Abstract
Bacterial pathogens frequently use protein secretion to mediate interactions with their hosts. Here we found that a virulence locus (HSI-I) of Pseudomonas aeruginosa encodes a protein secretion apparatus. The apparatus assembled in discrete subcellular locations and exported Hcp1, a hexameric protein that forms rings with a 40 angstrom internal diameter. Regulatory patterns of HSI-I suggested that the apparatus functions during chronic infections. We detected Hcp1 in pulmonary secretions of cystic fibrosis (CF) patients and Hcp1-specific antibodies in their sera. Thus, HSI-I likely contributes to the pathogenesis of P. aeruginosa in CF patients. HSI-I–related loci are widely distributed among bacterial pathogens and may play a general role in mediating host interactions.
Pseudomonas aeruginosa is an opportunistic pathogen that chronically infects the lungs of >80% of cystic fibrosis patients and is the primary cause of morbidity and mortality in these patients (1). A distinguishing feature of the bacterium is its high degree of versatility, which provides P. aeruginosa sufficient phenotypic plasticity to form both acute and chronic infections in humans (2, 3). The choice between these disparate life-styles is governed by global virulence regulators, including RetS (regulator of exopolysaccharide and type III secretion) (4) and LadS (lost adherence sensor) (5). RetS and LadS reciprocally regulate virulence determinants such as type III secretion, which is RetS-activated and LadS-repressed, as well as exopolysaccharide production, which is RetS-repressed and LadS-activated. These virulence factors are important in acute and chronic infections, respectively.
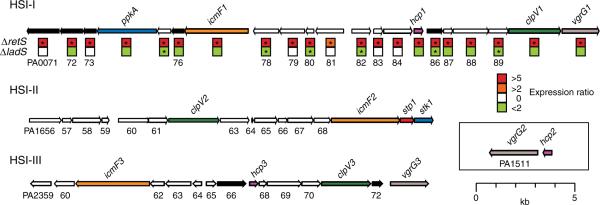
In addition to characterized virulence pathways, microarray analyses indicated that RetS and LadS reciprocally regulated a functionally uncharacterized virulence locus (Fig. 1). Consistent with its regulatory patterns by RetS and LadS, the virulence locus was required for chronic P. aeruginosa infection of the rat lung (Fig. 1) (6). This RetS- and LadS-regulated locus is highly homologous to a group of genes found in many Gram-negative proteobacteria that have been termed the IcmF-associated homologous protein (IAHP) cluster (7). P. aeruginosa encodes two other IAHP-related loci elsewhere in its genome; however, these loci were not regulated by either RetS or LadS and have no known role in virulence (Fig. 1). An overview of the distribution and genetic constituents of IAHP loci is shown (table S1).
Fig. 1.
Overview of P. aeruginosa HSI genes and reciprocal regulation of HSI-I by RetS and LadS. Conserved hypothetical HSI ORFs not discussed in the text (white) and ORFs that lie within the predicted HSI operons (black) are labeled with their genome annotation ORF number [assigned on the basis of (7)]. Predicted paralogous ORFs with prior characterization and those characterized in this study are colored consistently in each locus. The boxed insert shows the position of the hcp2/vgrG2 locus encoded elsewhere in the genome. The boxes beneath HSI-I genes summarize transcriptional profiling data from two prior studies (4, 5). The asterisks denote measurements that meet the statistical significance threshold defined in each study.
