, and absent or underrepresented subunits in black. dNSAF values used to generate the heat map are provided in Supplemental Table S2.
The replacement of TAF6 by SAF6 within SAGA is likely to have functional consequences for the DNA-binding activity of SAGA, because sequence analysis indicates that the sequence conservation between SAF6 and TAF6 sequences is limited to the HFD and does not include the region of TAF6 that contains DNA-binding activity (Shao et al. 2005). Thus, the replacement of TAF6 by SAF6 in dSAGA is likely to convey differential specificity of DNA binding by SAGA relative to TFIID in flies. One particular aspect of this differential DNA-binding specificity might involve the downstream core promoter element (DPE), which is present at many TATA-less promoters in Drosophila and other metazoans, but not in yeast (Burke and Kadonaga 1997; Juven-Gershon et al. 2008). TAF6 binds directly to the DPE (Burke and Kadonaga 1997). Whereas yeast promoters lack DPEs and contain TAF6 as a common subunit of both TFIID and SAGA, metazoans such as Drosophila, in which a subset of promoters contains DPE motifs, confine TAF6 to TFIID. Analysis of the promoters of SAGA-regulated genes confirms that these are less likely to contain a DPE motif compared with the rest of the genome (Supplemental Table S3), supporting the hypothesis that, without TAF6, dSAGA is not preferentially targeted to DPE-containing promoters.
The HFD of SAF6 interacts with TAF9
As discussed above, TAF6 forms part of a TAF octamer complex within ySAGA that bears remarkable similarities to the histone octamer (Selleck et al. 2001). Moreover, the HFDs of TAF6 and TAF9, which are most similar to those of histones H4 and H3, respectively, interact via hydrophobic contacts within this octamer complex to form a heterodimer (Xie et al. 1996). Structural studies have shown that the TAF6/TAF9 complex exists as a heterotetramer that is similar to the (H3/H4)2 heterotetrameric core of the histone octamer (Xie et al. 1996).
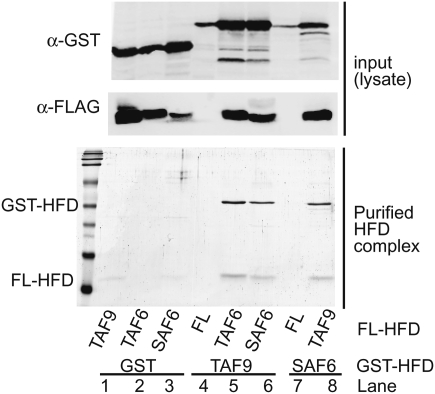
Alignment of TAF6 and TAF6L from multiple species with Drosophila SAF6 shows that, within the HFD of SAF6, 18 of the 28 residues involved in heterodimer contacts between TAF6 and TAF9 are conserved with TAF6 (Supplemental Fig. S1; Xie et al. 1996). Since SAF6 replaces TAF6 within dSAGA, we asked whether the HFD of SAF6 could interact with TAF9. The HFDs of SAF6, TAF6, and TAF9 were coexpressed in Escherichia coli as pairwise combinations of GST- and Flag-tagged proteins. Heterodimer complexes were purified by sequential glutathione-Sepharose chromatography, followed by Flag-agarose chromatography. This approach showed that GST-TAF9 copurifies with both Flag-TAF6 and Flag-SAF6 (Fig. 4, lanes 5,6). Furthermore, GST-SAF6 reciprocally copurifies with Flag-TAF9 (Fig. 4, lane 8). However, the HFDs of TAF9 and SAF6 do not copurify through these chromatographic steps in controls containing only a single HFD in the presence of either Flag or GST alone (Fig. 4, lanes 1–4,7). Thus, the HFD of SAF6 is sufficient to interact with the HFD of TAF9 in vitro. This suggests that SAF6 could incorporate into the TAF octamer structure within dSAGA via interactions with the HFD of TAF9.
Figure 4.
