Abstract
In budding yeast, Cdc13, Stn1, and Ten1 form a heterotrimeric complex (CST) that is essential for telomere protection and maintenance. Previous bioinformatics analysis revealed a putative oligonucleotide/oligosaccharide-binding (OB) fold at the N terminus of Stn1 (Stn1N) that shows limited sequence similarity to the OB fold of Rpa2, a subunit of the eukaryotic ssDNA-binding protein complex replication protein A (RPA). Here we present functional and structural analyses of Stn1 and Ten1 from multiple budding and fission yeast. The crystal structure of the Candida tropicalis Stn1N complexed with Ten1 demonstrates an Rpa2N–Rpa3-like complex. In both structures, the OB folds of the two components pack against each other through interactions between two C-terminal helices. The structure of the C-terminal domain of Saccharomyces cerevisiae Stn1 (Stn1C) was found to comprise two related winged helix–turn–helix (WH) motifs, one of which is most similar to the WH motif at the C terminus of Rpa2, again supporting the notion that Stn1 resembles Rpa2. The crystal structure of the fission yeast Schizosaccharomyces pombe Stn1N–Ten1 complex exhibits a virtually identical architecture as the C. tropicalis Stn1N–Ten1. Functional analyses of the Candida albicans Stn1 and Ten1 proteins revealed critical roles for these proteins in suppressing aberrant telomerase and recombination activities at telomeres. Mutations that disrupt the Stn1–Ten1 interaction induce telomere uncapping and abolish the telomere localization of Ten1. Collectively, our structural and functional studies illustrate that, instead of being confined to budding yeast telomeres, the CST complex may represent an evolutionarily conserved RPA-like telomeric complex at the 3′ overhangs that works in parallel with or instead of the well-characterized POT1–TPP1/TEBPα–β complex.
Keywords: Telomere, telomere-binding protein, telomerase, homologous recombination
Telomeres, the specialized nucleoprotein structures located at linear eukaryotic chromosomal termini, are essential for chromosome stability and are maintained by the special reverse transcriptase named telomerase (Ferreira et al. 2004; Bianchi and Shore 2008; Palm and de Lange 2008). Telomeric DNAs are typically repetitive in nature and terminate in 3′ overhangs (G tails) that are bound by distinct protein complexes in different organisms. In ciliated protozoa, a dimeric protein complex (TEBPα and TEBPβ) is responsible for G-tail recognition and protection (Gottschling and Zakian 1986). In fission yeast and humans, the TEBPα homolog POT1 provides the major G-tail-binding activity and associates with the respective TEBPβ homolog (Tpz1 in Schizosaccharomyces pombe, and TPP1 in humans) (Baumann and Cech 2001; Wang et al. 2007; Xin et al. 2007; Miyoshi et al. 2008). Interestingly, the G tails of budding yeast telomeres are apparently protected by an altogether distinct, nonhomologous complex named CST (Cdc13–Stn1–Ten1) (Garvik et al. 1995; Grandin et al. 2001; Petreaca et al. 2006; Gao et al. 2007). Nevertheless, all of these proteins appear to contain one or more OB (oligosaccharide/oligonucleotides-binding) folds, testifying to the versatility of this domain in single-strand nucleic acid recognition (Bochkarev and Bochkareva 2004). Many of the G-tail-interacting proteins are essential for cell viability, and hypomorphic alleles of genes encoding these proteins have been shown to induce a variety of telomere aberrations—including catastrophic telomere loss, uncontrolled telomere elongation, telomere C-strand degradation, and telomere fusions—thus underscoring their fundamental importance in telomere protection (Bertuch and Lundblad 2006; Cooper and Hiraoka 2006; Palm and de Lange 2008).
Initially, components of the CST complex were thought to be unique to budding yeast, and in particular to organisms without POT1 homologs. In other words, the POT1–TPP1 and CST complex are postulated to represent two mutually exclusive means of G-tail protection. However, recent studies have uncovered Stn1 and Ten1 homologs in a multitude of POT1-containing organisms, and have implicated the S. pombe Stn1 and Ten1 as well as Arabidopsis thaliana Stn1 in telomere capping (Martin et al. 2007; Song et al. 2008). Moreover, the S. pombe Stn1 and Ten1 proteins exhibit no evident interaction with Pot1, suggesting that they can function independently of the major G-tail-binding activity (Martin et al. 2007). Indeed, the Saccharomyces cerevisiae Stn1 and Ten1, when overexpressed, are capable of mediating Cdc13-independent protection of telomeres (Petreaca et al. 2006). Even though Stn1 or Ten1 alone apparently recognizes telomere G tails with low affinity, available evidence suggests that they can be recruited to telomeres through an interaction with the B subunit of the DNA polymerase α–primase complex Pol12 (Grossi et al. 2004; Petreaca et al. 2006). Altogether, these observations hint at a more prevalent role for Stn1 and Ten1, possibly as components of an alternative telomere end protective complex that functions in parallel to the POT1-containing complex.
Recent bioinformatic analysis points to potential structural similarities between Stn1 and Rpa2 (Gao et al. 2007). The validity of the Stn1–Rpa2 analogy was supported by a domain-swapping experiment in which the N-terminal OB fold-like domain of Stn1 was shown to function in place of the Rpa2 OB fold. In addition, similar to Rpa2 and Rpa3, the N terminus of Stn1 interacts with Ten1 in vitro and in vivo (Petreaca et al. 2006; Gao et al. 2007). Both Rpa2 and Rpa3 are subunits of a trimeric, nonspecific ssDNA-binding complex (replication protein A [RPA]) that mediates critical and diverse DNA transactions throughout the genome (Wold 1997; Bochkarev and Bochkareva 2004). Their potential similarities to Stn1 and Ten1 thus raise the intriguing possibility that the CST complex represents a chromosome locus-specific RPA complex. While highly provocative, this hypothesis awaits experimental confirmation. In addition, many questions with regard to the structure, function, and conservation of the CST complex remain unresolved. In this study, we provide structural and functional analyses of the Stn1 and Ten1 protein from multiple budding and fission yeast. Our atomic resolution structures of several complexes and a protein domain provide direct confirmation of structural similarity between components of the CST and the RPA complexes, and reveal a detailed molecular view of the Stn1–Ten1 interaction interface. Our functional studies underscore the importance of Stn1–Ten1 interaction in telomere protection, and reveal critical functions for these proteins in suppressing aberrant telomerase and recombination activities at telomeres.
Results
Identification of the CST complex genes in budding yeast Candida and Saccharomyces genomes
The branches of budding yeast exemplified by Candida albicans have apparently undergone rapid evolutionary divergence with respect to its telomere sequence and telomere-related proteins (McEachern and Blackburn 1994; Teixeira and Gilson 2005). For instance, unlike S. cerevisiae, many Candida spp. have long (up to 25-base-pair [bp]), distinct and regular telomere repeat units. Moreover, the putative telomere maintenance proteins of Candida spp. (e.g., Rap1) have been observed to exhibit significant structural divergence from their Saccharomyces counterparts (Biswas et al. 2003). Indeed, until recently, homologs of the CST complex were difficult to identify in these genomes, raising interesting questions concerning their telomere protection mechanisms (Teixeira and Gilson 2005).
To initiate a comparative analysis of telomere end protection mechanisms in this unusual group of budding yeast, we systemically searched the NCBI and Broad Institute databases for homologs of Cdc13, Stn1, and Ten1 using available sequences as queries. This exercise resulted in the identification of plausible homologs of each CST component in all completely sequenced Candida and Saccharomyces genomes (Supplemental Fig. S1). In keeping with the theme of rapid evolutionary divergence, we found that many Cdc13 homologs in Candida spps. are considerably smaller and evidently lack the N-terminal half of their S. cerevisiae counterpart, thus partly accounting for the prior difficulties in their detection. To ascertain the functions of these homologs in telomere regulation, we attempted to generate C. albicans strains that are null for CDC13, STN1, or TEN1 by sequential deletion of the two alleles (Fonzi and Irwin 1993; Enloe et al. 2000). Perhaps, not surprisingly, we were unable to generate a cdc13-null strain, suggesting that this gene, like its S. cerevisiae homolog, is essential for cell viability (Garvik et al. 1995). In contrast, we were able to obtain multiple isolates of stn1- and ten1-null strains, indicating that these genes are not essential in C. albicans. The availability of the null strains allowed us to investigate in detail the functions and mechanisms of Stn1 and Ten1 in C. albicans.
C. albicans Stn1 and Ten1 are important for telomere maintenance
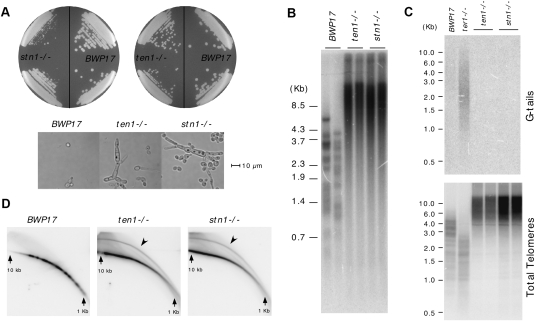
Both the stn1- and ten1-null mutants grow more slowly than the parental BWP17 strain (Fig. 1A). Microscopic examination revealed an abundance of filamentous cells in liquid cultures; quantitation indicated an ∼20-fold increase in the percentage of such cells (data not shown). Although the reasons for this aberrant growth morphology are not understood, similar aberrations have been described for other C. albicans DNA repair mutants, suggesting a shared underlying mechanism (Andaluz et al. 2006; Legrand et al. 2007). Consistent with a role for Stn1 and Ten1 in telomere regulation, we observed extremely long and heterogeneous telomeres in multiple isolates of both null mutants (Fig. 1B; data not shown). Long and heterogeneous telomeres were detected at the earliest time point following the derivation of the mutants (∼100 generations), and were stably maintained for at least 150 generations thereafter (Supplemental Fig. S2). In contrast to the parental BWP17, whose telomeres range in size from ∼1 to 5 kb, the stn1 and ten1 mutants possess extremely long (>20-kb) and short (<1-kb) telomeres, consistent with loss of the homeostatic mechanism that normally regulates telomere length.
Figure 1.