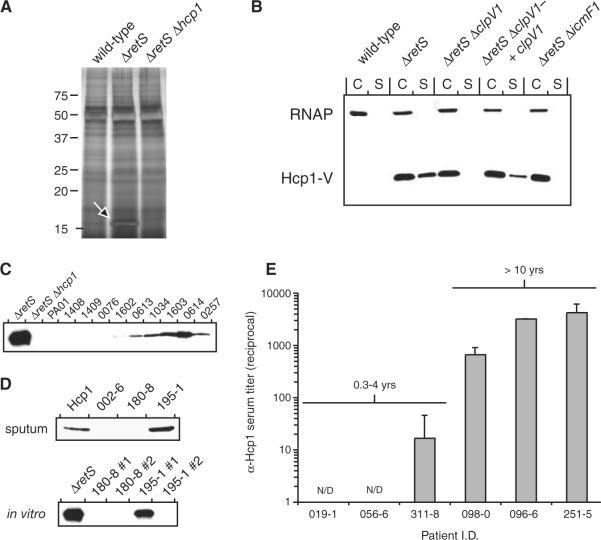
The IAHP-related locus of Vibrio cholerae, which the authors have designated a type VI secretion system, mediates cytotoxicity in phagocytic cells and is required for the extracellular secretion of four proteins lacking canonical hydrophobic amino-terminal signal sequences (8). We postulated that the RetS- and LadS–regulated IAHP locus in P. aeruginosa could play a similar role in extracellular protein targeting. To test this hypothesis, we activated expression of the locus in P. aeruginosa PAO1 by deleting retS. Comparison of the supernatant fractions of ΔretS and wild-type revealed that a small protein (Mr ~18 kD) was abundantly secreted by ΔretS (Fig. 2A). In-gel digestion followed by tandem mass spectrometry identified the protein as Hcp1 (PA0085), a result that was confirmed by deleting hcp1 in ΔretS (Fig. 2A). Notably, Hcp1 does not have a recognizable signal sequence (9) and is orthologous to one of the four proteins secreted by V. cholerae in an IAHP-dependent manner (8). Also, the gene encoding Hcp1 resides in the IAHP-related virulence locus regulated by RetS and LadS (Fig. 1). We will thus refer to this locus as P. aeruginosa Hcp1 secretion island I (HSI-I in Fig. 1).
Fig. 2.
P. aeruginosa HSI-I is required for secretion of Hcp1 and is active in cystic fibrosis infections. (A) Hcp1 is hypersecreted by ΔretS. SDS–polyacrylamide gel electrophoresis analysis of concentrated culture supernatants from various P. aeruginosa strains. The arrow highlights the position of secreted Hcp1 in ΔretS. (B) Immunoblot analysis of HSI-I–dependent secretion of Hcp1-V. In addition to the genetic alterations indicated, each strain contains hcp1-V. Equal quantities of cell (C) and supernatant (S) fractions were probed with antibodies specific for the β-subunit of RNA polymerase (RNAP) and the VSV-G epitope. (C) Immunoblot analysis of Hcp1 secretion by control strains and a panel of CF patient clinical isolates. (D) Immunoblot analysis of Hcp1 in sputum from CF patients (upper blot). Sputum sample 002-6 is from a CF patient not infected with P. aeruginosa; sputum sample 180-8 is from a CF patient infected with two P. aeruginosa strains that do not secrete Hcp1 (lower blot); and sputum sample 195-1 is from a CF patient infected with a P. aeruginosa strain that actively secretes Hcp1 and a second that does not (lower blot). (E) ELISA analysis of sera from CF patients for antibody response against Hcp1.
To assess more quantitatively the production and localization of Hcp1, we constructed a C-terminal chromosomal fusion of the vesicular stomatitis virus glycoprotein (VSV-G) epitope to hcp1 (hcp1-V). The VSV-G epitope did not affect Hcp1 overexpression or secretion in ΔretS (Fig. 2B). Detection of Hcp1 in ΔretS supernatants was not due to cell lysis, as determined by an intracellular protein control (Fig. 2B). The fraction of secreted Hcp1-V relative to intracellular Hcp1-V was lower in wild-type than in ΔretS (fig. S1),which suggests that, in addition to repression of hcp1, RetS negatively regulates Hcp1 secretion.
Next, we tested the dependence of Hcp1 secretion by ΔretS on HSI-I components by generating a deletion of icmF1 in the ΔretS hcp1-V background and examining the cellular localization of Hcp1 (Fig. 2B). IcmF is required for Legionella pneumophila type IVB-dependent secretion of SidC (10), and the V. cholerae IcmF homolog (VasK) is required for Hcp secretion (8); thus, we hypothesized that icmF1 would be essential for HSI-I-dependent Hcp1 secretion. Although deletion of icmF1 did not lower cellular Hcp1-V levels relative to ΔretS, secreted Hcp1-V levels were dramatically reduced (Fig. 2B). This result supports the requirement of an intact HSI-I for Hcp1 secretion.