The HFDs of TAF9, TAF6, and SAF6 were coexpressed in E. coli as pairwise combinations of GST and Flag fusion proteins. Soluble bacterial lysate was prepared from each of the following pairwise combinations: GST and Flag-TAF9 (lane 1), GST and Flag-TAF6 (lane 2), GST and Flag-SAF6 (lane 3), GST-TAF9 and Flag (lane 4), GST-TAF9 and Flag-TAF6 (lane 5), GST-TAF9 and Flag-SAF6 (lane 6), GST-SAF6 and Flag (lane 7), and GST-SAF6 and Flag-TAF9 (lane 8). (Top panel) Soluble lysate (input; 0.02%) was probed with antibodies against GST or Flag. (Bottom panel) HFD complexes were purified by sequential glutathione-Sepharose chromatography followed by Flag-agarose chromatography, separated by SDS-PAGE, and stained with Coomassie.
SAF6 is essential for SAGA coactivator function independent of enzymatic activity
The interaction of SAF6 and TAF9 suggests that SAF6 might incorporate into the TAF octamer within dSAGA and thus play a TAF-mediated role within the complex. Hence, SAF6 provides a means of examining TAF function within SAGA independent of TFIID. Mutations disrupting SAF6 might therefore affect the expression of SAGA-regulated genes and/or the catalytic activities of the complex if the TAF components within SAGA are critical for these processes.
To distinguish these possibilities, we sought to generate a mutation in saf6 that would disrupt expression of the protein. We identified a nonlethal P-element insertion in the 5′ untranslated region (UTR) of saf6: EY05869. Imprecise excision of EY05869 generated a 303-base-pair (bp) deletion [saf6Δ303] that removed 15 nucleotides (nt) of the first exon of saf6 as well as a large region of the adjacent gene, CG3639 (Supplemental Fig. S2). We generated a genomic rescue construct containing the adjacent uncharacterized gene, CG3639, together with its upstream and downstream regulatory regions, including 116 nt of the first exon of saf6. This construct [CG3639+] completely rescues CG3639 expression in larvae homozygous for the saf6Δ303 deletion (Fig. 5B; Supplemental Fig. S2). The saf6Δ303 deletion is a likely null allele because it removes the predicted promoter region, translation initiation codon, and part of the first exon. No SAF6 transcripts are detected by RT–PCR analysis in larvae homozygous for saf6Δ303. The saf6Δ303; CG3639+ genotype will be referred to hereafter as saf6.
Figure 5.
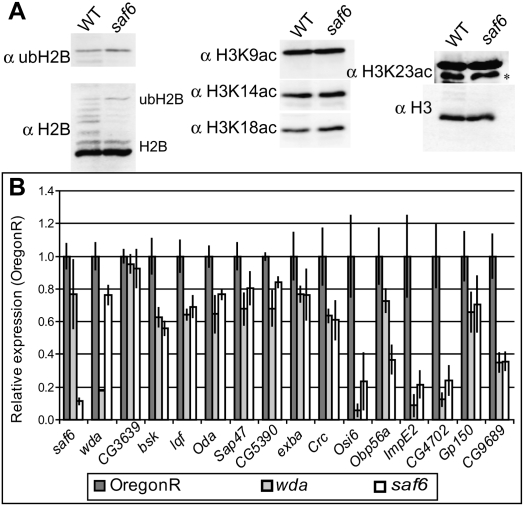
(A) Histones were acid-extracted from wild-type (OregonR) or saf6 second instar larvae and analyzed by Western blotting using antibodies against ubH2B, H2B, H3K9ac, H3K14ac, H3K18ac, H3K23ac, or H3. (*) Cross-reactive band. (B) Quantitative PCR was performed on cDNA isolated from OregonR (n = 3), saf6 (n = 3), and wda (n = 3) stage 14–16 embryos.
If SAF6 is important for either of the SAGA enzymatic activities, the global levels of acetylated or ubiquitinated histones should be altered in the saf6 mutant. We isolated histones from homozygous saf6 second instar larvae and compared them with histones from OregonR second instar larvae by Western blotting. Mutant saf6 larvae show no significant increase in global levels of ubH2B, and exhibit similar levels of H3 Lys 9 acetylation (H3K9ac) both globally (Fig. 5A) and at SAGA-regulated genes (Supplemental Fig. S3). This contrasts with other SAGA mutations: ada2b and wda significantly decrease H3K9ac, whereas nonstop and sgf11 mutations increase levels of ubH2B (Guelman et al. 2006; Weake et al. 2008). Furthermore, in contrast to wda embryos (Guelman et al. 2006), saf6 mutant animals show no decrease in H3K9ac levels, as observed by immunostaining of stage 15–16 embryos (data not shown). This implies that SAF6 is not critical for catalytic activities of SAGA, and suggested that SAGA remains intact and properly targeted in the absence of SAF6. Confirming this, antibodies against Gcn5 and Ada3 coimmunoprecipitate the SAGA-specific subunit Ada2b in extracts prepared from saf6 embryos (Supplemental Fig. S4). Additionally, there is no significant loss of Gcn5 at SAGA-regulated genes in the saf6 mutant (Supplemental Fig. S3).