Phenotypes of the C. albicans stn1−/− and ten1−/− mutants. (A) The slow-growing and filamentous morphology of the stn1−/− and ten1−/− mutants are displayed. (B) Chromosomal DNAs isolated from the parental BWP17, the stn1−/−, and the ten1−/− mutants were subjected to Southern analysis of the telomere terminal restriction fragments. The mutant samples were from two independently constructed null strains that have undergone ∼100 cell divisions following construction. (C, top) Chromosomal DNAs isolated from the parental BWP17, the stn1−/−, and the ten1−/− mutant were subjected to in-gel hybridization analysis of the level of G tails. (Bottom) Subsequently, the DNAs were denatured in the gel and reanalyzed using the same probe. As a positive control, the DNA from a ter1−/− strain, which was demonstrated previously to exhibit an increase in G-tail signal, was analyzed in parallel. (D) Chromosomal DNAs isolated from the parental BWP17, the stn1−/−, and the ten1−/− mutants were subjected to two-dimensional gel electrophoresis in order to resolve linear and circular telomeric DNA (marked by arrowheads).
The extremely long and heterogeneous telomeres suggest that telomeres in stn1 and ten1 are deprotected. Two other frequent consequences of deprotection are the accumulation of G tails and extrachromosomal telomeric circles (t-circles), which can be detected by in-gel hybridization and two-dimensional gel electrophoresis, respectively. Interestingly, we found no evidence of G-tail accumulation, but rather high levels of t-circles in the mutants (Fig. 1C,D). Quantitative analysis indicates that ∼10% of telomeric hybridization signals in the mutants reside in circular DNAs. In comparison, much less than 1% of the telomeric DNA in the parental BWP17 strain is in circular form. Notably, all of the growth and telomere abnormalities in the stn1 and ten1 mutants are suppressed by the reintegration of a wild-type copy of the respective genes, confirming that these phenotypes are due to loss of Stn1 and Ten1 (Supplemental Fig. S3; data not shown). We conclude that both the STN1 and TEN1 genes in C. albicans are necessary for the maintenance of proper telomere length and structure.
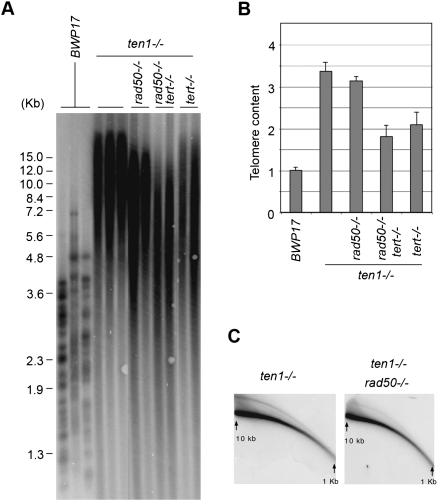
While the accumulation of t-circles in the mutants suggests that some recombination pathway is abnormally active at telomeres, the extremely long telomeres of the mutants could also arise from uncontrolled telomerase activity. To assess the contributions of these two pathways, we attempted to obtain mutants that combine deletion of TEN1 with that of TERT and/or a recombination gene. Notably, we were unable to delete RAD52 in the ten1−/− background, suggesting that knocking out both genes might result in synthetic lethality (data not shown). Thus, we used deletion of RAD50 to gauge the contribution of recombination to telomere aberrations in the ten1 mutant. As expected, deletion of either TERT or RAD50 in the wild-type strain background had little effect on telomere length during early passages (Supplemental Fig. S4). On the other hand, loss of TERT or RAD50 in the ten1 mutant each reduced significantly the extreme telomere elongation, while the ten1 tert rad50 triple mutant exhibited the mildest telomere lengthening (Fig. 2A). The average telomere lengths of the various mutants correlated with their overall telomeric DNA content (Fig. 2B). Thus, both recombination and telomerase are partly responsible for the telomere length deregulation observed in the ten1 mutant. The residual telomere lengthening of the triple mutant may be accounted for by the existence of Rad50-independent recombination pathways, which have been well described in other budding yeast (McEachern and Haber 2006). Consistent with this notion, we found that the level of t-circles in the ten1 mutant were reduced (by approximately fourfold) but not abolished by RAD50 deletion (Fig. 2C). We conclude that the Stn1–Ten1 complex mediates important functions in the suppression of aberrant telomerase and recombination activities at C. albicans telomeres. Chromatin immunoprecipitation (ChIP) analysis confirmed the association of Ten1 with telomeric DNA in vivo, indicating that the complex is likely to be acting directly at telomeres (Supplemental Fig. S5). On the other hand, we were unable to detect significant binding of a Stn1–Ten1 complex from Candida to telomeric DNA in vitro, suggesting that the complex may require additional protein–protein interactions in order to associate with telomeres in vivo (data not shown).
Figure 2.
The role of telomerase and recombination in generating the aberrant telomere structures of the C. albicans ten1−/− mutant. (A) Chromosomal DNAs isolated from various combination mutants (as indicated at the top) were subjected to Southern analysis of the telomere restriction fragments. For each genotype, two or three independently derived clones were analyzed. (B) Telomere hybridization signals from experiments such as those shown in A were normalized against the RAD52 hybridization signals from the same samples and were plotted; shown are means ± SD from two to three independent experiments. (C) Chromosomal DNAs from the ten1−/− and the ten1−/− rad50−/− mutants were subjected to two-dimensional gel electrophoresis to assess the levels of t-circles.
Overall, our observations with regard to the function of the Stn1–Ten1 complex in C. albicans echo earlier findings in other budding yeast. Specifically, hypomorphic CST mutations have been shown to result in abnormal telomerase and recombination activities at telomeres in both S. cerevisiae and Kluyveromyces lactis (Grandin et al. 2001; Iyer et al. 2005; Petreaca et al. 2006; Puglisi et al. 2008). A point mutant allele of STN1 in K. lactis, in particular, exhibits extremely long and heterogeneous telomeres that are (at least partly) telomerase-independent (Iyer et al. 2005). The close phenotypic resemblance of this K. lactis mutant to the C. albicans stn1 and ten1 mutants argues for a substantial degree of mechanistic conservation in budding yeast. On the other hand, some features of the C. albicans systems are clearly unique. For example, both STN1 and TEN1 are dispensable for cell viability, allowing the consequences of complete gene deletions to be analyzed in the absence of other genetic changes. Also unusual was our failure to observe G-tail accumulation, which is a frequent consequence of hypomorphic CST mutations in budding yeast. Yet these differences do not necessarily imply fundamentally different mechanisms of telomere protection by the CST in Candida. Most prior studies of the CST complex were conducted in haploid yeast, which differs physiologically from the obligate diploid C. albicans used in our analysis. Moreover, failure to observe G tails may be due to their transience rather than absence. One can imagine, for instance, that G tails were generated by C-strand degradation in the C. albicans stn1 and ten1 mutants, but were more efficiently repaired by recombination or fill-in synthesis. Further studies will be necessary to determine whether the apparent differences between C. albicans and other budding yeast reflect some fundamental mechanistic divergence.
In many respects, the phenotypes of the C. albicans stn1 and ten1 mutants mimic those of ALT (alternative lengthening of telomeres) cancer cells, which are also characterized by telomere length heterogeneity, elevation of t-circles, and telomere maintenance through recombination (Neumann and Reddel 2006; Nabetani and Ishikawa 2009). Thus, our findings suggest that one possible pathway for attaining the ALT status was through deprotection of G tails. Interestingly, a recent study in K. lactis argues that deficiency of Rap1 (the major double-strand telomere-binding protein in budding yeast) can lead to similar phenotypes (Bechard et al. 2009). It is tempting to speculate that aberrations in some telomere protein component may be a necessary condition for the activation of the ALT pathway.
Structure determination of the Candida tropicalis Stn1–Ten1 complex
Sequence alignment and secondary structure predictions of Stn1 proteins have revealed previously in members of this conserved family a putative N-terminal OB fold domain that is most similar to the OB fold of Rpa2 (Gao et al. 2007). Notably, an Rpa2 chimera that carries the predicted OB fold of Stn1 in place of its own OB fold can rescue the lethal phenotype of an rpa2-Δ yeast strain (Gao et al. 2007). Stn1 interacts with Ten1 both in vivo and in vitro (Gao et al. 2007), and sequence analysis supports the existence of an OB fold in Ten1 (data not shown). These results led to the hypothesis that Stn1 binds to Ten1 to form an Rpa2–Rpa3-like complex at telomeres (Gao et al. 2007). However, there is no detectable sequence similarity between Ten1 and Rpa3 protein families (Gao et al. 2007). Furthermore, it is unknown how Stn1 interacts with Ten1 and whether this interaction resembles that between Rpa2 and Rpa3. Thus, validation of the hypothesis that Stn1–Ten1 represents a telomere-specific Rpa2–Rpa3 complex requires structural characterization of the Stn1–Ten1 complex.
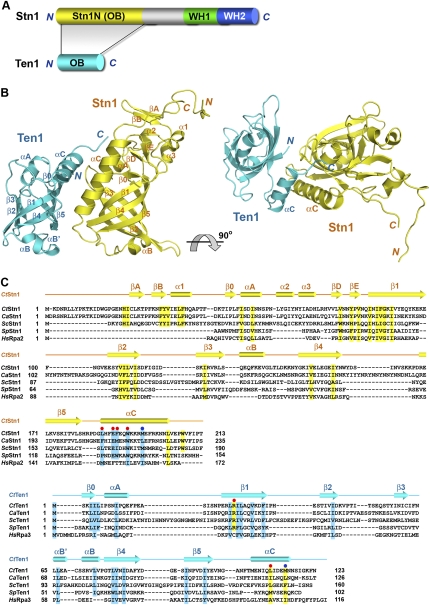
Complexes consisting of Ten1 and the N-terminal domain of Stn1 (Stn1N) from several different budding yeast species including S. cerevisiae, C. albicans, and C. tropicalis were prepared and used in crystallization trials (Fig. 3A). After extensive screening, the C. tropicalis Stn1N–Ten1 complex was found to generate crystals suitable for structural determination. The complex was crystallized in space group P41212 with two complexes per asymmetric unit (Supplemental Table S1). The structure was solved by single-wavelength anomalous dispersion (SAD) with mercury (MeHgAc) derivative crystals, and refined to 2.4 Å resolution. The high-quality experimental electron density map enabled us to fit and refine most of the complex except several N- and C-terminal residues of Stn1N.
Figure 3.
