Abstract
Vibrio parahaemolyticus is an important pathogen causing food-borne disease worldwide. An 80-kb pathogenicity island (Vp-PAI), which contains two tdh (thermostable direct hemolysin) genes and a set of genes for the type III secretion system (T3SS2), is closely related to the pathogenicity of this bacterium. However, the regulatory mechanisms of Vp-PAI's gene expression are poorly understood. Here we report that two novel ToxR-like transcriptional regulatory proteins (VtrA and VtrB) regulate the expression of the genes encoded within the Vp-PAI region, including those for TDH and T3SS2-related proteins. Expression of vtrB was under control of the VtrA, as vector-expressed vtrB was able to recover a functional protein secretory capacity for T3SS2, independent of VtrA. Moreover, these regulatory proteins were essential for T3SS2-dependent biological activities, such as in vitro cytotoxicity and in vivo enterotoxicity. Enterotoxic activities of vtrA and/or vtrB deletion strains derived from the wild-type strain were almost absent, showing fluid accumulation similar to non-infected control. Whole genome transcriptional profiling of vtrA or vtrB deletion strains revealed that the expression levels of over 60 genes were downregulated significantly in these deletion mutant strains and that such genes were almost exclusively located in the Vp-PAI region. These results strongly suggest that VtrA and VtrB are master regulators for virulence gene expression in the Vp-PAI and play critical roles in the pathogenicity of this bacterium.
Introduction
Vibrio parahaemolyticus is a gram-negative marine bacterium that causes acute gastroenteritis in humans associated with the consumption of raw or undercooked seafood [1], [2]. In some cases, infection by this pathogen results in primary septicemia and wound infections [3], [4]. Most of the clinical isolates of V. parahaemolyticus isolated from patients with diarrhea exhibit beta-hemolysis on a special blood agar plate (Wagatsuma agar), whereas environmental isolates barely do so [5]. This hemolysis is called the Kanagawa phenomenon (KP), which has been considered to be a useful marker to distinguish pathogenic from non-pathogenic strains [6], [7]. Thermostable direct hemolysin (TDH) is responsible for KP and purified TDH shows a number of biological effects, such as erythrocyte lysis, cytotoxicity and induction of fluid accumulation in an ileal loop model [6], [8], [9], [10], [11], [12], [13], [14], [15], [16], [17]. Thus, TDH has been considered a major virulence factor of V. parahaemolyticus.
Whole genome sequencing of a KP-positive V. parahaemolyticus strain RIMD2210633 revealed that this strain contains two sets of gene clusters for Type III Secretion System (T3SS), one on each of its two chromosomes (termed T3SS1 and T3SS2, respectively) [18]. Recently, comparative genomic analysis using microarray revealed that an 80-kb pathogenicity island (Vp-PAI) on chromosome II is conserved exclusively in KP-positive pathogenic strains and not in KP-negative strains [19], [20]. Vp-PAI contains not only two tdh genes (tdhA and tdhS) but also the T3SS2 gene cluster. This is highly associated with KP-positive strains and is also involved in the enterotoxicity of this bacterium [19], [21], [22]. Therefore, Vp-PAI has been considered to be related to the pathogenicity of V. parahaemolyticus in humans. Despite having an important role in pathogenicity in humans, the regulatory mechanism of genes expression from Vp-PAI is poorly understood.
In this study, we show that two putative DNA-binding proteins encoded within the Vp-PAI region, which have a winged-helix-turn-helix (WHTH) DNA-binding domain of the OmpR family, control the expression of Vp-PAI's genes in a highly specific manner. Accordingly, they must play a critical role in the pathogenicity of V. parahaemolyticus.
Results
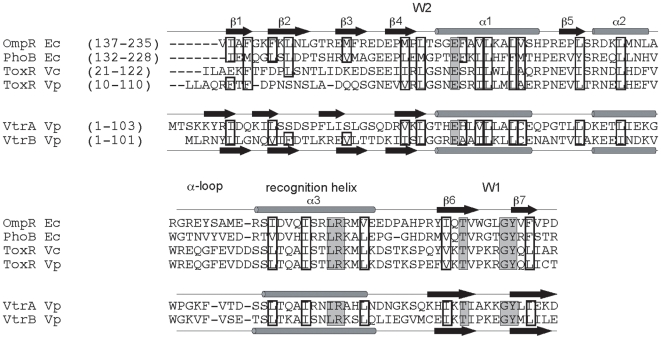
VPA1332 (VtrA) and VPA1348 (VtrB) Have a Winged-Helix-Turn-Helix (WHTH) DNA-Binding Domain of the OmpR Family
In our functional analysis of T3SS2 in V. parahaemolyticus, we noted that two open reading frames (ORFs) (VPA1332 and VPA1348), which share a degree of identity with the N-terminal end of V. cholerae and V. parahaemolyticus ToxR (32% and 34% identity with V. cholerae ToxR, and 45% and 32% identity with V. parahaemolyticus ToxR, respectively), were encoded in the Vp-PAI locus. ToxR is a transcription factor found in V. cholerae that regulates expression of the genes encoding cholera toxin (CT) and toxin-coregulated pilus (TCP) [23]. The N-terminal domain of ToxR encodes a WHTH DNA-binding domain, which is a typical characteristic of the OmpR family of proteins and is necessary for transcriptional regulation of ToxR regulons [24]. The WHTH domain consists of an amino-terminal four-stranded beta sheet, a central three-helical bundle and a carboxy-terminal two-stranded beta sheet. The predicted secondary structures of the N-terminal portions of VPA1332 and VPA1348 were also similar to the DNA-binding domains of OmpR and PhoB of E. coli (Fig. 1). Multiple sequence alignments of these proteins revealed that most of the amino acids forming hydrophobic cores were conserved in VPA1332 and VPA1348 and that highly conserved amino acids were identical to that of OmpR and PhoB. Therefore, VPA1332 and VPA1348 were termed VtrA (V. parahaemolyticus T3SS2 regulator A) and VtrB, respectively. Their possible roles as transcriptional regulators were examined in the following experiments.
Figure 1. VPA1332 (VtrA) and VPA1348 (VtrB) have a winged-helix-turn-helix DNA-binding domain of OmpR family.
Multiple sequence alignment and secondary structure assignments of DNA-binding and trans-activation domains of OmpR, PhoB, ToxR, VtrA, and VtrB proteins are shown. The amino acids that form the hydrophobic cores are highlighted with boxes. Highly conserved amino acids are highlighted with gray boxes.
VtrA and VtrB Regulate the Expression of the Genes for T3SS2-Related Proteins and TDH
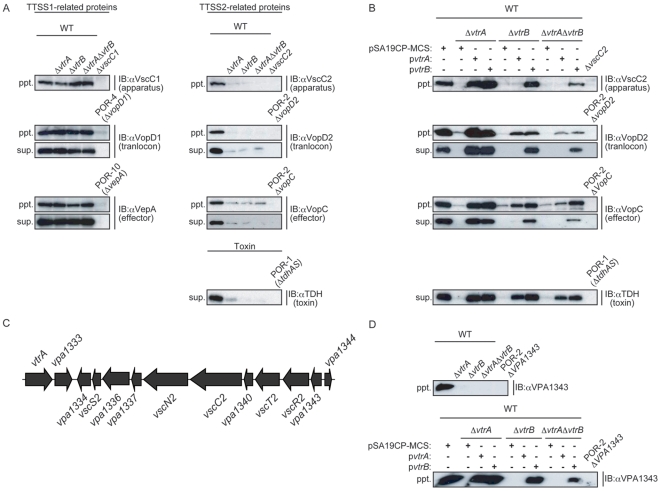
We first constructed vtrA and/or vtrB deletion strains from TDH-producing V. parahaemolyticus RIMD2210633 and then examined their effects on the production of T3SS1- and T3SS2-related proteins and TDH by immunoblotting. As shown in Fig. 2A, deletion of the vtrA or vtrB genes did not affect on the production of T3SS1-related proteins (VscC1, T3SS1 apparatus protein; VopD1, T3SS1 translocon protein and VepA, T3SS1 effector protein), whereas either deletion mutant produced a marked decrease in T3SS2-related proteins (VscC2, T3SS2 apparatus protein; VopD2, T3SS2 translocon protein VopC, T3SS2 effector protein) and in TDH both in bacterial pellets and supernatants. The amounts of T3SS2-related proteins and TDH were recovered fully in both the bacterial pellets and supernatants by complementation of each gene (Fig. 2B). Interestingly, vector-expressed vtrB (pvtrB) could also completely restore the production of T3SS2-related proteins and TDH for the WTΔvtrA strain in both the bacterial pellets and supernatants. Although vector-expressed vtrA (pvtrA) could recover the production of TDH in the supernatant and VopD2 and VopC proteins of the bacterial pellet of the WTΔvtrB strain, no VscC2 protein was found in the bacterial pellet, and neither of the VopD2 and VopC proteins could be detected in the supernatant. In a similar fashion, in a double deletion mutant strain (WTΔvtrAΔvtrB), complementation with vtrB (pvtrB) led to recovery of all proteins in both the bacterial pellet and the supernatant, whereas VopD2 and VopC proteins in the supernatant and VscC2 protein in the bacterial pellet were not detected by complementation with vtrA (pvtrA). Unlike VopD2, VopC, and TDH, VscC2 protein production seemed to be controlled strictly by vtrB. Hence, it was next determined whether a gene located on the same operon as the vscC2 gene was also regulated by VtrB. Production of the VP1343 protein, which was expected to be co-transcribed with vscC2 (Fig. 2C), was examined by immunoblotting. VP1343 protein was not detected in bacterial pellets from vtrA and/or vtrB deletion strains (Fig. 2D, upper panel). Similar to the VscC2 protein, vector-expressed vtrB could overcome a defect in VPA1343 production in vtrA deletion strains, such as WTΔvtrA and WTΔvtrAΔvtrB, whereas vector-expressed vtrA did not induce VPA1343 production in vtrB deletion strains (WTΔvtrB and WTΔvtrAΔvtrB) (Fig. 2D, lower panel). Together, these results suggest that vtrA and vtrB are necessary for the expression of genes encoding T3SS2-related proteins and TDH and that the operon containing vscC2 and vpa1343 is strictly controlled by vtrB. As this operon contains some genes homologous to the T3SS-apparatus (vscS2, vscN2, vscC2, vscT2 and vscR2) (Fig. 2C) that are essential for T3SS secretion, this could explain why, in vector-expressed vtrA, only T3SS2 secreted proteins (VopD2 and VopC proteins) in the bacterial pellets of the WTΔvtrB and WTΔvtrAΔvtrB strains.