Studies in yeast have shown that SAGA has important functions as a coactivator that are independent of its enzymatic activity. At SAGA-regulated promoters, SAGA is required for recruitment of the general transcription machinery independent of the HAT Gcn5 (Dudley et al. 1999; Bhaumik and Green 2001). Specifically, Spt3 within ySAGA interacts with TBP and is required for TBP recruitment to the promoters of some inducible genes (Eisenmann et al. 1992; Sterner et al. 1999; Larschan and Winston 2001; Mohibullah and Hahn 2008). Furthermore, the ySAGA subunit Spt8 binds TBP directly, and activator-recruited SAGA has been implicated in transferring TBP to the TATA box (Warfield et al. 2004; Sermwittayawong and Tan 2006; Mohibullah and Hahn 2008). Our MudPIT data support a limited interaction between SAGA and TBP/TFIID in Drosophila (Fig. 3).
The existence of a coactivator function of dSAGA that is independent of enzymatic activity is supported by our observations that SAF6 is essential for Drosophila development. Mutant saf6 larvae die during the second larval instar stage and, in addition, no viable saf6−/− adult progeny are obtained when the saf6 allele is crossed to a deficiency spanning the SAF6 gene. In order to test for defects in SAGA coactivator function, we examined saf6 mutants for defects in SAGA-mediated gene regulation. RNA was isolated from saf6 and wda mutant stage 14–16 embryos, and the expression of SAGA-regulated genes was examined using quantitative RT–PCR (Fig. 5B). RNA was isolated from wild-type OregonR embryos for comparison. Mutant individuals of both genotypes show no apparent gross morphological defects during embryogenesis (data not shown), despite the decrease in H3K9ac already observed by stage 16 of embryogenesis in wda individuals (Guelman et al. 2006). However, in both saf6 and wda mutants, expression of a subset of SAGA-regulated genes that include the JUN kinase basket and the Notch signaling pathway component liquid facets is down-regulated relative to the wild type. saf6 mutant animals have near wild-type levels of WDA transcripts, indicating that these defects in gene expression do not result from down-regulation of WDA. Transcripts corresponding to the gene adjacent to SAF6, CG3639, which is rescued in the saf6 mutant animals by a genomic expression construct, are similar in all three genotypes. Thus, while saf6 mutants fail to alter SAGA recruitment or histone acetylation, both globally and at SAGA-regulated genes, they show defects in SAGA-regulated gene expression similar to those observed for wda mutants. These data indicate that SAF6, and likely the TAF octamer in SAGA, primarily play a role in the coactivator functions of the SAGA complex and are not essential for the histone-modifying activities or recruitment of the complex.
Materials and methods
Additional methods are provided in the Supplemental Material.
Affinity purification and MudPIT analysis
Stable S2 cell lines expressing Ada2B (isoform B; NP_001027151.1), Sgf29 (CG30390; NP_726051.1), SAF6 (CG3883; NP_608545.1), CG6459 (NP_611243.1), WDA, Ada1, and TAF6 (FBgn0010417; NP_524161.2) in the pRmHa3-CHA2FL2 vector were generated as described previously (Guelman et al. 2006).
HAT assays
HAT assays were performed using Flag-purified HAT complexes and 500 ng of HeLa core histones as substrate (Eberharter et al. 1998).
Coexpression and purification of HFD proteins
The HFDs of TAF6 (residues 1–82), SAF6 (residues 123–204), and TAF9 (residues 11–95; NP_523391.3) were subcloned into pGex6P1 or pET28bNFlag and coexpressed and purified from E. coli.