Overview of the C. tropicalis Stn1N–Ten1 complex structure. (A) Domain organization of the Stn1 and Ten1 polypeptide chains. In Stn1, the N-terminal OB fold is colored in yellow, the middle linker region is shown in gray, and the two C-terminal WH motifs are shown in green and blue, respectively. Ten1 is colored in cyan. The shaded area between Stn1 and Ten1 indicates that the Stn1–Ten1 interaction is mediated by the two OB folds of the proteins. (B) Ribbon diagram of two orthogonal views of the Stn1N–Ten1 complex. Stn1N and Ten1 are colored in yellow and cyan, respectively. The secondary structure elements are labeled. The Stn1N–Ten1 complex at right is rotated by 90° about a horizontal axis relative to the complex at left. (C) Amino acid sequence alignment of Stn1N and Ten1. (Top panel) Sequence alignment of the N-terminal OB fold regions of the budding yeast Stn1 family members together with the OB folds of S. pombe Stn1 and human Rpa2. (Bottom panel) Sequence alignment of the budding yeast Ten1 family members together with S. pombe Ten1 and human Rpa3. The alignments with Rpa2 and Rpa3 are based on the crystal structure of the Rpa2N–Rpa3 complex (Bochkarev et al. 1999). Secondary structure assignments from our Stn1N–ten1 crystal structure are shown as colored cylinders (α helices) and arrows (β strands) above the aligned sequences. Red dots denote the C. tropicalis residues important for the Stn1N–ten1 interaction in the yeast two-hybrid assay, whereas blue dots denote the less important residues. Conserved hydrophobic residues in Stn1N/Rpa2N and Ten1/Rpa3 are highlighted in yellow and cyan, respectively.
The Stn1N–Ten1 complex structure
The Stn1N–Ten1 complex structure reveals a 1:1 stoichiometry between Stn1N and Ten1, consistent with the observed molecular weight of the complex as determined by gel filtration chromatography(∼37.5 kDa) (Supplemental Fig. S6). The crystal structure shows that each protein indeed comprises a single OB fold, consisting of a highly curved five-stranded β barrel, as expected from previous primary sequence analysis (Fig. 3B). In addition to the central β barrel, there are several structural features common to the OB folds of Stn1N and Ten1. First, both proteins contain a C-terminal helix αC, which contributes most of the contact interface between Stn1N and Ten1 (Fig. 3B). Second, short α helices (αB in Stn1N, and αB′ and αB in Ten1) that cover the bottom of the β barrels of the OB folds are found between strands β3 and β4 (Fig. 3B). Third, an N-terminal helix, αA, closes the other end of the β barrel, and the position of this helix is stabilized by a short-strand β0, which interacts with strand β1 in an anti-parallel orientation (Fig. 3B).
Besides helix αA and strand β0, Stn1N contains a unique segment N-terminal to the core of the OB fold (Fig. 3B; Supplemental Fig. S7). This segment, which consists of residues 1–45 (located N-terminal to β0), folds into a β hairpin (βA and βB) and a short helix (α1) (Fig. 3B; Supplemental Fig. S7). Another unique feature of Stn1N is the connection between helix αA and the β barrel, which contains a 27-residue insertion (residues 57–83) that comprises two short helices (α2 and α3) and another short β hairpin (βD and βE). These two extra elements fold together into a unique motif to cap the top of the OB fold of Stn1 (henceforth referred to as the “cap” motif of Stn1) (Fig. 3B; Supplemental Fig. S7). Notably, the C-terminal tails following helix αC (residues 204–213) of both Stn1N molecules in the asymmetric unit are well ordered and make hydrophobic contacts with the cap region (Supplemental Fig. S7). In particular, the aromatic side chain of W208 is nested in a hydrophobic pocket formed by Y32, L36, F37, and Y80 (Supplemental Fig. S7). All of these residues are highly conserved in the Stn1 family members (Fig. 3C). Consistent with this observation, efforts to prepare an Stn1N fragment without the C-terminal tail (residues 2–205) yielded little soluble protein, suggesting that this tail is important for the correct folding of Stn1 (data not shown).
The structural conservation between Stn1N–Ten1 and Rpa2N–Rpa3
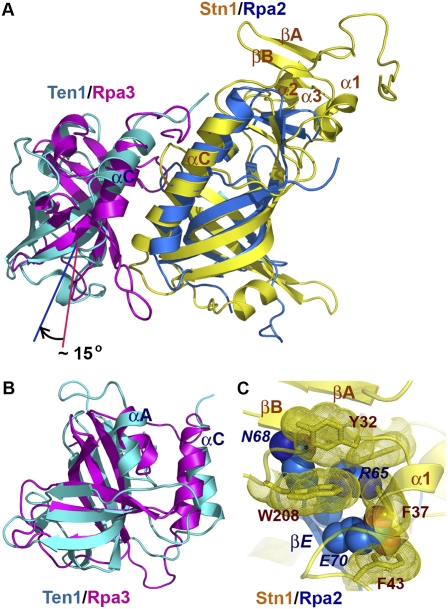
The crystal structure of Stn1N–Ten1 closely resembles that of the Rpa2N–Rpa3 complex (Fig. 4A; Bochkarev et al. 1999). An unbiased search for structurally homologous proteins using the Dali server (Holm and Sander 1991) revealed that the structure of the Stn1 OB fold is most similar to that of the OB fold of Rpa2 (Bochkarev et al. 1999), consistent with previous sequence alignment predictions (Fig. 3C; Gao et al. 2007). The two OB folds can be superimposed with a root-mean-square deviation (RMSD) of 2.4 Å for 119 equivalent Cα pairs (Fig. 4A). Notably, the structurally highly conserved region includes not only the central β barrel of the OB fold, but also peripheral α helices (αA and αC) and β strands (βD, βE, and β0) in the N- and C-terminal extension regions, suggesting that Stn1 and Rpa2 are structurally homologous proteins (Fig. 4A). Unlike Stn1 and Rpa2, bioinformatics analysis failed to detect any substantial sequence similarity between Ten1 and Rpa3 (Gao et al. 2007). However, comparison of the structures of Ten1 and Rpa3 clearly reveals a high degree of structural similarity (Fig. 4B). In fact, Rpa3 is one of the top solutions revealed by Dali that are structurally most similar to Ten1, with an RMSD of 2.8 Å for 96 equivalent Cα atoms. This close structural similarity is rather unexpected, given that the sequences of the OB folds of Ten1 and Rpa3 share only 7% identity (Fig. 3C). In addition to similarities between the individual components, the Stn1N–Ten1 and the Rpa2–Rpa3 complexes exhibit another common feature; in both cases, the two subunits heterodimerize mainly through hydrophobic contacts mediated by the two C-terminal αC helices (Fig. 4A). Taken together, we concluded that Stn1–Ten1 is structurally similar to and evolutionarily related to the Rpa2–Rpa3 complex.
Figure 4.
The C. tropicalis Stn1N–Ten1 complex is structurally similar to Rpa2N–Rpa3. (A) Superposition of the Stn1N–Ten1 complex on the crystal structure of the human Rpa2N–Rpa3 complex (Bochkarev et al. 1999). Stn1N and Ten1 are colored in yellow and cyan and Rpa2N and Rpa3 are shown in blue and magenta. The superposition is based on the structures of Stn1N and Rpa2N. Ten1 and Rpa3 are not aligned well, and Ten1 rotates ∼15° relative to the orientation of Rpa3. (B) Overlay of Ten1 and Rpa3 based on the OB fold β barrels of the proteins. (C) Superposition of Stn1N and Rpa2N based on the OB fold β barrels shows collisions between the cap motif of Stn1N and the N-terminal β hairpin (βD–βE) of Rpa2N. Residues in Stn1N are drawn as a stick model with dotted surface. Residues in Rpa2N are shown as a space-filling model. Stn1N and Rpa2N are colored as in A. Labels for residues in Rpa2 are in italics, to differentiate them from residues in Stn1.
Notwithstanding the high degree of overall structural conservation, there are substantial differences between the Stn1–Ten1 and the Rpa2–Rpa3 complexes. Most notably, the relative orientations between the two components are different in the two complexes. When both complex structures are overlaid based on the OB folds of Stn1 and Rpa2, Ten1 exhibits an ∼15° rotation relative to the position of Rpa3 (Fig. 4A). Second, compared with Rpa2, Stn1 contains an extra N-terminal extension (βA, βB, and α1) and a 12-residue insertion before strand βD (α2 and α3) (Fig. 4C). Additionally, significant sequence and structural disparities are evident in most of the connecting loop regions. For example, Stn1 has a long loop (12 residues), L45, between strands β4 and β5 that packs against helix α2 in the N-terminal cap motif. In contrast, strands β4 and β5 of Rpa2 are connected by a short two-residue turn.
These structural differences provide a plausible explanation for the published findings on domain exchange between Stn1 and Rpa2 (Gao et al. 2007). As noted before, the specific interactions between Stn1 and Ten1 and between Rpa2 and Rpa3 primarily involve the hydrophobic contacts between the two αC helices C-terminal to the OB folds (Figs. 3B, 4A; Bochkarev et al. 1999). Thus, the chimeric Rpa2-OBStn1 protein, which carries the OB fold of Stn1 in place of the Rpa2 OB fold and still contains helix αC of Rpa2, retains the ability to bind Rpa3 and rescues the inviability of an rpa2-Δ yeast strain (Gao et al. 2007). In contrast, due to the incompatibility between the two αC helices of Stn1 and Rpa3, the rpa2-Δ strain could not be rescued by high-level expression of Stn1 (Gao et al. 2007). For the same reason, the chimeric Rpa2-OBStn1 protein could not interact with Ten1 to rescue an stn1-Δ strain (Gao et al. 2007). Furthermore, the N-terminal cap motif of Stn1 (βA, βB, and αA) is expected to collide with strands βD and βE of Rpa2 if the OB fold of Stn1 is replaced with that of Rpa2 (Fig. 4C). Hence, the chimeric Stn1-OBRpa2 is unlikely to fold into a stable and functional protein, explaining the failure of Stn1-OBRpa2 to rescue the stn1-Δ mutant (Gao et al. 2007).