Figure 2. VtrA and VtrB regulate the expression levels of T3SS2-related proteins and TDH.
A. Loss of vtrA and vtrB diminished the expression of T3SS2-related proteins and TDH. Western blot analysis of bacterial pellets (ppt.) and secreted proteins (sup.) from isogenic mutants of wild-type (WT) V. parahaemolyticus. Lane 1, wild-type V. parahaemolyticus (WT); lane 2, vtrA deletion strain (WTΔvtrA); lane 3, vtrB deletion strain (WTΔvtrB); lane 4, vtrA and vtrB double deletion strain (WTΔvtrAΔvtrB). Samples from indicated strains were loaded in lane 5 to confirm the specificity of each antibody. Blots were probed with anti-VscC1, anti-VopD1, anti-VepA, anti-VscC2, anti-VopD2, anti-VopC, and anti-TDH polyclonal antibodies. B. Vector-induced vtrB could restore the secretory capacity of T3SS2 independent of vtrA. Western blot analyses of bacterial pellets (ppt.) and secreted proteins (sup.) from indicated strains are shown. Blots were probed with anti-VscC2, anti-VopD2, anti-VopC, and anti-TDH polyclonal antibodies. C. Genetic organization of the DNA region containing vscC2 and vpa1343 of V. parahaemolyticus RIMD2210633. D. VPA1343 protein expression was strictly regulated by VtrB. Western blot analysis of bacterial pellets (ppt.) from isogenic mutants of wild-type (WT) V. parahaemolyticus (upper panel) and their complemented strains (lower panel). Blots were probed with anti-VPA1343 polyclonal antibodies.
Expression of VtrB Is Controlled Directly by VtrA
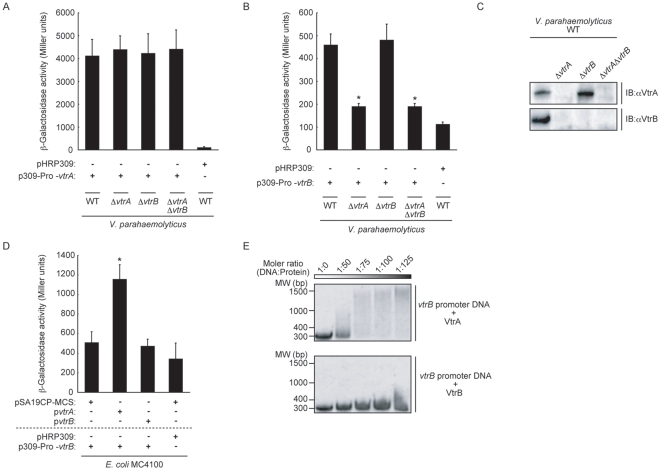
Based on the above observations that vector-induced vtrB could restore the production of TDH and T3SS2-related protein even though both vtrB and vtrA are essential for the production of these proteins (Fig. 2), we next examined the possibility that vtrA might regulate vtrB expression. For this, we used vtrA-lacZ or vtrB-lacZ transcriptional fusion reporters. Neither vtrA nor vtrB gene deletion had any influence on vtrA-lacZ transcription in V. parahaemolyticus (Fig. 3A). In contrast, vtrB-lacZ transcription decreased dramatically in the vtrA deletion strains (Fig. 3B). Immunoblotting of VtrA and VtrB proteins in the vtrA and/or vtrB deletion strains revealed that VtrA protein production occurs regardless of the expression of vtrB, whereas deletion of vtrA caused a decrease in production of the VtrB protein (Fig. 3C). This transcriptional activation of VtrA against vtrB gene transcription was also observed in E. coli, as vtrB-lacZ transcription was significantly induced only when VtrA was produced (Fig. 3D). Direct binding of the VtrA DNA binding domain to vtrB promoter DNA was then examined by a gel shift assay. A shift in electrophoretic mobility of vtrB promoter DNA was observed at a low concentration of the VtrA DNA binding domain (Fig. 3E, upper panel), whereas only a weak shift was seen at the highest concentration of VtrB DNA binding domain (Fig. 3E, lower panel). These results indicate that VtrA activates vtrB gene transcription by direct binding to its promoter.
Figure 3. VtrB expression is under the control of VtrA.
A. Neither vtrA nor vtrB was involved in the transcription of vtrA. V. parahaemolyticus strains carrying the vtrA-lacZ transcriptional fusion vector were assayed for β-galactosidase activity. The bars show the average of three separate experiments, and the standard deviations are indicated by error bars. B. Transcription of vtrB was decreased in vtrA deletion strains. V. parahaemolyticus strains carrying the vtrB-lacZ transcriptional fusion vector were assayed for β-galactosidase activity. The bars show the average of three separate experiments, and the standard deviations are indicated by error bars. C. Deletion of vtrA caused a decrease in the production of VtrB. Immunoblot analysis of VtrA and VtrB protein expression in vtrA and vtrB mutant strains are shown. Lane 1, wild-type V. parahaemolyticus (WT); lane 2, vtrA mutant strain (WTΔvtrA); lane 3, vtrB mutant strain (WTΔvtrB); lane 4, vtrA and vtrB double mutant strain (WTΔvtrAΔvtrB). Blots were probed with anti-VtrA (upper panel) and anti-VtrB (lower panel) polyclonal antibodies. D. Effects of vtrA and vtrB expression on vtrB transcription in E. coli. E. coli MC4100 carrying vtrB-lacZ transcriptional fusion vector were assayed for β-galactosidase activity. The bars show the average of three separate experiments, and the standard deviations are indicated by error bars. E. Binding of purified VtrA DNA binding domain to the upstream region of vtrB is shown by an electrophoretic mobility shift assay. Each lane contains the same amount of upstream region of vtrB (30 nM) and various concentrations (0, 1.5, 2.25, 3.0, 4 µM) of VtrA DNA binding domain (upper panel) or VtrB DNA binding domain (lower panel). The molecular ratios are indicated in the top line.