To address whether secretion of Hcp1 occurs naturally in P. aeruginosa, we used Hcp1-specific antibodies to examine Hcp1 secretion in a collection of CF patient P. aeruginosa isolates (Fig. 2C, table S2). Hcp1 secretion varied among isolates: Robust secretion was detected in some isolates, yet was below the limit of detection in others. Mass spectrometric analysis was used to confirm the secretion of Hcp1 from clinical isolates.
Because our analysis of Hcp1 secretion in CF isolates required their growth in vitro, which might artificially stimulate Hcp1 secretion, we assayed Hcp1 secretion directly in CF patient sputum. Hcp1 was readily detected in sputum from a patient infected with P. aeruginosa actively secreting Hcp1 in vitro (195–1 no. 1 in Fig. 2D and fig. S2); however, the protein was not detected in sputum from an uninfected patient (002–6) or a patient infected with P. aeruginosa strains not secreting Hcp1 (180–8 in Fig. 2D). To determine whether Hcp1 secretion might have immunological consequences, we assayed for Hcp1-specific antibodies in the sera of six CF patients. At the time of sampling, three of these patients had maintained chronic P. aeruginosa infections lasting more than 10 years and three had been infected with P. aeruginosa for 0.3 to 4 years. A robust and specific Hcp1 response was detected in the three patients chronically infected with P. aeruginosa, whereas only a weak response was detected in one of the patients infected for a shorter duration (Fig. 2E and table S2). Thus, Hcp1 is actively secreted within the lungs of CF patients who have had long-term infection, which supports a role for HSI-I in chronic P. aeruginosa infections.
The IAHP-related gene clusters include an open reading frame (ORF), clpV, encoding a protein with strong homology to the AAA+ family protein ClpB (Fig. 1, and table S1, fig. S2). ClpB is required for thermotolerance and translocates aggregated proteins in an energy-dependent manner through its central channel (11). Because AAA+ family adenosine triphosphatases (ATPases) are hypothesized to provide the force for substrate translocation in many bacterial secretion systems (12, 13), we speculated that P. aeruginosa ClpV1 may function similarly for the purpose of secretion rather than disaggregation. Indeed, the IAHP ClpV proteins of enteropathogenic Escherichia coli and Salmonella typhimurium form hexameric structures that have ATPase activity, but lack protein disaggregase activity (14).
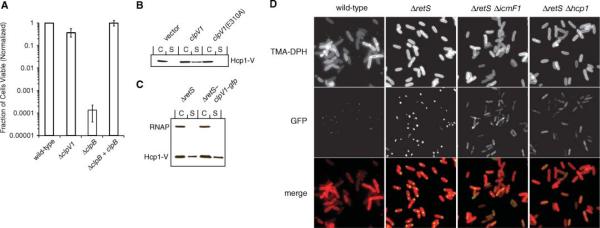
To distinguish the functions of ClpB and ClpV1 in P. aeruginosa, we determined the thermotolerance of ΔclpB and ΔclpV1. Consistent with our hypothesis, deletion of clpB resulted in thermosensitivity, whereas deletion of clpV1 did not affect thermotolerance (Fig. 3A). Furthermore, deletion of clpV1 in ΔretS hcp1-V abrogated Hcp1-V secretion without altering Hcp1-V levels in the cellular fraction (Fig. 2B). To determine whether the adenosine triphosphate (ATP) hydrolytic activity of ClpV1 is essential for the mechanism of Hcp1 secretion, we inactivated the ATP hydrolysis activity of the AAA-1 domain of ClpV1 by introducing a mutation into the Walker B motif [Glu310 replaced by Ala (E310A)]. Mutation of the analogous residue of ClpB (E279A) significantly decreased its activity, both in vitro and in vivo (15). When ClpV1E310A was expressed at levels similar to that of ClpV1 in ΔclpV1 ΔretS, the mutant protein failed to complement the defect in Hcp1 secretion (Fig. 3B). Thus, ATP hydrolysis by the AAA-1 domain of ClpV1 is required for Hcp1 secretion. We propose that ClpV1 functions critically as the energy source facilitating HSI-I–dependent Hcp1 secretion.