The Stn1N–Ten1 interaction
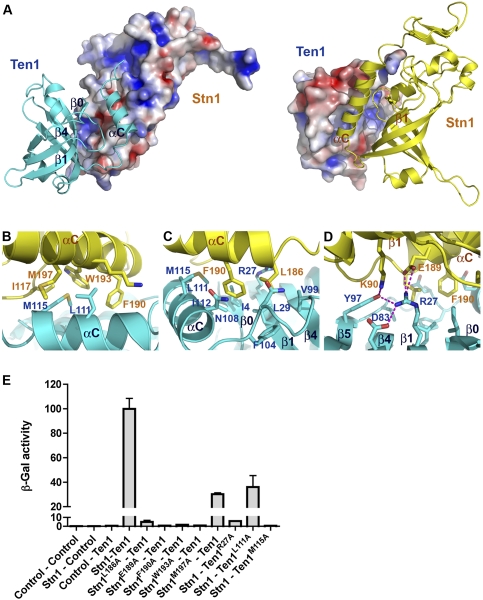
The interface between Stn1N and Ten1 in the crystal structure is relatively flat and hydrophobic (Fig. 5A). The interactions are mediated primarily by the amphipathic αC helices of both proteins and one side of the Ten1 β barrel (Fig. 5A), burying 1060 and 1128 Å2 of solvent-accessible surface on Stn1N and Ten1, respectively. The angle between the axes of the two αC helices of Stn1 and Ten1 is ∼60°. As a consequence, only the crossover regions of the helices make extensive contacts with each other; hydrophobic residues from Stn1 (F190, W193, and M197) and Ten1 (L111 and M115) interdigitate with one another to form the core of the hydrophobic interface (Fig. 5B). At the N-terminal end of the αC helix of Stn1, the side chain of Stn1 L186 is positioned into a hydrophobic pocket of Ten1 formed by residues from helix αC, loop L5C (between β5 and αC), and strands β0, β1, and β4 (Fig. 5C). The β barrel of Stn1 makes much less direct contact with Ten1 and contributes only one hydrogen-bonding interaction between Stn1 K90 and Ten1 Y97 (Fig. 5A,D).
Figure 5.
The C. tropicalis Stn1N–Ten1 interface. (A) The hydrophobic interface between Stn1N and Ten1. (Left) Stn1N is in surface representation and colored according to its electrostatic potential (positive potential, blue; negative potential, red). Ten1 is in ribbon representation. (Right) Ten1 is in electrostatic surface representation, while Stn1N is in ribbon. The orientation of the complex is rotated by 180° about a vertical axis relative to the complex in the left panel. (B–D) Hydrophobic interactions (B,C) and electrostatic interactions (D) between Stn1N and Ten1. Side chains of residues important for interaction are shown as stick models and are colored as in A. The intermolecular hydrogen bonds are shown as dashed magenta lines. (E) Effects of the Stn1 and Ten1 mutations on the Stn1–Ten1 interaction in a yeast two-hybrid assay. Interaction of LexA–Stn1 with GAD–Ten1 was measured as β-galactosidase activity. Data are the average of three independent β-galactosidase measurements normalized to the wild-type Stn1–Ten1 interaction, arbitrarily set to 100.
In addition to hydrophobic contacts, hydrogen-bonding interactions appear also to strengthen the interface and contribute to the specificity of the Stn1–Ten1 complex. There are six intermolecular hydrogen bonds at the Stn1–Ten1 interface, all located at the periphery. Specifically, at the N-terminal end of the Stn1 αC helix, the carboxylate side chain of E189 makes two salt bridge interactions with the amino group of R27 in the Ten1 β1 strand (Fig. 5D). The R27 side chain also accepts two intramolecular hydrogen bonds from D83 and Y97 of Ten1 (Fig. 5D). Moreover, the side chain amino group of K90 of Stn1 donates another hydrogen bond to Y97 of Ten1 (Fig. 5D). Together, this elaborate electrostatic interaction network extends the contact interface area and helps to stabilize the relative orientation of Stn1 and Ten1 in the complex. Notably, both E189 of Stn1 and R27 of Ten1 are highly conserved in both families of proteins (Fig. 3C), consistent with their important roles in Stn1–Ten1 complex formation, as revealed by the crystal structure.
To corroborate our structural analysis, we examined whether missense mutations of residues at the interface of Stn1 and Ten1 could weaken or disrupt the Stn1–Ten1 interaction using yeast two-hybrid assay. Consistent with the crystal structure, we found that substitution of a hydrophobic residue (L186, F190, or W193) of Stn1 on the interface with alanine was sufficient to abolish its interaction with Ten1 (Fig. 5E). Similarly, Ten1 mutation M115A on the other side of the interface also impaired the interaction (Fig. 5E). In contrast, alanine substitutions of M197 of Stn1 and L111 of Ten1 at the interface still maintained the interaction (∼30%–40% of the wild-type level), suggesting that the side chains of these two residues are not crucial for the Stn1–Ten1 complex formation (Fig. 5E). Notably, disruption of the electrostatic interactions between E189 of Stn1 and R27 of Ten1 by alanine mutation of either residue was sufficient to abolish the Stn1–Ten1 interaction (Fig. 5E). Collectively, we conclude that both the hydrophobic and the electrostatic contacts observed in the crystal structure are important for the interaction between Stn1 and Ten1.
Functional analysis of the Stn1N–Ten1 interaction
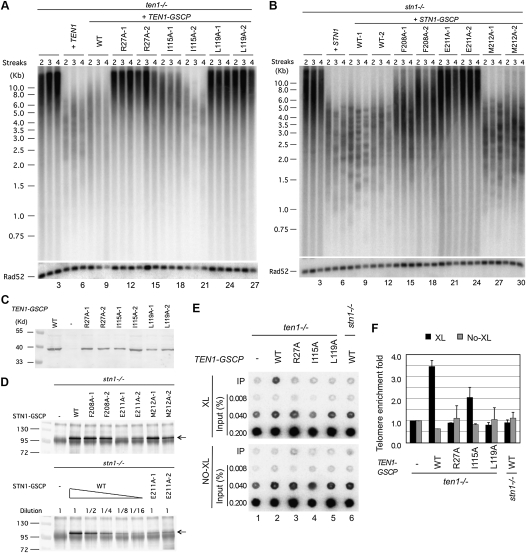
To assess the in vivo roles of the Stn1–Ten1 interaction in telomere regulation, we introduced several site-specific mutations in C. albicans STN1 and TEN1 designed to disrupt their contact interface based on the C. tropicalis Stn1N–Ten1 complex structure, and analyzed the phenotypes of the resulting mutants. To facilitate biochemical and genetic studies, each mutant allele was fused at its C terminus to a GSCP (Gly6–SBP–CBP–protein A) tag, which had little effect on the function of the wild-type gene in telomere regulation (Fig. 6A [cf. lanes 4–6 and 7–9], B [cf. lanes 4–6 and 7–12]). All three ten1 mutant proteins (R27A, I115A, and L119A [equivalent to C. tropicalis Ten1 R27A, L111A, and M115A]) as well as two of the stn1 mutant proteins (F208A and M212A [equivalent to C. tropicalis Stn1 L186A and F190A]) were expressed at near wild-type levels, suggesting that, in general, residues at the Stn1–Ten1 interface are not required for protein stability (Fig. 6C,D). The only exception was Stn1 E211A (equivalent to C. tropicalis Stn1 E189A), which was detected at ∼20% of the wild-type level (Fig. 6D, bottom panel). On the other hand, most mutants except Stn1 M212A exhibited significant loss of function with regard to telomere length regulation (Fig. 6A,B). Indeed, three of these mutants (Ten1 R27A, Ten1 L119A, and Stn1 E211A) manifested phenotypes that were as severe as the respective null mutant (Fig. 6A [lanes 10–15,22–27], B [lanes 19–24]). Thus, the interaction between Stn1 and Ten1 is evidently critical for telomere length regulation.
Figure 6.
The effects of point mutations designed to disrupt the C. albicans Stn1–Ten1 interaction on protein levels, telomere length regulation, and protein–telomere association. (A) Chromosomal DNAs were isolated from the ten1−/− mutant and various reconstituted strains after two to four streaks (∼50–100 generations) on plates and were subjected to telomere restriction fragment analysis.(Bottom) As loading controls, the telomere probe was stripped and the blot was rehybridized with a RAD52 fragment. (B) Chromosomal DNAs were isolated from the stn1−/− mutant and various reconstituted strains after two to four streaks (∼50–100 generations) on plates and were subjected to telomere restriction fragment analysis. (Bottom) As loading controls, the telomere probe was stripped and the blot was rehybridized with a RAD52 fragment. (C) Extracts were prepared from strains containing different GSCP-tagged Ten1 mutant proteins and were subjected directly to Western analysis using antibodies against protein A. (D, top) Extracts from strains bearing different GSCP-tagged Stn1 mutant proteins were subjected to affinity pull-down with IgG-Sepharose, followed by Western analysis using antibodies against protein A. (Bottom) To compare the levels of GSCP-tagged Stn1 and Stn1–E211A, we subjected the wild-type extract to serial dilutions prior to the assays. (E) ChIP analysis was performed using strains containing tagged wild-type or mutant Ten1 with (top) or without (bottom) formaldehyde cross-linking. Following immunoprecipitation, the levels of telomeric DNA in the immunoprecipitation and input samples were assessed by hybridization with a telomere-specific probe. (F) The relative enrichment of telomere DNA in the immunoprecipitation samples were quantified by dividing the immunoprecipitation signals against the input signals and were plotted; data are means ± SD of two independent experiments.
Next, the effects of mutations on the telomere association of Ten1–GSCP were assessed using ChIP. As shown in Figure 6, E and F, telomere DNA is precipitated from cell extracts in a GSCP tag-dependent and cross-linker-dependent manner, supporting the association of Ten1 with telomeres in vivo. This association was abolished in the stn1−/− background, suggesting that Ten1 is recruited to telomeres through its interaction with Stn1 (Fig. 6E, cf. lanes 2 and 6,). Moreover, all three point mutants of Ten1 exhibited partial to complete loss of telomere association in a manner that is entirely consistent with the severity of the telomere length defects (Fig. 6E,F, lanes 2–5). In particular, the L115A mutant, which retained partial function in suppressing telomere elongation, also exhibited the mildest telomere association defect (Fig. 6A,F). Notably, the equivalent mutation in C. tropicalis Ten1, L111A, also retained partial Stn1-binding ability in yeast two-hybrid assay (Fig. 5E). These findings reinforce the notion that the interaction between Stn1 and Ten1 is necessary for both Ten1 recruitment and telomere regulation. Unfortunately, while Stn1-GSCP also appears to be telomere-associated, the relatively low signal-to-noise ratios of the Stn1 ChIP results prevented us from obtaining reliable data on the telomere association of the stn1 mutants (data not shown). Further studies will be necessary to determine how the interactions between Stn1 and Ten1 influence the localization of Stn1.