VtrA and VtrB Play Critical Roles in T3SS2-Dependent Cytotoxicity
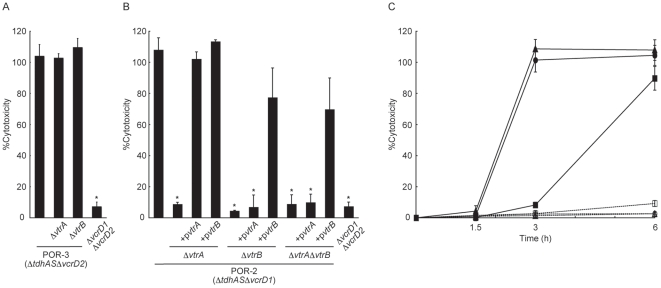
One characteristic of T3SSs in V. parahaemolyticus is their ability to cause cytotoxic effects on Caco-2 cells in vitro [25], [26]. Therefore, we next examined the role of vtrA and vtrB in T3SS1- and T3SS2-mediated cytotoxicity. Deletion of the vtrA or the vtrB gene in the tdhAS- and T3SS2-deficient strain POR-3 (POR-3ΔvtrA and POR-3ΔvtrB, respectively) had no effect on T3SS1-dependent cytotoxicity (Fig. 4A). By contrast, deletion of the vtrA and/or the vtrB from the tdhAS- and T3SS1-deficient strain POR-2 (POR-2ΔvtrA, POR-2ΔvtrB, and POR-2ΔvtrAΔvtrB, respectively) caused a decrease in cytotoxicity similar to that of the ΔvcrD1ΔvcrD2 strain, which is deficient in both T3SS1 and T3SS2 (Fig. 4B). A vector-expressed vtrB was able to overcome the defect in cytotoxicity of vtrA deletion strains (POR-2ΔvtrA and POR-2ΔvtrAΔvtrB). This result is in accordance with the previous results, showing that vector-expressed vtrB could recover the diminished secretory capacity of T3SS2 in vtrA deletion strains as shown in Fig. 2B and C. In contrast, complementation with vtrA restored cytotoxic capacity in the POR-2ΔvtrA strain but not in any of the POR-2ΔvtrB or POR-2ΔvtrAΔvtrB strains, which is also in agreement with the results shown in Fig. 2. Finally, the effect of vtrA and vtrB overexpression on T3SS2-dependent cytotoxicity was determined (Fig. 4C). Overexpression either vtrA or vtrB resulted in dramatic accelerations in cytotoxic activity from a tdhAS- and T3SS1-deficient strain (POR-2), whereas no effect was observed from overexpression in a tdhAS- and T3SS1/T3SS2-deficient strain (ΔvcrD1ΔvcrD2). These results indicate that both vtrA and vtrB are essential for T3SS2-dependent cytotoxicity.
Figure 4. VtrA and VtrB are not necessary for T3SS1-dependent cytotoxicity but necessary for T3SS2-dependent cytotoxicity.
A. vtrA and vtrB are not necessary for T3SS1-dependent cytotoxicity. Caco-2 cells were infected for 6 h with isogenic strains of POR-3 (ΔtdhASΔvcrD2). Bar 1: POR-3 (ΔtdhASΔvcrD2); bar 2: POR-3ΔvtrA; bar 3: POR-3ΔvtrB; bar 4: ΔvcrD1ΔvcrD2 (ΔtdhASΔvcrD1ΔvcrD2). Cytotoxicity was evaluated by the amount of LDH released. Error bars represent standard deviations for results from triplicate experiments. B. vtrA and vtrB are essential for T3SS2-dependent cytotoxicity. Caco-2 cells were infected for 6 h with isogenic mutant strains of POR-2 (ΔtdhASΔvcrD1). Bar 1: POR-2 (ΔtdhASΔvcrD1); bar 2: POR-2ΔvtrA (ΔtdhASΔvcrD1ΔvtrA); bar 3: POR-2ΔvtrA expressing vtrA (POR-2ΔvtrA+pvtrA); bar 4: POR-2ΔvtrA expressing vtrB (POR-2ΔvtrA+pvtrB); bar 5: POR-2ΔvtrB (ΔtdhASΔvcrD1ΔvtrB); bar 6; POR-2ΔvtrB expressing vtrA (POR-2ΔvtrB+pvtrA); bar 7: POR-2ΔvtrB expressing vtrB (POR-2ΔvtrB+pvtrB); bar 8: POR-2ΔvtrAΔvtrB (ΔtdhASΔvcrD1ΔvtrAΔvtrB); bar 9: POR-2ΔvtrAΔvtrB expressing vtrA (POR-2ΔvtrAΔvtrB+pvtrA); bar 10: POR-2ΔvtrAΔvtrB expressing vtrB (POR-2ΔvtrAΔvtrB+pvtrB); bar 11: ΔvcrD1ΔvcrD2 (ΔtdhASΔvcrD1ΔvcrD2). Cytotoxicity was evaluated by the amount of LDH released. Error bars represent standard deviations for results from triplicate experiments. Asterisks indicate significant differences from the results obtained with the parent strain (*P<0.05). C. Overexpressing of vtrA and vtrB promoted T3SS2-dependent cytotoxicity. Caco-2 cells were infected for 1.5–6 h with V. parahaemolyticus. Cytotoxicity was evaluated by the amount of LDH released. POR-2 (ΔtdhASΔvcrD1) with control vector (pSA19CP-MCS) (filled squares, solid line), POR-2 expressing vtrA (filled circles, solid line), POR-2 expressing vtrB (filled triangles, solid line), ΔvcrD1ΔvcrD2 (ΔtdhASΔvcrD1ΔvcrD2) with control vector (pSA19CP-MCS) (open squares, dashed line), ΔvcrD1ΔvcrD2 expressing vtrA (open circles, dashed line), and ΔvcrD1ΔvcrD2 expressing vtrB (open triangles, dashed line). Error bars represent standard deviations for results from triplicate experiments. Asterisks indicate significant differences from the results obtained with the parent strain (*P<0.05).
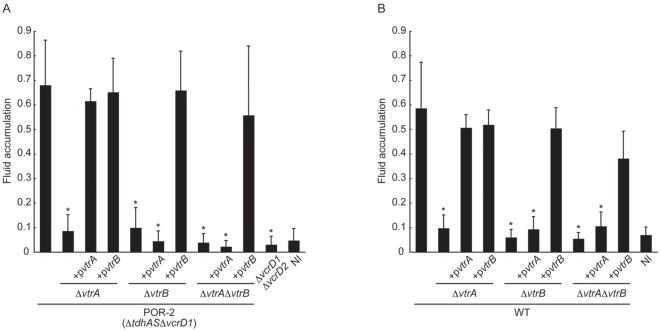
VtrA and VtrB Play Critical Roles in V. parahaemolyticus-Induced Enterotoxicity
To investigate the contribution of vtrA and vtrB to the enterotoxicity of V. parahaemolyticus, we examined the T3SS2-dependent enterotoxic activity of the vtrA and vtrB deletion strains using the rabbit ileal loop model. As reported previously, POR-2, which is a tdhAS- and T3SS1-deficient strain, caused a high level of fluid accumulation [21], [26]. This was dramatically decreased in vtrA and/or vtrB deletion strains (POR-2ΔvtrA, POR-2ΔvtrB, and POR-2 ΔvtrA ΔvtrB) and was similar to that of the ΔvcrD1ΔvcrD2 strain and non-infected (NI) control (Fig. 5A). The decrease in enterotoxicity was restored by trans-complementation of each gene. As with T3SS2-dependent cytotoxicity shown in Fig. 4B, a defect in enterotoxicity of vtrA deletion strains (POR-2ΔvtrA and POR-2ΔvtrAΔvtrB) was restored by vector-expressed vtrB (Fig. 5A). Similar to T3SS2-dependent enterotoxicity, both vtrA and vtrB greatly contributed to the wild-type V. parahaemolyticus-induced enterotoxicity, which has functional TDH and both T3SS1 and T3SS2 (Fig. 5B). The fluid accumulation that resulted after challenge with vtrA and/or vtrB deletion strains (WTΔvtrA, WTΔvtrB, and WTΔvtrAΔvtrB) was almost none, very similar to the NI control. Vector-expressed vtrB was able to restore the enterotoxicity to its full potential not only in the vtrB deletion strain, but also in vtrA deletion strains. These results indicate strongly that both vtrA and vtrB play critical roles in V. parahaemolyticus-induced enterotoxicity.
Figure 5. VtrA and VtrB have a critical role in V. parahaemolyticus-induced enterotoxicity.