Western blots
The following antibodies were used: Gcn5 (rabbit, 1:3000); Ada2b (guinea-pig; 1:1000) (Kusch et al. 2003); GST-Z5 (rabbit, 1:5000; Santa Cruz Biotechnologies); Flag-HRP (mouse, 1:5000; Sigma); H3 (rabbit, 1:3000; Abcam); H3K9ac (rabbit, 1:1000; Abcam), H3K14ac (rabbit, 1:1000; Upstate Biotechnologies); H3K18ac (rabbit, 1:1000; Upstate Biotechnologies); H3K23ac (rabbit, 1:1000; Abcam); H2B (rabbit, 1:1000; Upstate Biotechnologies), ubH2B (mouse, 1:100) (Minsky et al. 2008).
Acknowledgments
We thank N. Minsky and M. Oren for the ubH2B antibody, K. Wagner and J. Haug for cytometry, T. Parmely and V. Neubauer for cell culture, B. Sanderson for quantitative PCR, A. Mosley for heat maps, and A. Garrett and M. Gogol for bioinformatics. This work was supported by grant R37GM047867-18S1 from the NIGMS to J.L.W. and S.M.A., and funding from the Stowers Institute.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1846409.
Supplemental material is available at http://www.genesdev.org.
References
- Baker SP, Grant PA. The SAGA continues: Expanding the cellular role of a transcriptional co-activator complex. Oncogene. 2007;26:5329–5340. doi: 10.1038/sj.onc.1210603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaumik SR, Green MR. SAGA is an essential in vivo target of the yeast acidic activator Gal4p. Genes & Dev. 2001;15:1935–1945. doi: 10.1101/gad.911401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke TW, Kadonaga JT. The downstream core promoter element, DPE, is conserved from Drosophila to humans and is recognized by TAFII60 of Drosophila. Genes & Dev. 1997;11:3020–3031. doi: 10.1101/gad.11.22.3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carre C, Szymczak D, Pidoux J, Antoniewski C. The histone H3 acetylase dGcn5 is a key player in Drosophila melanogaster metamorphosis. Mol Cell Biol. 2005;25:8228–8238. doi: 10.1128/MCB.25.18.8228-8238.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley AM, Rougeulle C, Winston F. The Spt components of SAGA facilitate TBP binding to a promoter at a post-activator-binding step in vivo. Genes & Dev. 1999;13:2940–2945. doi: 10.1101/gad.13.22.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberharter A, John S, Grant PA, Utley RT, Workman JL. Identification and analysis of yeast nucleosomal histone acetyltransferase complexes. Methods. 1998;15:315–321. doi: 10.1006/meth.1998.0635. [DOI] [PubMed] [Google Scholar]
- Eisenmann DM, Arndt KM, Ricupero SL, Rooney JW, Winston F. SPT3 interacts with TFIID to allow normal transcription in Saccharomyces cerevisiae. Genes & Dev. 1992;6:1319–1331. doi: 10.1101/gad.6.7.1319. [DOI] [PubMed] [Google Scholar]
- Florens L, Washburn MP. Proteomic analysis by multidimensional protein identification technology. Methods Mol Biol. 2006;328:159–175. doi: 10.1385/1-59745-026-X:159. [DOI] [PubMed] [Google Scholar]
- Florens L, Carozza MJ, Swanson SK, Fournier M, Coleman MK, Workman JL, Washburn MP. Analyzing chromatin remodeling complexes using shotgun proteomics and normalized spectral abundance factors. Methods. 2006;40:303–311. doi: 10.1016/j.ymeth.2006.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guelman S, Suganuma T, Florens L, Weake V, Swanson SK, Washburn MP, Abmayr SM, Workman JL. The essential gene wda encodes a WD40 repeat subunit of Drosophila SAGA required for histone H3 acetylation. Mol Cell Biol. 2006;26:7178–7189. doi: 10.1128/MCB.00130-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry KW, Wyce A, Lo WS, Duggan LJ, Emre NC, Kao CF, Pillus L, Shilatifard A, Osley MA, Berger SL. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes & Dev. 2003;17:2648–2663. doi: 10.1101/gad.1144003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller M, Chen X, Pringle MJ, Suchorolski M, Sancak Y, Viswanathan S, Bolival B, Lin TY, Marino S, Fuller MT. Testis-specific TAF homologs collaborate to control a tissue-specific transcription program. Development. 2004;131:5297–5308. doi: 10.1242/dev.01314. [DOI] [PubMed] [Google Scholar]