Structural conservation between the C-terminal domains of Stn1 and Rpa2
Besides the N-terminal OB fold, sequence alignment revealed another conserved domain at the C terminus of Stn1 (henceforth referred to as Stn1C) (Fig. 3A; Supplemental Fig. S1). Stn1C interacts with both Cdc13 and Pol12 (Grossi et al. 2004; Puglisi et al. 2008). Notably, the C-terminal region of Rpa2 is also known to be a globular domain that contains a winged helix–turn–helix (WH) motif (Mer et al. 2000). This motif is composed of three α helices flanked by three β strands (Mer et al. 2000). Rpa2WH interacts with a myriad of protein factors essential for DNA replication, recombination, and repair (Fanning et al. 2006). Based on the observation that both Stn1C and Rpa2WH are located at the C termini and both mediate protein–protein interactions, we hypothesized that Stn1C might adopt an Rpa2WH-like WH fold conformation. However, no obvious sequence similarity could be detected between Stn1C and Rpa2WH. In addition, the size of Stn1C (∼200 amino acids) is almost three times that of Rpa2WH (∼70 amino acids) (Mer et al. 2000). Thus, it is unclear whether the structural similarity between Stn1 and Rpa2 could be extended to their C-terminal regions. To address this question, various Stn1C constructs from S. cerevisiae, C. albicans, and C. tropicalis were expressed and purified for structural characterization. After optimization by limited proteolysis and mass spectrometry analysis (data not shown), we succeeded in crystallizing S. cerevisiae Stn1C (residues 311–494) and determining its structure by SAD method using MeHgAc derivative crystals at a resolution of 2.1 Å (Fig. 7A; Supplemental Table S2). The calculated electron density map allowed unambiguous tracing of most of Stn1C except a disordered loop (residues 472–479).
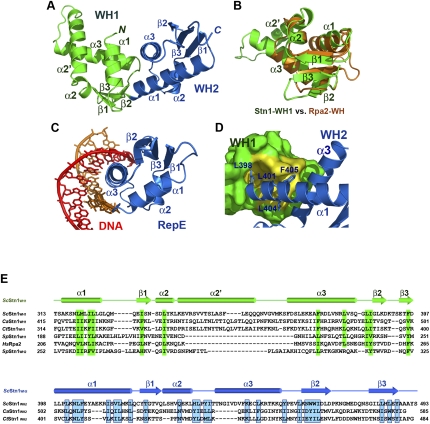
Figure 7.
Crystal structure of the C-terminal domain of S. cerevisiae Stn1. (A) Ribbon diagram of ScStn1C. The WH1 and WH2 motifs of ScStn1N are colored as in Figure 3A. The secondary structure elements are labeled. The dotted line represents the disordered loop (472–479) between strands β2 and β3 of WH2. Although sharing the same topology, the two WH motifs are quite different in structure. (B) Superposition of the WH1 motif of ScStn1 (in green) on the NMR (nuclear magnetic resonance) structure of the WH motif of Rpa2 (in orange) (Mer et al. 2000). Except for a large insertion between α2 and α3, the rest of Stn1WH1 closely resembles the WH motif of Rpa2. (C) Ribbon diagram of the RepE–DNA complex (Komori et al. 1999). The orientation of the WH motif of RepE is the same as the WH2 of ScStn1C in A. (D) The hydrophobic interactions between the WH1 and WH2 motifs of ScStn1C. There are no linker residues between WH1 and WH2, so that ScStnC folds into a globular and compact structure. WH1 is shown in surface representation and is colored in green, except for the WH2-interacting surface, shown in yellow. WH2 is in ribbon representation. Side chains of residues in WH2 important for the WH1–WH2 interaction are shown in stick representations. (E) Amino acid sequence alignment of the C-terminal WH1 and WH2 motifs of budding yeast Stn1 family members together with the WH motifs of S. pombe Stn1 and human Rpa2. The alignment with Rpa2 is based on the NMR structure of the Rpa2C–UNG2 complex (Mer et al. 2000). Secondary structure assignments from our ScStn1C crystal structure are shown. Conserved hydrophobic residues in WH1 and WH2 are highlighted in green and blue blocks, respectively. In contrast to the WH motifs in budding yeasts, both WH1 and WH2 of SpStn1 are similar to Rpa2WH.
Unexpectedly, Stn1C is composed of two topologically similar WH motifs that are related to each other by a noncrystallographic dyad, although no such similarity was expected from its primary sequence (Fig. 7A). Notably, the folding of the first WH motif, Stn1WH1, is indeed structurally similar to RPA32WH (Fig. 7B). The RMSD between the two WH motifs is 1.8 Å for 58 Cα atom pairs (Fig. 7B). One unique feature of Stn1WH1 is a large insertion (a 17-residue α2′ helix and an eight-residue L2′3 loop) between helices α2 and α3 (Fig. 7B,E). In contrast, α2 and α3 of Rpa2WH are connected by a short five-residue loop (Fig. 7B,E). This marked local variance explains the failure to detect similarity between the WH motifs of Stn1 and Rpa2 by bioinformatics analysis. Nevertheless, the striking structural resemblance between Stn1WH1 and Rpa2WH further supports the notion that Stn1 is an Rpa2-like telomeric protein.
Other than sharing a similar topology, the structure of Stn1WH2 is rather different from that of Stn1WH1 (Fig. 7A). Instead, Stn1WH2 is most similar to the DNA-binding WH motifs of the pur operon repressor (Bera et al. 2003) and RepE replication initiator (Komori et al. 1999). However, comparison of the crystal structures of Stn1C and the RepE–DNA complex indicates that Stn1WH2 would be unlikely to bind DNA due to the occlusion of its putative target site by Stn1WH1 (cf. Fig. 7A,C). This was further confirmed by an electrophoretic mobility shift assay in which Stn1C failed to exhibit binding to double-stranded telomeric DNAs even at a very high protein concentration (100 μM) (data not shown).
Several features of Stn1 appear to fix the relative orientation between the WH1 and WH2 motif and allow Stn1C to adopt a compact and globular structure resembling a single folded unit. First, the N terminus of Stn1WH2 is immediately adjacent to the end of Stn1WH1; there is no linker residue between β3 of WH1 and α1 of WH2 (Fig. 7A,E). In addition, L401, L404, and F405 in WH2 make contact with a hydrophobic surface formed by the WH1 helix α1 (Fig. 7D). Finally, the side chain of Leu398 in WH2 inserts into a deep hydrophobic pocket of WH1, further stabilizing the relative disposition of the two motifs (Fig. 7D). The twisted architecture of Stn1C gives rise to a large surface area for potential interactions with other proteins such as Cdc13 and Pol12, as suggested by earlier genetic studies (Grossi et al. 2004; Puglisi et al. 2008).
Crystal structure of fission yeast S. pombe Stn1N–Ten1 complex
The budding yeast CST complex has long been considered an evolutionary exception, as most other eukaryotic organisms use the POT1–TPP1 or a POT1–TPP1-like complex to bind G tails and protect telomeres (Gray et al. 1991; Horvath et al. 1998; Baumann and Cech 2001; Lei et al. 2003, 2004; Wang et al. 2007; Miyoshi et al. 2008). Recent studies have challenged this view. Putative Stn1 and Ten1 orthologs have been identified in a plethora of organisms ranging from fission yeast and plants to humans (Martin et al. 2007; Song et al. 2008; Miyake et al. 2009; Surovtseva et al. 2009; Wan et al. 2009). This suggests that the CST complex may be another conserved complex at the telomere G tails besides the well-characterized POT1–TPP1 complex. However, the sequences of the S. pombe Stn1 and Ten1 proteins are only weakly similar to those of the budding yeast proteins (Martin et al. 2007). Thus, whether SpStn1 and SpTen1 represent true homologs of the budding yeast proteins is unclear. To resolve this question, we reconstituted and crystallized the complex between full-length SpTen1 and the N-terminal putative OB fold of SpStn1 (SpStn1N, residues 2–186) and determined its structure at 1.65 Å resolution by SAD method using Se-Met-substituted proteins (Supplemental Table S3).
The crystal structure of the SpStn1N–SpTen1 complex reveals that both SpStn1N and SpTen1 are indeed made of an OB fold, and the complex adopts a three-dimensional architecture similar to the C. tropicalis Stn1N–Ten1 complex (Fig. 8A). The OB folds are closely conserved, with a Cα RMSD values of 2.2 Å between the OB folds of SpStn1N and CtStn1N and 2.0 Å between the OB folds of SpTen1 and CtTen1. Given C. tropicalis Stn1–Ten1 is an Rpa2–Rpa3-like complex, it is not unexpected that the structure of the SpStn1–SpTen1 complex also closely resembles that of Rpa2N–Rpa3. In fact, SpStn1N is structurally more similar to Rpa2N than to CtStn1; the Cα RMSD is only 1.6 Å between SpStn1N and Rpa2N.
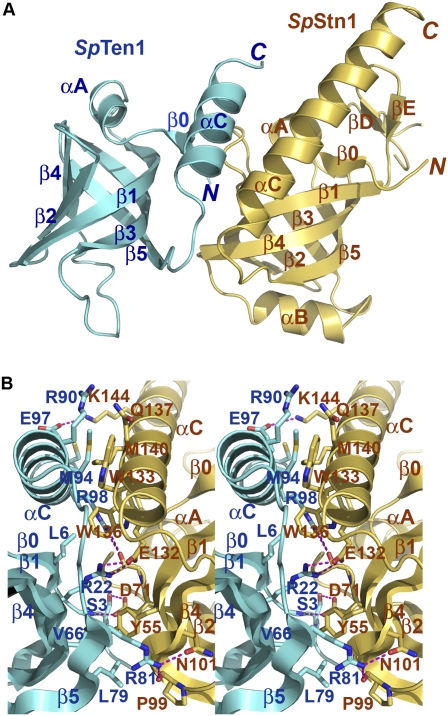
Figure 8.
Crystal structure of the S. pombe Stn1N–Ten1 complex. (A) Ribbon diagram of the SpStn1N–SpTen1 complex. SpStn1N and SpTen1 are colored in pale yellow and sky blue, respectively. The orientation of the complex is the same as that of the left C. tropicalis Stn1N–Ten1 complex in Figure 3B. (B) Stereo view of the SpStn1N–SpTen1 interface. SpStn1N- and SpTen1-interacting residues are presented as stick models. SpStn1N and SpTen1 are colored as in A. The intermolecular hydrogen bonds are shown as dashed magenta lines.
The interface between SpStn1N and SpTen1 involves both hydrophobic and electrostatic interactions (Fig. 8B). Compared with the C. tropicalis Stn1N–Ten1 complex, the most conserved feature is the hydrophobic packing between the two αC helices of SpTen1N and SpTen1, which appears to be the major driving force for complex formation (Fig. 8B). Unlike the C. tropicalis Stn1N–Ten1, electrostatic interactions contribute more to the SpStn1N–SpTen1 interface. There are a total of nine intermolecular electrostatic interactions between SpStn1N and SpTen1 (Fig. 8B). Except for the one between the side chains of SpStn1N E132 and SpTen1 R22, most of these electrostatic interactions are not present in the C. tropicalis Stn1N–Ten1 complex (Figs. 3C, 4D, 8B). Thus, the weak similarities between the Stn1 and Ten1 protein of fission yeast and budding yeast at the primary sequence level can be explained in part by the evolution of distinct interacting residues at the subunit interface.