A. VtrA and VtrB are essential for T3SS2-dependent enterotoxicity. The enterotoxic activity levels of isogenic mutants of POR-2 (ΔtdhASΔvcrD1) and complemented strains in rabbit ileal loops were examined. Bar 1, POR-2 (ΔtdhASΔvcrD1); bar 2, POR-2ΔvtrA (ΔtdhASΔvcrD1ΔvtrA); bar 3, POR-2ΔvtrA expressing vtrA (POR-2ΔvtrA+pvtrA); bar 4, POR-2ΔvtrA expressing vtrB (POR-2ΔvtrA+pvtrB); bar 5, POR-2ΔvtrB (ΔtdhASΔvcrD1ΔvtrB); bar 6, POR-2ΔvtrB expressing vtrA (POR-2ΔvtrB+pvtrA); bar 7, POR-2ΔvtrB expressing vtrB (POR-2ΔvtrB+pvtrB); bar 8, POR-2ΔvtrAΔvtrB (ΔtdhASΔvcrD1ΔvtrAΔvtrB); bar 9, POR-2ΔvtrAΔvtrB expressing vtrA (POR-2ΔvtrAΔvtrB+pvtrA); bar 10, POR-2ΔvtrAΔvtrB expressing vtrB (POR-2ΔvtrAΔvtrB+pvtrB); bar 11, ΔvcrD1ΔvcrD2 (ΔtdhASΔvcrD1ΔvcrD2); bar 12, non-infected (NI) control. Results were measured as the amount of accumulated fluid (in milliliters) per length (in centimeters) of ligated rabbit small intestine. Error bars represent standard deviations for results from triplicate experiments. Asterisks indicate significant differences from the results obtained with the parental strain (P<0.05). B. VtrA and VtrB are essential for V. parahaemolyticus-induced enterotoxicity. The enterotoxic activity of isogenic mutants of wild-type V. parahaemolyticus (WT) and complemented strains in rabbit ileal loops were examined. Bar 1, wild-type (WT); bar 2, WTΔvtrA; bar 3, WTΔvtrA expressing vtrA (WTΔvtrA+pvtrA); bar 4, WTΔvtrA expressing vtrB (WTΔvtrA+pvtrB); bar 5, WTΔvtrB; bar 6, WTΔvtrB expressing vtrA (WTΔvtrB+pvtrA); bar 7, WTΔvtrB expressing vtrB (WTΔvtrB+pvtrB); bar 8, WTΔvtrAΔvtrB; bar 9, WTΔvtrAΔvtrB expressing vtrA (WTΔvtrAΔvtrB+pvtrA); bar 10, WTΔvtrAΔvtrB expressing vtrB (WTΔvtrAΔvtrB+pvtrB); bar 11, NI control. Error bars represent standard deviations for results from triplicate experiments. Asterisks indicate significant differences from the results obtained with the parental strain (P<0.05).
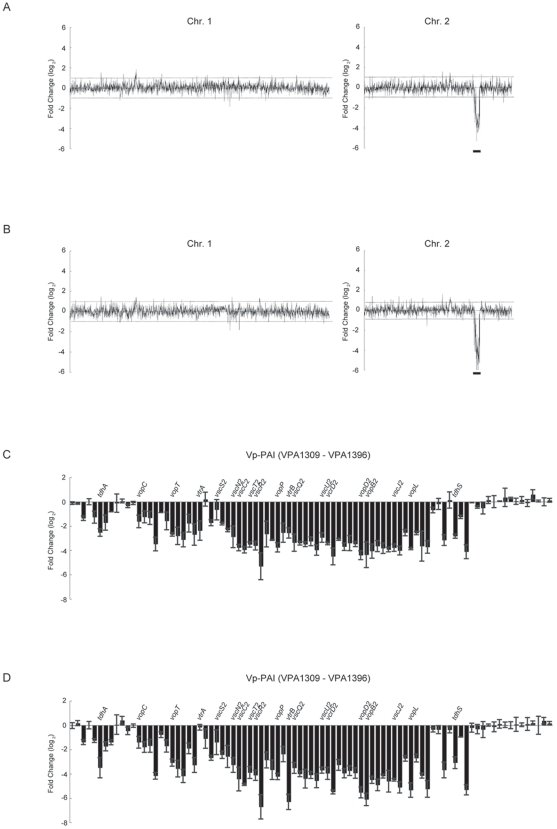
VtrA and VtrB Specifically Regulate Genes Encoded in the Vp-PAI Region
To identify transcriptional targets of VtrA and VtrB on the complete chromosomes of V. parahaemolyticus, genome-wide transcriptional profiles of vtrA or vtrB deletion strains were compared with that of the wild-type strain (Fig. 6 and Table 1). Overview of the transcriptome microarray analysis revealed that most gene expressions were unaffected by deletion of the vtrA or the vtrB genes respectively (Fig. 6A, B, respectively). However, it was also obvious that genes located on a particular region of chromosome 2 were remarkably down-regulated in both vtrA and vtrB deletion strains (Fig. 6A, B; lined). Interestingly, this region is included in the Vp-PAI region (vpa1309-vpa1396) [19], [20] and the transcription of both tdhAS genes and T3SS2-related genes were also decreased significantly in each of the mutant strains (Fig. 6C, D). The pattern of gene expression profile was almost similar between these two mutant strains. Expression of vtrA was not affected by vtrB gene deletion (–1.1 -fold change), which is in accordance with our previous observations that vtrB gene deletion did not have any effect on vtrA gene expression, as assessed by the reporter gene assay of vtrA and immunoblotting of VtrA (Fig. 3A, C). Only a few other genes encoded outside of the Vp-PAI region were affected (Table 1). These results suggest that these two proteins regulate gene expression in the Vp-PAI region in highly specific manners.
Figure 6. Whole-genome transcriptional profiling of vtrA and vtrB deletion strain.
Genome-wide transcript analysis of the VtrA and VtrB regulons is shown. Gene expression was determined by comparing cDNA generated from WTΔvtrA (A) or WTΔvtrB (B) in exponential phase grown in LB medium with 0.5% NaCl with that from the WT strain. The Vp-PAI region is indicated by a bold line. Effect of the vtrA (C) or vtrB (D) deletion on expression of genes located within Vp-PAI (vpa1309-vpa1396). Representative gene functions are indicated at the top.
Table 1. Microarray analysis of VtrA and VtrB regulon in V. parahaemolyticus.
| Fold changea | |||
| Identification | ORF Description | vtrA | vtrB |
| Downregulated genes (VtrA or VtrB-activated) | |||
| VP0047 | peptide ABC transporter, ATP-binding protein | −1.6 | −2.0 |
| VP1880 | L-serine dehydratase 1 | −1.4 | −2.2 |
| VP1904 | methyl-accepting chemotaxis protein | −3.6 | −3.6 |
| VP2015 | putative cytochrome c | −2.1 | −3.4 |
| VP2016 | hypothetical protein | −2.4 | −2.4 |
| VP2362 | outer membrane protein OmpK precursor | −2.0 | −2.0 |
| VP2679 | ribosomal large subunit pseudouridine synthase A | −1.5 | −2.0 |
| VPA1311 | hypothetical protein | −2.6 | −2.6 |
| VPA1313 | hypothetical protein | −2.4 | −2.4 |
| VPA1314 | thermostable direct hemolysin A | −5.8 | −11.3 |
| VPA1315 | hypothetical protein | −3.3 | −3.3 |
| VPA1321 | cytotoxic necrotizing factor | −3.1 | −3.1 |
| VPA1322 | putative zinc finger protein | −2.4 | −3.5 |
| VPA1323 | hypothetical protein | −2.5 | −3.2 |
| VPA1324 | hypothetical protein | −11.2 | −17.8 |
| VPA1326 | hypothetical protein | −3.0 | −3.3 |
| VPA1327 | putative exoenzyme T | −6.5 | −8.4 |
| VPA1328 | hypothetical protein | −7.0 | −11.9 |
| VPA1329 | putative traA protein | −8.7 | −18.0 |
| VPA1330 | hypothetical protein | −3.4 | −3.7 |
| VPA1331 | putative OspC2 | −6.5 | −9.6 |
| VPA1332 | VtrA protein | −5.2 | −1.1 |
| VPA1334 | hypothetical protein | −3.3 | −6.4 |
| VPA1336 | hypothetical protein | −3.5 | −5.4 |
| VPA1337 | hypothetical protein | −5.0 | −5.0 |
| VPA1338 | putative ATPase YscN | −7.4 | −9.6 |
| VPA1339 | putative type III secretion system EscC protein | −13.7 | −21.5 |
| VPA1340 | hypothetical protein | −15.5 | −29.2 |
| VPA1341 | putative Spa29, component of the Mxi-Spa secretion machinery | −11.5 | −14.8 |
| VPA1342 | putative Type III secretion protein Spa24 | −12.3 | −17.3 |
| VPA1343 | hypothetical protein | −39.7 | −104.7 |
| VPA1344 | hypothetical protein | −6.3 | −7.3 |
| VPA1345 | hypothetical protein | −8.9 | −12.7 |
| VPA1346 | putative targeted effector protein YopP | −13.5 | −18.5 |
| VPA1347 | hypothetical protein | −5.9 | −5.2 |
| VPA1348 | VtrB protein | −5.9 | −78.8 |
| VPA1349 | putative Type III secretion protein Spa33 | −10.4 | −11.4 |
| VPA1350 | hypothetical protein | −10.7 | −16.0 |
| VPA1351 | hypothetical protein | −11.3 | −21.0 |
| VPA1352 | hypothetical protein | −9.6 | −17.0 |
| VPA1353 | putative outer membrane protein | −15.7 | −23.7 |
| VPA1354 | putative type III secretion system EscU protein | −7.7 | −12.9 |
| VPA1355 | putative type III secretion system EscV protein | −10.8 | −15.5 |
| VPA1356 | hypothetical protein | −22.7 | −44.8 |
| VPA1357 | hypothetical protein | −8.6 | −7.3 |
| VPA1358 | putative dimethyladenosine transferase | −13.0 | −15.4 |
| VPA1359 | hypothetical protein | −10.6 | −13.3 |
| VPA1360 | hypothetical protein | −11.2 | −15.2 |
| VPA1361 | hypothetical protein | −21.2 | −46.1 |
| VPA1362 | putative secreted protein EspD | −20.6 | −68.5 |