Fig. 3.
ClpV1 is not involved in thermotolerance and localizes to discrete foci in a manner dependent on IcmF and Hcp1. (A) Thermotolerance assay of P. aeruginosa strains bearing a clpB or clpV1 deletion. Cell viability before and after a 25-min heat pulse at 55°C was determined by colony-forming units. The thermotolerance of each strain was normalized relative to wild-type. (B and C) Immunoblot analysis of Hcp1-V secretion by ClpV1 and the AAA-1 mutant ClpV1E310A (B) and in ΔretS clpV1-gfp (C). (D) Fluorescence microscopy of indicated strains also bearing clpV1-gfp. TMA-DPH is a membrane dye used to highlight the outline of the cells.
As an essential component of the secretion apparatus encoded by HSI-I, we reasoned that the function of ClpV1 should be reflected in its subcellular localization. To visualize ClpV1, we generated a strain carrying a chromosomal C-terminal fusion of the green fluorescent protein (GFP) to ClpV1 (ClpV1-GFP) in the ΔretS background (ΔretS clpV1-gfp). Hcp1-V secretion was not affected by fusing GFP to ClpV1, and the fusion protein remained intact (Fig. 3C and fig. S3). ClpV1-GFP localized to single discrete foci in the majority of ΔretS clpV1-gfp cells (Fig. 3D). Although less intense, a similar pattern of punctate localization was observed in the wild-type background (Fig. 3D).
To establish whether the punctate localization of ClpV1-GFP was functionally relevant, we deleted icmF1 in the ΔretS clpV1-gfp background and measured the effect on ClpV1-GFP localization. Deletion of icmF1 dramatically reduced the number of cells with discrete ClpV1-GFP foci. Instead, the fluorescent signal in this mutant strain was most often evenly distributed across the cell and occasionally detected as weak foci (Fig. 3D). We next questioned whether Hcp1 might also be required for proper localization of ClpV1-GFP. To address this, we generated an hcp1 deletion in the ΔretS clpV1-gfp background and assessed ClpV1-GFP localization. Deletion of hcp1 in these cells resulted in a diffuse localization pattern of ClpV1-GFP similar to that observed in the ΔretS ΔicmF1 background, although the frequency of residual punctate foci was decreased in cells lacking Hcp1 relative to those lacking IcmF1 (Fig. 3D). Neither the hcp1 nor icmF1 mutations in the ΔretS clpV1-gfp background affected the stability or expression level of ClpV1-GFP (fig. S3). Thus, IcmF1 and Hcp1 are required for punctate localization of ClpV1-GFP. We interpret the punctate localization pattern of ClpV1-GFP to indicate the presence of an HSI-I–encoded secretion apparatus. IcmF1 appears to be required for efficient assembly of the apparatus, whereas Hcp1 is absolutely required for assembly. Given that wild-type P. aeruginosa fails to secrete Hcp1 but maintains ClpV1-GFP localization, the presence of Hcp1, but not its secretion, appears to be required for punctate ClpV1-GFP localization.
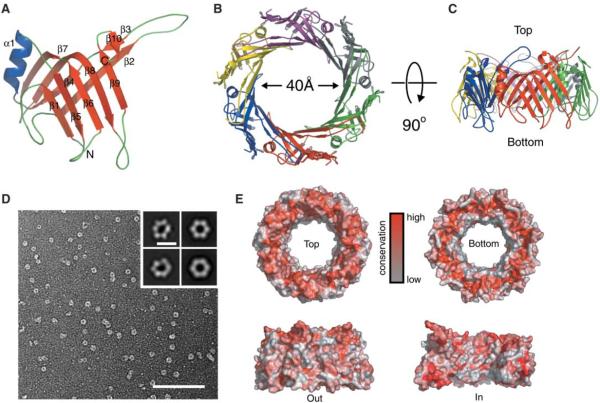
Although Hcp is highly conserved among Gram-negative proteobacteria (table S1), it shares little sequence homology with proteins of known structure. In an effort to gain insight into the function of Hcp, we determined the x-ray crystal structure of P. aeruginosa Hcp1 to a resolution of 1.95 Å. Hcp1 crystallized in the P6 space group with three nearly identical monomers in the asymmetric unit (Fig. 4A and table S3). The three Hcp1 monomers formed two closely related hexameric rings [0.25 Å root mean square deviation]: a true hexamer with six-fold symmetry (chain B in the Protein Data Bank, code 1Y12) and a pseudohexamer with three-fold symmetry (chains A and C) (Fig. 4, B and C). The Hcp1 ring interior is a 24-stranded β barrel ~40 Å in diameter.