Similar to budding yeast Stn1, SpStn1 also contains a C-terminal domain (SpStn1C). We performed a secondary structure prediction for SpStn1C using the program PredictProtein (Rost et al. 2004), which accurately predicted the positions of the α helices and β strands in the two WH motifs of ScStn1C (data not shown). The putative secondary structural elements in SpStn1C were then aligned with those present in ScStn1 and Rpa2 (Fig. 7E). This analysis identified two presumed WH motifs in SpStn1C (Fig. 7E). However, unlike budding yeast Stn1, both WH motifs in SpStn1 show a similar distribution of α helices and β strands that coincides well with CtStn1WH1 and Rpa2WH, suggesting that SpStn1 has two similar Rpa2-like WH motifs (Fig. 7E). Nonetheless, the detection of Rpa2-like WH motifs in SpStn1, together with the overall structural similarity between the fission yeast and budding yeast Stn1N–Ten1 complexes, strongly supports the notion that an Rpa2–Rpa3-like complex is also conserved at fission yeast telomeres.
Discussion
Our structural analyses demonstrate that both the budding yeast and the fission yeast Stn1–Ten1 complexes share the same three-dimensional architecture as the Rpa2–Rpa3 complex despite minimal sequence similarity, thus providing the first direct confirmation of structural similarity between components of the CST and the RPA complexes. The reliability of our structures was further corroborated by mutational analyses of Stn1 and Ten1, which underscored the importance of functional heterodimerization between Stn1 and Ten1 for telomere localization of Ten1 and telomere length regulation. Thus, our findings provide a foundation for leveraging insights from the analysis of RPA to the study of the CST complex.
Budding yeast was believed to have evolved a very different set of telomeric proteins to protect and maintain chromosome ends. Hence, the budding yeast CST complex has been considered to serve as the functional equivalent of the POT1–TPP1 complex in fission yeast and other POT1-containing organisms. However, putative homologs of the CST proteins have been identified recently in both plants and humans (Casteel et al. 2009; Miyake et al. 2009; Surovtseva et al. 2009), suggesting that this telomere regulatory complex is probably more widespread in nature than previously believed, even in organisms that use POT1 for telomere protection. On the other hand, the almost complete lack of sequence similarity between the CST components from budding yeast and POT1-containing organisms raised serious doubts concerning the structural and functional conservation of these proteins in these two groups of organisms. These doubts are now substantially alleviated by our structural data showing that the budding and fission yeast Stn1–Ten1 complexes share similar three-dimensional structures. As a consequence, insights from our structural studies are expected to provide a platform for functional studies of at least two components of the CST complexes in a wide range of organisms, including humans. In support of this notion, multiple sequence alignment indicates that the critical Glu–Arg interactions that we uncovered in the budding and fission yeast Stn1–Ten1 complexes (Stn1 E189–Ten1 R27 in C. tropicalis and Stn1 E132–Ten1 R22 in S. pombe) are likely to be conserved in both plants and mammals (data not shown). Nevertheless, it would be premature to extrapolate from the current findings to other features of the CST complexes. In particular, whether the remaining components of the CST complexes in different organisms (i.e., Cdc13 in yeast and Ctc1 in plants and humans) (Miyake et al. 2009; Surovtseva et al. 2009) resemble one another and whether they exhibit similarities to Rpa1 are largely unresolved. Clarifying these and other key issues in CST structure, assembly, and mechanisms will require detailed structural and functional analyses of the entire complex.
Materials and methods
Strains and plasmids
The C. albicans strain BWP17 (ura3Δ∷λimm434/ura3Δ∷λimm434 his1∷hisG/his1∷hisG arg4∷hisG/arg4∷hisG) was used as the parental strain (Wilson et al. 1999). The derivations of mutant strains are described below.
Construction of mutant Candida strains
The deletion strain stn1−/− was generated by subjecting BWP17 to two rounds of transformation and 5-FOA selection using a stn1∷hisG-URA3-hisG cassette (containing ∼700 bp of STN1 upstream and ∼700 bp of downstream sequence). Similarly, the deletion strain ten1−/− was generated by subjecting BWP17 to two rounds of transformation and 5-FOA selection using a ten1∷hisG-URA3-hisG cassette (containing ∼900 bp of TEN1 upstream and ∼900 bp of downstream sequence). The reconstituted strains stn1−/−∷STN1 and ten1−/−∷TEN1 were obtained by transforming the deletion strains with the pGEM-URA3-STN1 and pGEM-URA3-TEN1 integrating plasmid linearized by HpaI and HindIII digestion, respectively. The pGEM-URA3-STN1 plasmid contains a 3.1-kb fragment spanning the STN1 gene, while the pGEM-URA3-TEN1 plasmid contains a 2.1-kb fragment spanning the TEN1 gene, each cloned into the SalI and SacI sites of pGEM-URA3 (Wilson et al. 1999). Derivatives of the plasmids were used to introduce epitope-tagged STN1 and TEN1 into the deletion strains, as follows. The C terminus of each gene was mutated by QuikChange to introduce an AvrII and a BspEI restriction site, thus allowing the introduction of the GSCP tag, which contains a Gly8 linker, a streptavidin-binding peptide, a calmodulin-binding peptide, and a protein A tag (the complete sequence is available upon request). Alanine substitution mutants of tagged STN1 and TEN1 were generated by the same mutagenesis protocol.
The tert−/− ten1−/− and tert−/− ten1−/− rad50−/− mutants were constructed sequentially starting with a tert−/− mutant (Steinberg-Neifach and Lue 2006) using the aforementioned ten1∷hisG-URA3-hisG cassette and a rad50∷hisG-URA3-hisG cassette (containing ∼750 bp of RAD50 upstream and ∼700 bp of downstream sequence). C. albicans transformations and 5-FOA selections were carried out as described previously (Fonzi and Irwin 1993). Correct integrations of all disruption and reconstitution cassettes were confirmed by Southern analysis.
Analysis of telomeres and G-strand overhangs
Chromosomal DNAs were isolated by Smash and Grab as described previously except that the initial aqueous phase was subjected to one additional round of PCI (phenol/chloroform/isoamyl alcohol [25:24:1]) extraction to minimize nuclease contamination (Hoffman and Winston 1987). Standard telomere Southern analysis and the in-gel hybridization analysis were performed using established protocols (Hsu et al. 2007; Yu et al. 2008). The two-dimensional gel analysis was performed according to the protocol of Brewer and Fangman (1987) as modified by Cohen and Lavi (1996). Briefly, the first dimension (0.5% agarose) was run at 0.5 V/cm for 16 h in the absence of ethidium bromide (EtBr), while the second dimension (1.2% agarose) was run at 5 V/cm for 5 h in the presence of 0.3 μg/mL EtBr. The DNAs in the gels were transferred to nylon membrane and probed with labeled CaC2 oligonucleotides, as in the case of standard telomere Southern blots.
ChIP
ChIP was performed using a combination of previously described protocols with some additional modifications (Loayza and De Lange 2003; Yu et al. 2007). Cells were fixed with 1% formaldehyde for 30 min at 30°C and cross-linking was quenched with 125 mM glycine for 5 min at 30°C. Formaldehyde-fixed or untreated cells were resuspended in lysis buffer (50 mM HEPES at pH 7.5, 1 mM EDTA, 150 mM NaCl, protease inhibitors) and broken by glass beads. The lysates were sonicated 10 times for 5 sec each (constant duty cycle, 35%–40% output) to shear DNAs to a mean length of ∼600 bp. Extracts were adjusted to 1.6 mg/mL protein in 600 μL of lysis buffer and then diluted with 600 μL of immunoprecipitation dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl at pH 8.0, 450 mM NaCl, protease inhibitors). Five percent of each cell extract was set aside and used as the input sample. The remainder was subjected to immunoprecipitation using 20 μL of IgG-Sepharose beads for 2 h at 4°C. Immunoprecipitation samples were washed for 5 min with rotation in the following buffers; one time with Buffer A (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl at pH 8.0, 400 mM NaCl), four times with Buffer B (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl at pH 8.0, 600 mM NaCl), one time with Buffer C (0.25 M LiCl, 1% NP-40, 1% Na-Deoxycholate, 1 mM EDTA, 10 mM Tris-HCl at pH 8.0), and one time with TE. All wash buffers contain protease inhibitors. Immunoprecipitation samples were eluted in 500 μL of 1% SDS and 0.1 M NaHCO3 and cross-links were reversed for 5 h at 65°C. Samples were treated with RNase A and proteinase K, extracted with phenol/chloroform, precipitated with ethanol, and resuspended in 100 μL of water. The DNA samples were then applied to Hybond-N using a dot blot apparatus, and the membrane was probed with 32P-labeled CaC2 (CATCCGTACACCAAGAAGTTAGACATCCGTACACCAAGAAGTTAGA) corresponding to two copies of the C. albicans telomeric repeat. Signals were quantified using ImageQuant software (Molecular Dynamics, Inc.).
Western and IP-Western
Western and IP-Western were performed as described previously using antibodies directed against protein A (Yu et al. 2008).
Yeast two-hybrid assay
The yeast two-hybrid assays were performed using the L40 strain harboring pBTM116 and pACT2 (Clontech) fusion plasmids. The colonies containing both plasmids were selected on −Leu −Trp plates. β-Galactosidase activities were measured by liquid assay (Moretti et al. 1994).
Protein expression and purification
The N-terminal domains of C. tropicalis Stn1 (residues 2–217) and S. pombe Stn1N (residues 2–186) were cloned into a GST fusion protein expression vector, pGEX6p-1 (GE healthcare). C. tropicalis Ten1 (residues 2–123), S. pombe Ten1 (residues 2–102), and S. cerevisiae Stn1 C-terminal domain (residues 311–493) were cloned into a modified pET28b vector with a Sumo protein fused at the N terminus after the His6 tag (Wang et al. 2007).