| VPA1363 | putative chaperone | −16.7 | −22.5 |
| VPA1364 | hypothetical protein | −12.6 | −30.0 |
| VPA1365 | putative two-component response regulator | −14.2 | −18.2 |
| VPA1366 | hypothetical protein | −15.3 | −24.3 |
| VPA1367 | putative type III secretion system lipoprotein precursor EprK | −13.8 | −22.4 |
| VPA1368 | hypothetical protein | −16.2 | −34.5 |
| VPA1369 | hypothetical protein | −5.8 | −6.6 |
| VPA1370 | hypothetical protein | −14.4 | −40.3 |
| VPA1371 | hypothetical protein | −6.0 | −6.5 |
| VPA1373 | hypothetical protein | −13.2 | −37.8 |
| VPA1376 | conserved hypothetical protein | −8.9 | −12.8 |
| VPA1378 | thermostable direct hemolysin S | −7.2 | −8.5 |
| VPA1380 | putative OspB protein | −17.3 | −39.5 |
| Upregulated genes (VtrA or VtrB-repressed) | |||
| VP0368 | mannitol operon repressor | 1.9 | 2.1 |
| VP0996 | putative 54 kDa polar flagellar sheath protein A | 2.0 | 2.0 |
| VPA0548 | putative protein F-related protein | 1.7 | 2.0 |
Fold change in gene transcripts between the wild-type and ΔvtrA or ΔvtrB mutant as determined by microarray analysis. Statistically significant changes (≥2-fold difference with P<0.05) are highlighted in bold as described in Materials and Methods .
Discussion
Vibrio parahaemolyticus is a gram-negative marine bacterium that causes acute gastroenteritis in humans [1], [2]. TDH has been considered a major virulence factor of gastroenteritis, because TDH is responsible for KP (a marker of pathogenic strains) and has cytotoxic and enterotoxic activities [8], [15], [27], [28], [29], [30], [31], [32]. Whole genome sequencing of this KP-positive strain revealed the presence of two sets of genes encoding for two separate type III secretion systems (T3SS1 and T3SS2)[18]. The T3SS1 gene cluster is found in both KP-negative and -positive strains, while the T3SS2 gene cluster is highly associated with KP-positive strains [21]. A functional characterization of T3SS2 has revealed that it is associated with cytotoxic activity against Caco-2 cells in vitro [25], [26]. Furthermore, the enterotoxicity observed for a tdhAS deletion mutant strain was not observed for a T3SS2-deficient mutant strain [21], [26], [33]. Therefore, T3SS2 is also thought to be related to the enterotoxicity of V. parahaemolyticus. Comparative genomic analysis using microarrays to analyze both pathogenic and non-pathogenic strains, revealed that only the genes in the 80-kb pathogenicity island (Vp-PAI) on chromosome II, including two tdh genes (tdhAS) and a set of type III secretion system (T3SS2), were detected only in the KP-positive pathogenic strains [19], [20]. Therefore, it has been considered that the genes encoded in the 80-kb pathogenicity island (Vp-PAI) play major roles in the pathogenicity of this bacterium. However, the regulatory mechanisms for such genes are poorly understood. In this study, we found that two novel ToxR-like transcriptional regulatory proteins (VtrA; VPA1332 and VtrB; VPA1348), which are encoded in the Vp-PAI region, played important roles in pathogenicity (enterotoxicity) of V. parahaemolyticus, controlling virulence genes in the Vp-PAI region (including tdh and T3SS2-related genes) expression. These findings strongly indicated that this pathogen equips refined virulence gene expression system to cause gastroenteritis in humans and that these regulators are key players in the virulence of this bacterium.
Recently, a T3SS2-related T3SS gene cluster was found in trh (TDH-related hemolysin)-positive (KP-negative) V. parahaemolyticus strain TH3996, which is also pathogenic to humans. These T3SS-related genes are highly associated among trh-positive strains [34]. The T3SS2-related T3SS gene cluster is also encoded in a flanking region of the trh gene on chromosome II, which is called Vp-PAITH3996, and not only TRH but also the T3SS-related genes in Vp-PAITH3996 are involved in the enterotoxicity of trh-positive strains [34]. Moreover, a T3SS2-related gene cluster was also found in non-O1, non-O139 V. cholerae strains, and it was required for colonization in the infant mouse model [35], [36]. Therefore, genes in the Vp-PAI region, especially those encoding for hemolysins and T3SS2, have been considered to be related to the pathogenicity of not only V. parahaemolyticus but also non-O1, non-O139 V. cholerae to humans. The T3SS gene set found in these strains contains a pair that is highly similar to the vtrA and vtrB genes, suggesting that these regulators might contribute to the virulence of these bacteria by controlling virulence gene expression levels.
The transcriptional activator ToxR controls the expression of the genes for CT, TCP, and outer membrane proteins in V. cholerae [23]. ToxR is an integral membrane protein and consists of three functional domains: cytoplasmic domain, transmembrane domain and periplasmic domain [37]. The N-terminal cytoplasmic domain of ToxR encodes an OmpR-like DNA-binding domain that is essential for transcriptional regulation of ToxR-regulated genes [37]. The transmembrane (TM) and periplasmic domains of ToxR are believed to act as sensors of environmental signals [38]. The N-terminal portions of VtrA and VtrB share sequence similarity with the DNA binding domain of ToxR. The TM-PRED program (http://www.ch.embnet.org/cgi-bin/TMPRED from parser; TM helix length between 17 and 33 residues; scores, >1,000) predicted that VtrA and VtrB would contain one TM region (VtrA, amino acids 134–153 and VtrB, amino acids 157–182), indicating that VtrA and VtrB are transmembrane transcriptional activators. The TM region of VtrB is located on its C-terminal end, whereas that of VtrA is located on the middle region as ToxR. Although the C-terminal region of VtrA did not show significant sequence homology with ToxR, it is possible that it might be involved in receiving and transmitting environmental signals to elicit VtrA-mediated regulation. This signal transduction could exert virulence in V. parahaemolyticus.
V. parahaemolyticus has a homolog of V. cholerae toxRS operon (Vp-ToxRS), and Vp-ToxR is involved in the production of TDH [39]. Recently, Nakano et al. reported that Hfq, which is conserved in a wide range of bacteria and modulates the stability and transcription of mRNAs, also regulates TDH expression in V. parahaemolyticus [40]. In our investigation, expression levels of vp-toxR and hfq were not significantly affected by deletion of vtrA and vtrB genes under our experimental conditions (GSE17242), suggesting that neither vp-toxR nor hfq is involved in TDH expression mediated by VtrA and VtrB. It is not surprising that VtrA and VtrB are regulons, because both Vp-ToxR and Hfq are global regulators and their regulons comprise many genes, including virulence-associated genes [23], [41]. Because VtrA and VtrB regulons were specifically clustered in the Vp-PAI region (Fig. 6 and Table 1), it is possible that they are more directly related to the control of pathogenicity than Vp-ToxR and Hfq.