Fig. 4.
Hcp1 forms a hexameric ring with a large internal diameter. (A) Ribbon representation of the Hcp1 monomer colored by secondary structure: β strands, red; α helices, blue; and loops, green. (B) Top view of a ribbon representation of the crystallographic Hcp1 hexamer. The individual subunits are colored differently to highlight their organization. (C) Edge-on view of the Hcp1 hexamer shown in (B). (D) Electron microscopy and single-particle analysis of Hcp1. Electron micrograph of Hcp1 negatively stained with 0.75% (w/v) uranyl formate. Scale bar, 100 nm. (Inset) (Left) Representative class averages and (right) the same averages after six-fold symmetrization. Inset scale bar, 10 nm. (E) Sequence conservation analysis of Hcp1. An alignment of 107 Hcp proteins in 43 Gram-negative bacteria was used to plot the relative degree of conservation at each amino acid on the surface of Hcp1 (see methods in supporting online material). Conservation is indicated by color, where red residues are highly conserved and white residues are poorly conserved.
To determine the oligomeric state of Hcp1 in solution, we analyzed the purified protein by analytical gel-filtration chromatography (fig. S4) and transmission electron microscopy (Fig. 4D). The predominant form of Hcp1 was a ring assembly with dimensions closely matching those observed in the crystal lattice (Fig. 4D). Furthermore, averaging of ~6000 particles revealed that the rings contained six clearly discernible subunits with approximate six-fold symmetry (Fig. 4D). Thus, the hexameric rings found in the Hcp1 crystal structure are physiologically relevant and represent the predominant form of the protein in solution. Given the large diameter of hexameric Hcp1, assembly of the particle is likely to occur following secretion.
To identify regions of the Hcp1 structure potentially important for its biological activity, we generated an alignment of 107 Hcp1 proteins from 43 bacterial species and plotted the degree of conservation of each residue onto the structure. This analysis revealed a nonuniform pattern of Hcp1 surface residue conservation: the most highly conserved residues are found on the top and bottom faces of the protein; residues located around the inner and outer circumferences are poorly conserved (Fig. 4E). A particularly well-conserved patch of residues occupies the cleft on the bottom face of Hcp1 (residues 15, 16, 26, 60, 63, 88, 89, 169, and 139). Many of these residues mediate critical subunit contacts, perhaps explaining their high degree of conservation. Others such as Asp26, Lys88,and Gln139, are not engaged in hexamer-stabilizing interactions; thus, their conservation suggests that they may modulate the function of Hcp1. We propose a model whereby Hcp1 associates with proteins on both faces in order to build a channel through which other macromolecules could be transported.
We have provided biochemical and genetic evidence that a virulence-associated genetic locus of P. aeruginosa, termed HSI-I, encodes a protein secretion apparatus. We further demonstrate that a ClpB-like AAA+ family protein, ClpV1, forms a core component of this apparatus and is likely to provide the energy for translocation of Hcp1. The pattern of HSI-I regulation by RetS and LadS and the prior findings, that the locus is essential in the chronic rat lung infection model (6), and our current data indicating that the locus actively secretes Hcp1 during chronic infection of CF patients, collectively suggest a role for HSI-I in chronic P. aeruginosa infections. Our findings also support efforts to develop vaccines and therapeutics targeting Hcp1 or components of the HSI-I–encoded apparatus as treatments for chronic P. aeruginosa infections. Given that IAHP-related loci are conserved among many Gram-negative proteobacterial pathogens and have been implicated in host interactions in several instances (8, 16, 17), the secretory systems they encode may represent yet another general mechanism by which these bacterial pathogens communicate with their hosts.