The C. tropicalis Stn1N–Ten1 complex was coexpressed in Escherichia coli BL21(DE3). After induction for 16 h with 0.1 mM IPTG at 25°C, the cells were harvested by centrifugation and the pellets were resuspended in lysis buffer (50 mM Tris-HCl at pH 8.0, 50 mM NaH2PO4, 400 mM NaCl, 3 mM imidazole, 10% glycerol, 1 mM PMSF, 0.1 mg/mL lysozyme, 2 mM 2-mercaptoethanol, homemade protease inhibitor cocktail). The cells were then lysed by sonication and the cell debris was removed by ultracentrifugation. The supernatant was mixed with Ni-NTA agarose beads (Qiagen) and rocked for 6 h at 4°C before elution with 250 mM imidazole. Then, Ulp1 protease was added to remove the His6-Sumo tag. The complex was then mixed with glutathione sepharose beads (GE Healthcare) and rocked for 8 h at 4°C before elution with 15 mM glutathione. Protease 3C was added to remove the GST tag. Finally, the Stn1N–Ten1 complex was further purified by passage through Mono-Q ion exchange column and by gel-filtration chromatography on a Hiload Superdex200 equilibrated with 25 mM Tris-HCl (pH 8.0), 150 mM NaCl, and 5 mM dithiothreitol (DTT). The purified Stn1–Ten1 complex was concentrated to 15 mg/mL and stored at −80°C.
S. cerevisiae Stn1C and the S. pombe Stn1N–ten1 complex were expressed in E. coli and purified following the same procedure as described above except that only one affinity chromatography step (Ni-NTA agarose) was used for ScStn1C.
Crystallization, data collection, and structure determination
C. tropicalis Stn1N–Ten1
Crystals were grown by sitting drop vapor diffusion method at 4°C. The precipitant/well solution contained 1 M MgSO4, 0.1 M sodium citrate (pH 5.6), and 10 mM DTT. Heavy-atom derivatives were obtained by soaking crystals in a solution containing 1.5 M MgSO4 and 0.3 mM of MeHgAc for 2–3 h and backsoaking for 1 h in 1.25 M MgSO4, 1.4 M NaHCO2, and 0.1 M sodium citrate (pH 5.6). Both native and heavy-atom derivative crystals were gradually transferred into a harvesting solution (0.25 M MgSO4, 5.25 M NaHCO2, 0.1 M sodium critrate at pH 5.6) before being flash-frozen in liquid nitrogen for storage and data collection under cryogenic conditions (100 K). Native and Hg-SAD (at Hg peak wavelength) data sets were collected at beamline 21ID-D at APS and processed using HKL2000 (Otwinowski and Minor 1997). Crystals belong to space group P41212 and contain two Stn1N–Ten1 complexes per asymmetric unit. Native crystals diffracted to 2.4 Å resolution with cell parameter a = b = 92.072 Å and c = 200.909 Å. Six mercury sites were located and refined, and the SAD phases were calculated using SHARP (De La Fortelle and Bricogne 1997). The initial SAD map was significantly improved by solvent flattening. A model was automatically built into the modified experimental electron density using ARP/WARP (Lamzin et al. 2001); the model was then further refined using simulated annealing and positional refinement in CNS (Brunger et al. 1998) with manual rebuilding, using program O (Jones et al. 1991).
S. cerevisiae Stn1C
Crystals were grown by the sitting drop vapor diffusion method at 4°C. The precipitant/well solution contained 80 mM HEPES (pH 7.0), 8% PEG6K, 1.6 M NaCl, and 10 mM DTT. Heavy-atom derivatives were obtained by soaking crystals in a solution containing 25% PEG6K, 0.5 M NaCl, 0.1 M HEPES (pH7.1), and 0.3 mM MeHgAc for 2–3 h and backsoaking for 1 h in 25% PEG6K, 10% glycerol, 0.5 M NaCl, and 0.1 M HEPES (pH 7.1). Both native and heavy-atom derivative crystals were transferred gradually into a harvesting solution (25% PEG6K, 25% glycerol, 0.5 M NaCl, 0.1 M HEPES at pH 7.1) before being flash-frozen in liquid nitrogen for storage and data collection under cryogenic conditions (100 K). Native and Hg-SAD (at Hg peak wavelength) data sets were collected at beamline 21ID-D at APS and processed using HKL2000 (Otwinowski and Minor 1997). ScStn1C crystal belongs to space group P43212 and contains one molecule in asymmetric unit. Native crystals diffracted 2.1 Å resolution with cell parameter a = b = 52.957 Å, c = 186.397 Å and contains one molecule in asymmetric unit. Two mercury sites were located and refined, and the SAD phases were calculated using SHARP (De La Fortelle and Bricogne 1997). Model building and refinement were carried out following the same procedure as those for the C. tropicalis Stn1N–Ten1 complex.
S. pombe Stn1N–Ten1
The native and the Se-Met-substituted S. pombe Stn1N–Ten1 complex crystals were obtained using hanging drop vapor diffusion method at 4°C. The precipitant/well solution contained 12% PEG4K, 12% isopropanol, 0.1 M sodium citrate (pH 5.6), and 5 mM DTT. Crystals were transferred gradually into a harvesting solution containing 25% PEG 4K, 16% isopropanol, 0.1 M sodium citrate (pH 5.6), and 25% glycerol before being flash-frozen in liquid nitrogen for storage and data collection under cryogenic conditions (100 K). Native and Se-Met-SAD (at Se peak wavelength) data sets were collected at beam line 21ID-F at APS and were processed using HKL2000 (Otwinowski and Minor 1997). S. pombe Sten1N–ten1 complex crystals belong to space group P41212 and contain one complex per asymmetric unit. Native data set diffracted to 1.65 Å resolution with unit cell parameters a = b= 93.871 Å and c = 56.273 Å. Seven selenium atoms were located and refined, and the SAD phases were calculated using SHARP (De La Fortelle and Bricogne 1997). Model building and refinement were carried out following the same procedure as those for the C. tropicalis Stn1N–Ten1 complex.
Accession numbers
The coordinates and structure factors of the C. tropicalis Stn1N–Ten1 complex, S. cerevisiae Stn1C, and the S. pombe Stn1N–Ten1 complex have been deposited in the RCSB Protein Data Bank under accession codes 3KF8, 3KEY, and 3KF6, respectively.
Acknowledgments
We thank Y. Chen and F. Wang for help at various stages of the project. This work was supported by NIH grants (GM 083015-01 to M.L. and GM069507 to N.L.), an American Cancer Society Research Scholar grant, and a Sidney Kimmel Scholar Award (to M.L.). M.L. is a Howard Hughes Medical Institute Early Career Scientist. N.F.L. thanks Drs. Ting-Fang Wang and Chung Wang (Academia Sinica, Taiwan) for support during his summer 2007 visit, on which occasion some of the works described here were initiated. Use of Life Sciences Collaborative Access Team Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor (grant 085P1000817). Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under contract number DE-AC02-06CH11357.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1851909.
Supplemental material is available at http://www.genesdev.org.
References
- Andaluz E, Ciudad T, Gomez-Raja J, Calderone R, Larriba G. Rad52 depletion in Candida albicans triggers both the DNA-damage checkpoint and filamentation accompanied by but independent of expression of hypha-specific genes. Mol Microbiol. 2006;59:1452–1472. doi: 10.1111/j.1365-2958.2005.05038.x. [DOI] [PubMed] [Google Scholar]
- Baumann P, Cech TR. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science. 2001;292:1171–1175. doi: 10.1126/science.1060036. [DOI] [PubMed] [Google Scholar]
- Bechard LH, Butuner BD, Peterson GJ, McRae W, Topcu Z, McEachern MJ. Mutant telomeric repeats in yeast can disrupt the negative regulation of recombination-mediated telomere maintenance and create an alternative lengthening of telomeres-like phenotype. Mol Cell Biol. 2009;29:626–639. doi: 10.1128/MCB.00423-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bera AK, Zhu J, Zalkin H, Smith JL. Functional dissection of the Bacillus subtilis pur operator site. J Bacteriol. 2003;185:4099–4109. doi: 10.1128/JB.185.14.4099-4109.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertuch AA, Lundblad V. The maintenance and masking of chromosome termini. Curr Opin Cell Biol. 2006;18:247–253. doi: 10.1016/j.ceb.2006.04.005. [DOI] [PubMed] [Google Scholar]
- Bianchi A, Shore D. How telomerase reaches its end: Mechanism of telomerase regulation by the telomeric complex. Mol Cell. 2008;31:153–165. doi: 10.1016/j.molcel.2008.06.013. [DOI] [PubMed] [Google Scholar]
- Biswas K, Rieger KJ, Morschhauser J. Functional analysis of CaRAP1, encoding the Repressor/activator protein 1 of Candida albicans. Gene. 2003;307:151–158. doi: 10.1016/s0378-1119(03)00456-6. [DOI] [PubMed] [Google Scholar]
- Bochkarev A, Bochkareva E. From RPA to BRCA2: Lessons from single-stranded DNA binding by the OB-fold. Curr Opin Struct Biol. 2004;14:36–42. doi: 10.1016/j.sbi.2004.01.001. [DOI] [PubMed] [Google Scholar]
- Bochkarev A, Bochkareva E, Frappier L, Edwards AM. The crystal structure of the complex of replication protein A subunits RPA32 and RPA14 reveals a mechanism for single-stranded DNA binding. EMBO J. 1999;18:4498–4504. doi: 10.1093/emboj/18.16.4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer BJ, Fangman WL. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell. 1987;51:463–471. doi: 10.1016/0092-8674(87)90642-8. [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Casteel DE, Zhuang S, Zeng Y, Perrino FW, Boss GR, Goulian M, Pilz RB. A DNA polymerase-α • primase cofactor with homology to replication protein A-32 regulates DNA replication in mammalian cells. J Biol Chem. 2009;284:5807–5818. doi: 10.1074/jbc.M807593200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S, Lavi S. Induction of circles of heterogeneous sizes in carcinogen-treated cells: Two-dimensional gel analysis of circular DNA molecules. Mol Cell Biol. 1996;16:2002–2014. doi: 10.1128/mcb.16.5.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper JA, Hiraoka Y. Fission yeast telomeres. In: de Lange T, et al., editors. Telomeres and telomerase. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2006. pp. 495–523. [Google Scholar]
- De La Fortelle E, Bricogne G. Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Meth Enzymol. 1997;276:472–494. doi: 10.1016/S0076-6879(97)76073-7. [DOI] [PubMed] [Google Scholar]
- Enloe B, Diamond A, Mitchell AP. A single-transformation gene function test in diploid Candida albicans. J Bacteriol. 2000;182:5730–5736. doi: 10.1128/jb.182.20.5730-5736.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanning E, Klimovich V, Nager AR. A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res. 2006;34:4126–4137. doi: 10.1093/nar/gkl550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira MG, Miller KM, Cooper JP. Indecent exposure: When telomeres become uncapped. Mol Cell. 2004;13:7–18. doi: 10.1016/s1097-2765(03)00531-8. [DOI] [PubMed] [Google Scholar]