Genome-wide transcriptional profiling of vtrA or vtrB deletion strains revealed that VtrA and VtrB regulons were specifically encoded in the Vp-PAI region (Fig. 6 and Table 1). Given our findings that the expression of vtrB is under control of VtrA (Fig. 3) and that vector-expressed vtrB could restore the defect in enterotoxicity of the WTΔvtrAΔvtrB strain (Fig. 5B), VtrB might determine this specific gene expression. The G+C content of the Vp-PAI region of V. parahaemolyticus is lower than the average G+C content of the small chromosome (ChrII) [18], [19]. Although a consensus sequence recognized by VtrB is unknown, it is possible that this characteristic of low G+C content in Vp-PAI might be one of the factors deciding the specificity of VtrB regulons. Given that the Vp-PAI sequence is unique to KP-positive pathogenic strains, plays an important role in the pathogenicity of V. parahaemolyticus and that VtrB has a critical role in the expression of genes from this region, VtrB may be considered a key player in the virulence of this bacterium. Therefore, it could be an “Achilles' heel” of this pathogen. It is possible that VtrB-specific drugs would perform well in the prevention and treatment of V. parahaemolyticus-induced illness.
Materials and Methods
Bacterial Strains and Plasmids
V. parahaemolyticus strain RIMD2210633 (KP positive, serotype O3:K6) [18] was used for constructing deletion mutants and in functional analysis. E. coli DH5α and SM10λpir were used for general manipulation of plasmids and mobilization of plasmids into V. parahaemolyticus. E. coli MC4100 was used for reporter gene assay. The strains and plasmids used in this study are listed in Table 2.
Table 2. Strains and plasmids used in this study.
| Strain or plasmid | Description | Source or reference |
| Vibrio parahaemolyticus | ||
| WT | RIMD2210633 (KP positive, serotype O3:K6) | [18] |
| POR-1 | ΔtdhAS derivative of WT | [21] |
| POR-2 | POR-1 knockout of vcrD1 (vp1696) gene | [21] |
| POR-3 | POR-1 knockout of vcrD2 (vpa1355) gene | [21] |
| ΔvcrD1ΔvcrD2 | POR-1 knockout of vcrD1 and vcrD2 gene | [25] |
| WTΔvtrA | KXV237 knockout of vtrA (vp1332) gene | This study |
| WTΔvtrB | KXV237 knockout of vtrB (vp1348) gene | This study |
| WTΔvtrA ΔvtrB | KXV237 knockout of vtrA and vtrB gene | This study |
| POR-2ΔvtrA | POR-2 knockout of vtrA (vp1332) gene | This study |
| POR-2ΔvtrB | POR-2 knockout of vtrB (vp1348) gene | This study |
| POR-2ΔvtrA ΔvtrB | POR-2 knockout of vtrA and vtrB gene | This study |
| POR-3ΔvtrA | POR-3 knockout of vtrA (vp1332) gene | This study |
| POR-3ΔvtrB | POR-3 knockout of vtrB (vp1348) gene | This study |
| ΔvscC1 | POR-1 knockout of vscC1 (vp1696) gene | [21] |
| POR-4 | POR-1 knockout of vopD1 (vp1656) gene | [43] |
| POR-10 | POR-1 knockout of vepA (vp1680) gene | [43] |
| ΔvscC2 | POR-1 knockout of vscC2 (vpa1339) gene | [21] |
| POR-2ΔvopD2 | POR-2 knockout of vopD2 (vp1361) gene | [26] |
| POR-2ΔvopC | POR-2 knockout of vopC (vpa1321) gene | [25] |
| POR-2Δvpa1343 | POR-2 knockout of vpa1343 gene | This study |
| Escherichia coli | ||
| DH5α | F− Φ80ΔlacZM15 Δ (lacZYA argF)U169 deoP recA1 endA1 hsdR17 (rK − mK −) | Laboratory collection |
| SM10λpir | thi thr leu tonA lacY supE recA::RP4-2-Tc::Mu λpir R6K | [24] |
| MC4100 | F– araD139 Δ (argF-lac) U169 rpsL150 (Strr) relA1 flbB5301 deoC1 ptsF25 rbsR | [44] |
| Plasmid | ||
| pHRP309 | lacZ transcriptional fusion vector, Gmr | [45] |
| p309-Pro-vtrA | Derivative of pHRP309, containing vtrA promoter | This study |
| p309-Pro-vtrB | Derivative of pHRP309, containing vtrB promoter | This study |
| pYAK1 | R6K-ori suicide vector containing sacB gene | [46] |
| pYAK1-ΔvtrA | Derivative of suicide vector pYAK1 for generating the vtrA deletion mutants | This study |
| pYAK1-ΔvtrB | Derivative of suicide vector pYAK1 for generating the vtrB deletion mutants | This study |
| pYAK1-Δvpa1343 | Derivative of suicide vector pYAK1 for generating the vpa1343 deletion mutants | This study |
| pSA19CP-MCS | Complement vector for V. parahaemolyticus, Cmr | [47] |
| pvtrA | Derivative of pSA19CP-MCS, containing vtrA gene | This study |
| pvtrB | Derivative of pSA19CP-MCS, containing vtrB gene | This study |
RNA Isolation
Bacterial strains were grown at 37°C in LB broth containing 0.5% NaCl to an OD600 of 1.0. Bacteria were harvested by centrifugation and the bacterial pellet was suspended with TRIzol Reagent (Invitrogen). After 1 h incubation at 4°C, one-fifth volume of chloroform was added to the suspension followed by recentrifugation. The aqueous layer was removed and a one-tenth volume of 3 M sodium acetate (pH 5.9) was added. Nucleic acids were precipitated with isopropanol and pelleted by centrifugation. The pellet was washed with 80% ethanol. Contaminating genomic DNA was removed from the RNA samples using Turbo DNA-free kits (Ambion). RNA was purified by acid phenol-chloroform extraction and ethanol precipitation. Finally, highly pure total RNA was further isolated using QIAGEN RNeasy Mini kits, according to the manufacturer's protocol.
DNA Microarray
A total of 20 µg of RNA was transcribed to DNA and labeled with aminoallyl dUTP using reverse transcriptase (Superscript III; Invitrogen) and random hexamers (TAKARA Bio) as primers. The aminoallyl-labeled DNA was purified by phenol chloroform extraction and ethanol precipitation. Precipitated DNA was resolved in 50 mM NaHCO3 (pH 9.0) and Cy3 or Cy5 monofunctional dye (GE Healthcare) was added to the solution. After 1 h incubation, unincorporated dye was removed using CentriSep spin columns (Princeton Separations, Inc.). Hybridization and detection of microarray signals was performed as described [19]. Equal volumes of Cy3- or Cy5-labeled probes from wild type and WT ΔvtrA or WT ΔvtrB V. parahaemolyticus strain were mixed with in hybridization solution (5×SSC buffer, 0.5% SDS, 0.1 mg/ml human Cot-1 DNA). Mixtures were heated for 5 min at 95°C, followed by ice incubation. The probe mixtures were applied to a microarray slides and covered with MAUI AO lids (BioMicro Systems). Microarray slides were incubated for 16 h at 55°C in a MAUI hybridization chamber. After hybridization, the microarray slides were washed and scanned using a Scan Array Express Lite (Perkin Elmer Life and Analytical Sciences). Each experiment was repeated in triplicates. Microarray data were analyzed using ScanArray Express software (Perkin Elmer Life and Analytical Sciences). The genes regulated by vtrA or vtrB were defined as genes that exhibited at least 2-fold difference on WTΔvtrA or WTΔvtrB in three experiments. All data were filtered for statistical significance (P<0.05) using t-tests in MultiExperiment Viewer (http://www.tm4.org/mev.html). Array results are available at the NCBI Gene Expression Omnibus database (GEO; http://www.ncbi.nlm.nih.gov/geo/) under the accession number GSE17242.
Immunoblot Analysis
V. parahaemolyticus strains were grown overnight in LB broth with 0.5% NaCl. Cultures were then diluted 1∶100 into LB broth with 0.5% NaCl and grown with shaking at 37°C for 5 h. After incubation, bacterial cultures were centrifuged and bacterial pellets solubilized with Laemmli buffer. Secreted proteins were harvested by precipitation with cold trichloroacetic acid to a final concentration of 10% (v/v) on ice for 60 min, followed by centrifugation at 48,000 g for 60 min. The pellets were rinsed in cold acetone and then solubilized in Laemmli buffer.
Samples for western blot analysis were separated by SDS–PAGE (10%, 10–20%, or 15–25% gradients of polyacrylamide; COSMO BIO). The transferred membrane was probed with anti-VscC1, anti-VopD1, anti-VepA, anti-VscC2, anti-VopD2, anti-VopC, anti-TDH, anti-VPA1342, anti-VtrA, or anti-VtrB rabbit polyclonal antibodies and then probed with horseradish peroxidase-conjugated goat anti-rabbit antibody (ZYMED). The blots were developed using enhanced chemiluminescence (ECL) western blotting kits (GE healthcare).