- Juven-Gershon T, Hsu JY, Theisen JW, Kadonaga JT. The RNA polymerase II core promoter—the gateway to transcription. Curr Opin Cell Biol. 2008;20:253–259. doi: 10.1016/j.ceb.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler A, Schneider M, Cabal GG, Nehrbass U, Hurt E. Yeast Ataxin-7 links histone deubiquitination with gene gating and mRNA export. Nat Cell Biol. 2008;10:707–715. doi: 10.1038/ncb1733. [DOI] [PubMed] [Google Scholar]
- Kurshakova MM, Krasnov AN, Kopytova DV, Shidlovskii YV, Nikolenko JV, Nabirochkina EN, Spehner D, Schultz P, Tora L, Georgieva SG. SAGA and a novel Drosophila export complex anchor efficient transcription and mRNA export to NPC. EMBO J. 2007;26:4956–4965. doi: 10.1038/sj.emboj.7601901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusch T, Guelman S, Abmayr SM, Workman JL. Two Drosophila Ada2 homologues function in different multiprotein complexes. Mol Cell Biol. 2003;23:3305–3319. doi: 10.1128/MCB.23.9.3305-3319.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larschan E, Winston F. The S. cerevisiae SAGA complex functions in vivo as a coactivator for transcriptional activation by Gal4. Genes & Dev. 2001;15:1946–1956. doi: 10.1101/gad.911501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KK, Swanson SK, Florens L, Washburn MP, Workman JL. Yeast Sgf73/Ataxin-7 serves to anchor the deubiquitination module into both SAGA and Slik(SALSA) HAT complexes. Epigenetics Chromatin. 2009;2:2. doi: 10.1186/1756-8935-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez E, Palhan VB, Tjernberg A, Lymar ES, Gamper AM, Kundu TK, Chait BT, Roeder RG. Human STAGA complex is a chromatin-acetylating transcription coactivator that interacts with pre-mRNA splicing and DNA damage-binding factors in vivo. Mol Cell Biol. 2001;21:6782–6795. doi: 10.1128/MCB.21.20.6782-6795.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minsky N, Shema E, Field Y, Schuster M, Segal E, Oren M. Monoubiquitinated H2B is associated with the transcribed region of highly expressed genes in human cells. Nat Cell Biol. 2008;10:483–488. doi: 10.1038/ncb1712. [DOI] [PubMed] [Google Scholar]
- Mohibullah N, Hahn S. Site-specific cross-linking of TBP in vivo and in vitro reveals a direct functional interaction with the SAGA subunit Spt3. Genes & Dev. 2008;22:2994–3006. doi: 10.1101/gad.1724408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosley AL, Florens L, Wen Z, Washburn MP. A label free quantitative proteomic analysis of the Saccharomyces cerevisiae nucleus. J Proteomics. 2009;72:110–120. doi: 10.1016/j.jprot.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muratoglu S, Georgieva S, Papai G, Scheer E, Enunlu I, Komonyi O, Cserpan I, Lebedeva L, Nabirochkina E, Udvardy A, et al. Two different Drosophila ADA2 homologues are present in distinct GCN5 histone acetyltransferase-containing complexes. Mol Cell Biol. 2003;23:306–321. doi: 10.1128/MCB.23.1.306-321.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy Z, Riss A, Romier C, le Guezennec X, Dongre AR, Orpinell M, Han J, Stunnenberg H, Tora L. The human SPT20-containing SAGA complex plays a direct role in the regulation of endoplasmic reticulum stress-induced genes. Mol Cell Biol. 2009;29:1649–1660. doi: 10.1128/MCB.01076-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogryzko VV, Kotani T, Zhang X, Schiltz RL, Howard T, Yang XJ, Howard BH, Qin J, Nakatani Y. Histone-like TAFs within the PCAF histone acetylase complex. Cell. 1998;94:35–44. doi: 10.1016/s0092-8674(00)81219-2. [DOI] [PubMed] [Google Scholar]