Supplementary Material
Acknowledgments
We thank A. Rietsch for comments on the manuscript and for providing valuable reagents and assistance with flow cytometry experiments, T. Doan and D. Rudner for assistance with fluorescence microscopy, M. Little for CF patient isolates, R. Melnyk and J. Collier for assistance with biophysical studies, A. Thanawastien for assistance with ELISA experiments, D. Hung, E. Cameron, S. Dove, and J. Thompson for critical reading of the manuscript, S. Pukatzki and A. Ma for sharing data, members of the Mekalanos laboratory for valuable discussions, F. Collart for providing the expression clone of Hcp1, and members of the Structural Biology Center at Argonne National Laboratory for their help in conducting experiments. This work was supported in part by grants to J.J.M. from the NIH (AI26289), to S.L. from the NIH (AI21451), and to A.J. from the NIH (GM62414 and GM074942) and the U.S. Department of Energy, Office of Biological and Environmental Research, under contract W-31-109-Eng-38. J.D.M. is a Damon Runyon Fellow supported by the Damon Runyon Cancer Research Foundation (DRG-1873-05).
Footnotes
Supporting Online Material www.sciencemag.org/cgi/content/full/312/5779/1526/DC1 Materials and Methods Figs. S1 to S4 Tables S1 to S3 References
References
- 1.Govan JR, Deretic V. Microbiol. Rev. 1996;60:539. doi: 10.1128/mr.60.3.539-574.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Furukawa S, Kuchma SL, O'Toole GA. J. Bacteriol. 2006;188:1211. doi: 10.1128/JB.188.4.1211-1217.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yahr TL, Greenberg EP. Mol. Cell. 2004;16:497. doi: 10.1016/j.molcel.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 4.Goodman AL, et al. Dev. Cell. 2004;7:745. doi: 10.1016/j.devcel.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 5.Ventre I, et al. Proc. Natl. Acad. Sci. U.S.A. 2006;103:171. [Google Scholar]
- 6.Potvin E, et al. Environ. Microbiol. 2003;5:1294. doi: 10.1046/j.1462-2920.2003.00542.x. [DOI] [PubMed] [Google Scholar]
- 7.Das S, Chaudhuri K. In Silico Biol. 2003;3:287. [PubMed] [Google Scholar]
- 8.Pukatzki S, et al. Proc. Natl. Acad. Sci. U.S.A. 2006;103:1528. doi: 10.1073/pnas.0510322103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Williams SG, Varcoe LT, Attridge SR, Manning PA. Infect. Immun. 1996;64:283. doi: 10.1128/iai.64.1.283-289.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.VanRheenen SM, Dumenil G, Isberg RR. Infect. Immun. 2004;72:5972. doi: 10.1128/IAI.72.10.5972-5982.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weibezahn J, et al. Cell. 2004;119:653. doi: 10.1016/j.cell.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 12.Akeda Y, Galan JE. Nature. 2005;437:911. doi: 10.1038/nature03992. [DOI] [PubMed] [Google Scholar]
- 13.Yeo HJ, Waksman G. J. Bacteriol. 2004;186:1919. doi: 10.1128/JB.186.7.1919-1926.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schlieker C, Zentgraf H, Dersch P, Mogk A. Biol. Chem. 2005;386:1115. doi: 10.1515/BC.2005.128. [DOI] [PubMed] [Google Scholar]
- 15.Mogk A, et al. J. Biol. Chem. 2003;278:17615. doi: 10.1074/jbc.M209686200. [DOI] [PubMed] [Google Scholar]
- 16.Bladergroen MR, Badelt K, Spaink HP. Mol. Plant Microbe Interact. 2003;16:53. doi: 10.1094/MPMI.2003.16.1.53. [DOI] [PubMed] [Google Scholar]
- 17.Parsons DA, Heffron F. Infect. Immun. 2005;73:4338. doi: 10.1128/IAI.73.7.4338-4345.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.