Reporter Gene Assays
E. coli MC4100 or V. parahaemolyticus strains, each harboring a reporter plasmid were grown for 1 h at 37°C in LB broth containing 1.0 or 0.5% NaCl. β-galactosidase activity was assayed in cell lysates by Miller's method using o-nitrophenyl-β-D-galactopyranoside (ONPG) as a substrate [42].
Electrophoretic Mobility Shift Assay (EMSA)
The promoter region of vtrB, containing a 284 bp upstream sequence of the start codon, was amplified by polymerase chain reaction (PCR). PCR products purified from agarose gels were then mixed with increasing concentrations of the purified DNA binding domains of VtrA (amino acids 1–133) and VtrB (amino acids 1–158) in a reaction buffer containing 10 µg/ml of bovine serum albumin (BSA). After 30 min incubation at room temperature, samples were separated by 5% polyacrylamide nondenatureing gels in TAE buffer at room temperature. DNA was stained with SYBR Green I Nucleic Acid Gel Stain (Lonza) and visualized with a LAS-4000 mini EPUV (Fujifilm) at 460 nm emission wevelength.
Cytotoxicity Assays
T3SS1 and T3SS2-dependent cytotoxicity assays were performed as described [25]. Briefly, Caco-2 cells were seeded at 3×104 cells per well in 96-well plates and cultured for 48 h to confluency. The cells were co-cultured for 1.5–6 h with phosphate buffered saline (PBS)-washed bacteria at a multiplicity of infection (MOI) of 10. The release of lactate dehydrogenase (LDH) into the medium was quantified using CytoTox96 (Promega). The LDH release (percent cytotoxicity) was calculated using the following equation: (optical density at 490 nm [OD490] of experimental release – OD490 of spontaneous release)/(OD490 of maximum release – OD490 of spontaneous release)×100. Spontaneous release was taken to be the amount of LDH released from the cytoplasm of uninfected cells, whereas the maximum release was the amount released by total lysis of uninfected cells.
Rabbit Ileal Loop Test
V. parahaemolyticus strains were grown overnight in LB broth with 3% NaCl. Cultures were then diluted 1∶100 into LB broth with 3% NaCl and grown with shaking for 5.5 h. After incubation, bacteria were harvested by centrifugation and suspended in LB broth with 0.5% NaCl. The bacterial suspensions (109 CFU) were injected into the ligated ileal loops of rabbits, and fluid accumulation in each loop was measured at 16 h after challenge. The result was expressed as the amount of accumulated fluid (in milliliters) per length (in centimeters) of ligated rabbit small intestine. All animal experiments were performed according to an experimental protocol approved by the Ethics Review Committee for Animal Experimentation of Research Institute for Microbial Diseases (Osaka University, Osaka, Japan).
Statistical Analysis
All data are presented as the mean and standard deviation of three determinations per experimental condition. The statistical significance was determined by one-way ANOVA followed by Dunnett's multiple comparison test, and P<0.05 was considered statistically significant.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by Grants-in-Aid for Young Scientists (21790419, http://kaken.nii.ac.jp/ja/p/21790419) and Scientific Research on Priority Areas Applied Genomics (17019058, http://kaken.nii.ac.jp/ja/p/17019058), and Matrix of Infection Phenomena from the Ministry of Education (18073003, http://kaken.nii.ac.jp/ja/p/18073003), Culture, Sports, Science and Technology of Japan. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Blake PA, Weaver RE, Hollis DG. Diseases of humans (other than cholera) caused by vibrios. Annu Rev Microbiol. 1980;34:341–367. doi: 10.1146/annurev.mi.34.100180.002013. [DOI] [PubMed] [Google Scholar]
- 2.Morris JG, Jr, Black RE. Cholera and other vibrioses in the United States. N Engl J Med. 1985;312:343–350. doi: 10.1056/NEJM198502073120604. [DOI] [PubMed] [Google Scholar]
- 3.Hlady WG, Klontz KC. The epidemiology of Vibrio infections in Florida, 1981–1993. J Infect Dis. 1996;173:1176–1183. doi: 10.1093/infdis/173.5.1176. [DOI] [PubMed] [Google Scholar]
- 4.Daniels NA, MacKinnon L, Bishop R, Altekruse S, Ray B, et al. Vibrio parahaemolyticus infections in the United States, 1973–1998. J Infect Dis. 2000;181:1661–1666. doi: 10.1086/315459. [DOI] [PubMed] [Google Scholar]
- 5.Miyamoto Y, Kato T, Obara Y, Akiyama S, Takizawa K, et al. In vitro hemolytic characteristic of Vibrio parahaemolyticus: its close correlation with human pathogenicity. J Bacteriol. 1969;100:1147–1149. doi: 10.1128/jb.100.2.1147-1149.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Honda T, Iida T. The pathogenicity of Vibrio parahaemolyticus and the role of the thermostable direct haemolysin and related haemolysins. Rev Med Microbiol. 1993;4:106–113. [Google Scholar]
- 7.Nishibuchi M, Kaper JB. Thermostable direct hemolysin gene of Vibrio parahaemolyticus: a virulence gene acquired by a marine bacterium. Infect Immun. 1995;63:2093–2099. doi: 10.1128/iai.63.6.2093-2099.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Honda T, Goshima K, Takeda Y, Sugino Y, Miwatani T. Demonstration of the cardiotoxicity of the thermostable direct hemolysin (lethal toxin) produced by Vibrio parahaemolyticus. Infect Immun. 1976;13:163–171. doi: 10.1128/iai.13.1.163-171.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Honda T, Takeda Y, Miwatani T, Kato K, Nimura Y. [Clinical features of patients suffering from food poisoning due to Vibrio parahaemolyticus - with special reference to changes in electrocardiograms]. Kansenshogaku Zasshi. 1976;50:216–223. doi: 10.11150/kansenshogakuzasshi1970.50.216. [DOI] [PubMed] [Google Scholar]
- 10.Sakurai J, Honda T, Jinguji Y, Arita M, Miwatani T. Cytotoxic effect of the thermostable direct hemolysin produced by Vibrio parahaemolyticus on FL cells. Infect Immun. 1976;13:876–883. doi: 10.1128/iai.13.3.876-883.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goshima K, Honda T, Hirata M, Kikuchi K, Takeda Y. Stopping of the spontaneous beating of mouse and rat myocardial cells in vitro by a toxin from Vibrio parahaemolyticus. J Mol Cell Cardiol. 1977;9:191–213. doi: 10.1016/0022-2828(77)90029-3. [DOI] [PubMed] [Google Scholar]
- 12.Tang GQ, Iida T, Yamamoto K, Honda T. Ca(2+)-independent cytotoxicity of Vibrio parahaemolyticus thermostable direct hemolysin (TDH) on Intestine 407, a cell line derived from human embryonic intestine. FEMS Microbiol Lett. 1995;134:233–238. doi: 10.1111/j.1574-6968.1995.tb07943.x. [DOI] [PubMed] [Google Scholar]
- 13.Tang G, Iida T, Yamamoto K, Honda T. Analysis of functional domains of Vibrio parahaemolyticus thermostable direct hemolysin using monoclonal antibodies. FEMS Microbiol Lett. 1997;150:289–296. doi: 10.1016/s0378-1097(97)00133-x. [DOI] [PubMed] [Google Scholar]
- 14.Fabbri A, Falzano L, Frank C, Donelli G, Matarrese P, et al. Vibrio parahaemolyticus thermostable direct hemolysin modulates cytoskeletal organization and calcium homeostasis in intestinal cultured cells. Infect Immun. 1999;67:1139–1148. doi: 10.1128/iai.67.3.1139-1148.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raimondi F, Kao JP, Fiorentini C, Fabbri A, Donelli G, et al. Enterotoxicity and cytotoxicity of Vibrio parahaemolyticus thermostable direct hemolysin in in vitro systems. Infect Immun. 2000;68:3180–3185. doi: 10.1128/iai.68.6.3180-3185.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Naim R, Yanagihara I, Iida T, Honda T. Vibrio parahaemolyticus thermostable direct hemolysin can induce an apoptotic cell death in Rat-1 cells from inside and outside of the cells. FEMS Microbiol Lett. 2001;195:237–244. doi: 10.1111/j.1574-6968.2001.tb10527.x. [DOI] [PubMed] [Google Scholar]