- Pankotai T, Komonyi O, Bodai L, Ujfaludi Z, Muratoglu S, Ciurciu A, Tora L, Szabad J, Boros I. The homologous Drosophila transcriptional adaptors ADA2a and ADA2b are both required for normal development but have different functions. Mol Cell Biol. 2005;25:8215–8227. doi: 10.1128/MCB.25.18.8215-8227.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi D, Larsson J, Mannervik M. Drosophila Ada2b is required for viability and normal histone H3 acetylation. Mol Cell Biol. 2004;24:8080–8089. doi: 10.1128/MCB.24.18.8080-8089.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Navarro S. Insights into SAGA function during gene expression. EMBO Rep. 2009;10:843–850. doi: 10.1038/embor.2009.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selleck W, Howley R, Fang Q, Podolny V, Fried MG, Buratowski S, Tan S. A histone fold TAF octamer within the yeast TFIID transcriptional coactivator. Nat Struct Biol. 2001;8:695–700. doi: 10.1038/90408. [DOI] [PubMed] [Google Scholar]
- Sermwittayawong D, Tan S. SAGA binds TBP via its Spt8 subunit in competition with DNA: Implications for TBP recruitment. EMBO J. 2006;25:3791–3800. doi: 10.1038/sj.emboj.7601265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao H, Revach M, Moshonov S, Tzuman Y, Gazit K, Albeck S, Unger T, Dikstein R. Core promoter binding by histone-like TAF complexes. Mol Cell Biol. 2005;25:206–219. doi: 10.1128/MCB.25.1.206-219.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner DE, Grant PA, Roberts SM, Duggan LJ, Belotserkovskaya R, Pacella LA, Winston F, Workman JL, Berger SL. Functional organization of the yeast SAGA complex: Distinct components involved in structural integrity, nucleosome acetylation, and TATA-binding protein interaction. Mol Cell Biol. 1999;19:86–98. doi: 10.1128/mcb.19.1.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suganuma T, Gutierrez JL, Li B, Florens L, Swanson SK, Washburn MP, Abmayr SM, Workman JL. ATAC is a double histone acetyltransferase complex that stimulates nucleosome sliding. Nat Struct Mol Biol. 2008;15:364–372. doi: 10.1038/nsmb.1397. [DOI] [PubMed] [Google Scholar]
- Tora L. A unified nomenclature for TATA box binding protein (TBP)-associated factors (TAFs) involved in RNA polymerase II transcription. Genes & Dev. 2002;16:673–675. doi: 10.1101/gad.976402. [DOI] [PubMed] [Google Scholar]
- Warfield L, Ranish JA, Hahn S. Positive and negative functions of the SAGA complex mediated through interaction of Spt8 with TBP and the N-terminal domain of TFIIA. Genes & Dev. 2004;18:1022–1034. doi: 10.1101/gad.1192204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weake VM, Lee KK, Guelman S, Lin CH, Seidel C, Abmayr SM, Workman JL. SAGA-mediated H2B deubiquitination controls the development of neuronal connectivity in the Drosophila visual system. EMBO J. 2008;27:394–405. doi: 10.1038/sj.emboj.7601966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Kokubo T, Cohen SL, Mirza UA, Hoffmann A, Chait BT, Roeder RG, Nakatani Y, Burley SK. Structural similarity between TAFs and the heterotetrameric core of the histone octamer. Nature. 1996;380:316–322. doi: 10.1038/380316a0. [DOI] [PubMed] [Google Scholar]
- Xu W, Edmondson DG, Evrard YA, Wakamiya M, Behringer RR, Roth SY. Loss of Gcn5l2 leads to increased apoptosis and mesodermal defects during mouse development. Nat Genet. 2000;26:229–232. doi: 10.1038/79973. [DOI] [PubMed] [Google Scholar]
- Zhang H, Kruk JA, Reese JC. Dissection of coactivator requirement at RNR3 reveals unexpected contributions from TFIID and SAGA. J Biol Chem. 2008;283:27360–27368. doi: 10.1074/jbc.M803831200. [DOI] [PMC free article] [PubMed] [Google Scholar]