- Fonzi WA, Irwin MY. Isogenic strain construction and gene mapping in Candida albicans. Genetics. 1993;134:717–728. doi: 10.1093/genetics/134.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H, Cervantes RB, Mandell EK, Otero JH, Lundblad V. RPA-like proteins mediate yeast telomere function. Nat Struct Mol Biol. 2007;14:208–214. doi: 10.1038/nsmb1205. [DOI] [PubMed] [Google Scholar]
- Garvik B, Carson M, Hartwell L. Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol Cell Biol. 1995;15:6128–6138. doi: 10.1128/mcb.15.11.6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschling DE, Zakian VA. Telomere proteins: Specific recognition and protection of the natural termini of Oxytricha macronuclear DNA. Cell. 1986;47:195–205. doi: 10.1016/0092-8674(86)90442-3. [DOI] [PubMed] [Google Scholar]
- Grandin N, Damon C, Charbonneau M. Ten1 functions in telomere end protection and length regulation in association with Stn1 and Cdc13. EMBO J. 2001;20:1173–1183. doi: 10.1093/emboj/20.5.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray JT, Celander DW, Price CM, Cech TR. Cloning and expression of genes for the Oxytricha telomere-binding protein: Specific subunit interactions in the telomeric complex. Cell. 1991;67:807–814. doi: 10.1016/0092-8674(91)90075-a. [DOI] [PubMed] [Google Scholar]
- Grossi S, Puglisi A, Dmitriev PV, Lopes M, Shore D. Pol12, the B subunit of DNA polymerase α, functions in both telomere capping and length regulation. Genes & Dev. 2004;18:992–1006. doi: 10.1101/gad.300004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman CS, Winston F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 1987;57:267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- Holm L, Sander C. Database algorithm for generating protein backbone and side-chain co-ordinates from a C α trace application to model building and detection of co-ordinate errors. J Mol Biol. 1991;218:183–194. doi: 10.1016/0022-2836(91)90883-8. [DOI] [PubMed] [Google Scholar]
- Horvath MP, Schweiker VL, Bevilacqua JM, Ruggles JA, Schultz SC. Crystal structure of the Oxytricha nova telomere end binding protein complexed with single strand DNA. Cell. 1998;95:963–974. doi: 10.1016/s0092-8674(00)81720-1. [DOI] [PubMed] [Google Scholar]
- Hsu M, McEachern MJ, Dandjinou AT, Tzfati Y, Orr E, Blackburn EH, Lue NF. Telomerase core components protect Candida telomeres from aberrant overhang accumulation. Proc Natl Acad Sci. 2007;104:11682–11687. doi: 10.1073/pnas.0700327104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer S, Chadha AD, McEachern MJ. A mutation in the STN1 gene triggers an alternative lengthening of telomere-like runaway recombinational telomere elongation and rapid deletion in yeast. Mol Cell Biol. 2005;25:8064–8073. doi: 10.1128/MCB.25.18.8064-8073.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaaed M. Improved methods for building protein models in electron density maps and the location of errors in these methods. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Komori H, Matsunaga F, Higuchi Y, Ishiai M, Wada C, Miki K. Crystal structure of a prokaryotic replication initiator protein bound to DNA at 2.6 A resolution. EMBO J. 1999;18:4597–4607. doi: 10.1093/emboj/18.17.4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamzin VS, Perrakis A, Wilson KS. The ARP/WARP suite for automated construction and refinement of protein models. In: Rossmann MG, Arnold E, editors. International tables for crystallography Volume F: Crystallography of biological macromolecules. Dordrecht, Kluwer Academic Publishers; The Netherlands: 2001. pp. 720–722. [Google Scholar]
- Legrand M, Chan CL, Jauert PA, Kirkpatrick DT. Role of DNA mismatch repair and double-strand break repair in genome stability and antifungal drug resistance in Candida albicans. Eukaryot Cell. 2007;6:2194–2205. doi: 10.1128/EC.00299-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei M, Podell ER, Baumann P, Cech TR. DNA self-recognition in the structure of Pot1 bound to telomeric single-stranded DNA. Nature. 2003;426:198–203. doi: 10.1038/nature02092. [DOI] [PubMed] [Google Scholar]
- Lei M, Podell ER, Cech TR. Structure of human POT1 bound to telomeric single-stranded DNA provides a model for chromosome end-protection. Nat Struct Mol Biol. 2004;11:1223–1229. doi: 10.1038/nsmb867. [DOI] [PubMed] [Google Scholar]
- Loayza D, De Lange T. POT1 as a terminal transducer of TRF1 telomere length control. Nature. 2003;423:1013–1018. doi: 10.1038/nature01688. [DOI] [PubMed] [Google Scholar]
- Martin V, Du LL, Rozenzhak S, Russell P. Protection of telomeres by a conserved Stn1-Ten1 complex. Proc Natl Acad Sci. 2007;104:14038–14043. doi: 10.1073/pnas.0705497104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEachern MJ, Blackburn EH. A conserved sequence motif within the exceptionally diverse telomeric sequences of budding yeasts. Proc Natl Acad Sci. 1994;91:3453–3457. doi: 10.1073/pnas.91.8.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEachern MJ, Haber JE. Break-induced replication and recombinational telomere elongation in yeast. Annu Rev Biochem. 2006;75:111–135. doi: 10.1146/annurev.biochem.74.082803.133234. [DOI] [PubMed] [Google Scholar]
- Mer G, Bochkarev A, Gupta R, Bochkareva E, Frappier L, Ingles CJ, Edwards AM, Chazin WJ. Structural basis for the recognition of DNA repair proteins UNG2, XPA, and RAD52 by replication factor RPA. Cell. 2000;103:449–456. doi: 10.1016/s0092-8674(00)00136-7. [DOI] [PubMed] [Google Scholar]
- Miyake Y, Nakamura M, Nabetani A, Shimamura S, Tamura M, Yonehara S, Saito M, Ishikawa F. RPA-like mammalian Ctc1–Stn1–Ten1 complex binds to single-stranded DNA and protects telomeres independently of the Pot1 pathway. Mol Cell. 2009;36:193–206. doi: 10.1016/j.molcel.2009.08.009. [DOI] [PubMed] [Google Scholar]
- Miyoshi T, Kanoh J, Saito M, Ishikawa F. Fission yeast Pot1–Tpp1 protects telomeres and regulates telomere length. Science. 2008;320:1341–1344. doi: 10.1126/science.1154819. [DOI] [PubMed] [Google Scholar]
- Moretti P, Freeman K, Coodly L, Shore D. Evidence that a complex of SIR proteins interacts with the silencer and telomere-binding protein RAP1. Genes & Dev. 1994;8:2257–2269. doi: 10.1101/gad.8.19.2257. [DOI] [PubMed] [Google Scholar]
- Nabetani A, Ishikawa F. Unusual telomeric DNAs in human telomerase-negative immortalized cells. Mol Cell Biol. 2009;29:703–713. doi: 10.1128/MCB.00603-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann A, Reddel R. Telomerase independent maintenance of mammalian telomeres. In: de Lange T, et al., editors. Telomeres and telomerase. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2006. pp. 163–198. [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. In: Carter CW Jr, Sweet RM, editors. Method in enzymology, vol. 26. Academic Press; San Diego: 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu Rev Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- Petreaca RC, Chiu HC, Eckelhoefer HA, Chuang C, Xu L, Nugent CI. Chromosome end protection plasticity revealed by Stn1p and Ten1p bypass of Cdc13p. Nat Cell Biol. 2006;8:748–755. doi: 10.1038/ncb1430. [DOI] [PubMed] [Google Scholar]
- Puglisi A, Bianchi A, Lemmens L, Damay P, Shore D. Distinct roles for yeast Stn1 in telomere capping and telomerase inhibition. EMBO J. 2008;27:2328–2339. doi: 10.1038/emboj.2008.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rost B, Yachdav G, Liu J. The PredictProtein server. Nucleic Acids Res. 2004;32:W321–W326. doi: 10.1093/nar/gkh377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Leehy K, Warrington RT, Lamb JC, Surovtseva YV, Shippen DE. STN1 protects chromosome ends in Arabidopsis thaliana. Proc Natl Acad Sci. 2008;105:19815–19820. doi: 10.1073/pnas.0807867105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg-Neifach O, Lue NF. Modulation of telomere terminal structure by telomerase components in Candida albicans. Nucleic Acids Res. 2006;34:2710–2722. doi: 10.1093/nar/gkl345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surovtseva YV, Churikov D, Boltz KA, Song X, Lamb JC, Warrington R, Leehy K, Heacock M, Price CM, Shippen DE. Conserved telomere maintenance component 1 interacts with STN1 and maintains chromosome ends in higher eukaryotes. Mol Cell. 2009;36:207–218. doi: 10.1016/j.molcel.2009.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira MT, Gilson E. Telomere maintenance, function and evolution: The yeast paradigm. Chromosome Res. 2005;13:535–548. doi: 10.1007/s10577-005-0999-0. [DOI] [PubMed] [Google Scholar]
- Wan M, Qin J, Songyang Z, Liu D. OB fold-containing protein 1 (OBFC1), a human homolog of yeast Stn1, associates with TPP1 and is implicated in telomere length regulation. J Biol Chem. 2009;284:26725–26731. doi: 10.1074/jbc.M109.021105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Podell ER, Zaug AJ, Yang Y, Baciu P, Cech TR, Lei M. The POT1–TPP1 telomere complex is a telomerase processivity factor. Nature. 2007;445:506–510. doi: 10.1038/nature05454. [DOI] [PubMed] [Google Scholar]
- Wilson RB, Davis D, Mitchell AP. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J Bacteriol. 1999;181:1868–1874. doi: 10.1128/jb.181.6.1868-1874.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wold MS. Replication protein A: A heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu Rev Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- Xin H, Liu D, Wan M, Safari A, Kim H, Sun W, O'Connor MS, Songyang Z. TPP1 is a homologue of ciliate TEBP-β and interacts with POT1 to recruit telomerase. Nature. 2007;445:559–562. doi: 10.1038/nature05469. [DOI] [PubMed] [Google Scholar]
- Yu EY, Steinberg-Neifach O, Dandjinou AT, Kang F, Morrison AJ, Shen X, Lue NF. Regulation of telomere structure and functions by subunits of the INO80 chromatin remodeling complex. Mol Cell Biol. 2007;27:5639–5649. doi: 10.1128/MCB.00418-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu EY, Wang F, Lei M, Lue NF. A proposed OB-fold with a protein-interaction surface in Candida albicans telomerase protein Est3. Nat Struct Mol Biol. 2008;15:985–989. doi: 10.1038/nsmb.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]