- 17.Takahashi A, Iida T, Naim R, Naykaya Y, Honda T. Chloride secretion induced by thermostable direct haemolysin of Vibrio parahaemolyticus depends on colonic cell maturation. J Med Microbiol. 2001;50:870–878. doi: 10.1099/0022-1317-50-10-870. [DOI] [PubMed] [Google Scholar]
- 18.Makino K, Oshima K, Kurokawa K, Yokoyama K, Uda T, et al. Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V cholerae. Lancet. 2003;361:743–749. doi: 10.1016/S0140-6736(03)12659-1. [DOI] [PubMed] [Google Scholar]
- 19.Izutsu K, Kurokawa K, Tashiro K, Kuhara S, Hayashi T, et al. Comparative genomic analysis using microarray demonstrates a strong correlation between the presence of the 80-kilobase pathogenicity island and pathogenicity in Kanagawa phenomenon-positive Vibrio parahaemolyticus strains. Infect Immun. 2008;76:1016–1023. doi: 10.1128/IAI.01535-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sugiyama T, Iida T, Izutsu K, Park KS, Honda T. Precise region and the character of the pathogenicity island in clinical Vibrio parahaemolyticus strains. J Bacteriol. 2008;190:1835–1837. doi: 10.1128/JB.01293-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park KS, Ono T, Rokuda M, Jang MH, Okada K, et al. Functional characterization of two type III secretion systems of Vibrio parahaemolyticus. Infect Immun. 2004;72:6659–6665. doi: 10.1128/IAI.72.11.6659-6665.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meador CE, Parsons MM, Bopp CA, Gerner-Smidt P, Painter JA, et al. Virulence gene- and pandemic group-specific marker profiling of clinical Vibrio parahaemolyticus isolates. J Clin Microbiol. 2007;45:1133–1139. doi: 10.1128/JCM.00042-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Skorupski K, Taylor RK. Control of the ToxR virulence regulon in Vibrio cholerae by environmental stimuli. Mol Microbiol. 1997;25:1003–1009. doi: 10.1046/j.1365-2958.1997.5481909.x. [DOI] [PubMed] [Google Scholar]
- 24.Miller VL, Mekalanos JJ. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kodama T, Rokuda M, Park KS, Cantarelli VV, Matsuda S, et al. Identification and characterization of VopT, a novel ADP-ribosyltransferase effector protein secreted via the Vibrio parahaemolyticus type III secretion system 2. Cell Microbiol. 2007;9:2598–2609. doi: 10.1111/j.1462-5822.2007.00980.x. [DOI] [PubMed] [Google Scholar]
- 26.Kodama T, Hiyoshi H, Gotoh K, Akeda Y, Matsuda S, et al. Identification of two translocon proteins of Vibrio parahaemolyticus type III secretion system 2. Infect Immun. 2008;76:4282–4289. doi: 10.1128/IAI.01738-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Honda T, Ni YX, Hata A, Yoh M, Miwatani T, et al. Properties of a hemolysin related to the thermostable direct hemolysin produced by a Kanagawa phenomenon negative, clinical isolate of Vibrio parahaemolyticus. Can J Microbiol. 1990;36:395–399. doi: 10.1139/m90-069. [DOI] [PubMed] [Google Scholar]
- 28.Niikawa T, Obara Y, Yamai S, Miyamoto Y. Purification of a hemolysin from Vibrio parahaemolyticus. Jpn J Med Sci Biol. 1972;25:197–200. [PubMed] [Google Scholar]
- 29.Zen-Yoji H, Hitokoto H, Morozumi S, Le Clair RA. Purification and characterization o;f a hemolysin produced by Vibrio parahaemolyticus. J Infect Dis. 1971;123:665–667. doi: 10.1093/infdis/123.6.665. [DOI] [PubMed] [Google Scholar]
- 30.Sakurai J, Matsuzaki A, Miwatani T. Purification and characterization of thermostable direct hemolysin of Vibrio parahaemolyticus. Infect Immun. 1973;8:775–780. doi: 10.1128/iai.8.5.775-780.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miyamoto Y, Obara Y, Nikkawa T, Yamai S, Kato T, et al. Simplified purification and biophysicochemical characteristics of Kanagawa phenomenon-associated hemolysin of Vibrio parahaemolyticus. Infect Immun. 1980;28:567–576. doi: 10.1128/iai.28.2.567-576.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nishibuchi M, Fasano A, Russell RG, Kaper JB. Enterotoxigenicity of Vibrio parahaemolyticus with and without genes encoding thermostable direct hemolysin. Infect Immun. 1992;60:3539–3545. doi: 10.1128/iai.60.9.3539-3545.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park KS, Ono T, Rokuda M, Jang MH, Iida T, et al. Cytotoxicity and enterotoxicity of the thermostable direct hemolysin-deletion mutants of Vibrio parahaemolyticus. Microbiol Immunol. 2004;48:313–318. doi: 10.1111/j.1348-0421.2004.tb03512.x. [DOI] [PubMed] [Google Scholar]
- 34.Okada N, Iida T, Park KS, Goto N, Yasunaga T, et al. Identification and characterization of a novel type III secretion system in trh-positive Vibrio parahaemolyticus strain TH3996 reveal genetic lineage and diversity of pathogenic machinery beyond the species level. Infect Immun. 2009;77:904–913. doi: 10.1128/IAI.01184-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tam VC, Serruto D, Dziejman M, Brieher W, Mekalanos JJ. A type III secretion system in Vibrio cholerae translocates a formin/spire hybrid-like actin nucleator to promote intestinal colonization. Cell Host Microbe. 2007;1:95–107. doi: 10.1016/j.chom.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 36.Dziejman M, Serruto D, Tam VC, Sturtevant D, Diraphat P, et al. Genomic characterization of non-O1, non-O139 Vibrio cholerae reveals genes for a type III secretion system. Proc Natl Acad Sci U S A. 2005;102:3465–3470. doi: 10.1073/pnas.0409918102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller VL, Taylor RK, Mekalanos JJ. Cholera toxin transcriptional activator toxR is a transmembrane DNA binding protein. Cell. 1987;48:271–279. doi: 10.1016/0092-8674(87)90430-2. [DOI] [PubMed] [Google Scholar]
- 38.Hung DT, Mekalanos JJ. Bile acids induce cholera toxin expression in Vibrio cholerae in a ToxT-independent manner. Proc Natl Acad Sci U S A. 2005;102:3028–3033. doi: 10.1073/pnas.0409559102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin Z, Kumagai K, Baba K, Mekalanos JJ, Nishibuchi M. Vibrio parahaemolyticus has a homolog of the Vibrio cholerae toxRS operon that mediates environmentally induced regulation of the thermostable direct hemolysin gene. J Bacteriol. 1993;175:3844–3855. doi: 10.1128/jb.175.12.3844-3855.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakano M, Takahashi A, Su Z, Harada N, Mawatari K, et al. Hfq regulates the expression of the thermostable direct hemolysin gene in Vibrio parahaemolyticus. BMC Microbiol. 2008;8:155. doi: 10.1186/1471-2180-8-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Valentin-Hansen P, Eriksen M, Udesen C. The bacterial Sm-like protein Hfq: a key player in RNA transactions. Mol Microbiol. 2004;51:1525–1533. doi: 10.1111/j.1365-2958.2003.03935.x. [DOI] [PubMed] [Google Scholar]
- 42.Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor, NY.: Cold Spring Harbor Laboratory; 1972. Experiment 48 Assay of β-Galactosidase. [Google Scholar]
- 43.Ono T, Park KS, Ueta M, Iida T, Honda T. Identification of proteins secreted via Vibrio parahaemolyticus type III secretion system 1. Infect Immun. 2006;74:1032–1042. doi: 10.1128/IAI.74.2.1032-1042.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Casadaban MJ. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J Mol Biol. 1976;104:541–555. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
- 45.Parales RE, Harwood CS. Construction and use of a new broad-host-range lacZ transcriptional fusion vector, pHRP309, for gram- bacteria. Gene. 1993;133:23–30. doi: 10.1016/0378-1119(93)90220-w. [DOI] [PubMed] [Google Scholar]
- 46.Kodama T, Akeda Y, Kono G, Takahashi A, Imura K, et al. The EspB protein of enterohaemorrhagic Escherichia coli interacts directly with alpha-catenin. Cell Microbiol. 2002;4:213–222. doi: 10.1046/j.1462-5822.2002.00176.x. [DOI] [PubMed] [Google Scholar]
- 47.Nomura T, Hamashima H, Okamoto K. Carboxy terminal region of haemolysin of Aeromonas sobria triggers dimerization. Microb Pathog. 2000;28:25–36. doi: 10.1006/mpat.1999.0321. [DOI] [PubMed] [Google Scholar]