Abstract
The CUG-BP and ETR-3 like factors (CELF) are a family of six highly conserved RNA-binding proteins that preferentially bind to UG-rich sequences. One of the key functions of these proteins is to mediate alternative splicing in a number of tissues, including brain, heart and muscle. To fully understand the function of CELF proteins, it is important to identify downstream targets of CELF proteins. In this communication, we report that neurofibromatosis type I (NF1) exon 23a is a novel target of CELF protein-mediated splicing regulation in neuron-like cells. NF1 regulates Ras signaling, and the isoform that excludes exon 23a shows 10 times greater ability to down-regulate Ras signaling than the isoform that includes exon 23a. Five of the six CELF proteins strongly suppress the inclusion of NF1 exon 23a. Over-expression or siRNA knockdown of these proteins in cell transfection experiments altered the levels of NF1 exon 23a inclusion. In vitro binding and splicing analyses demonstrate that CELF proteins block splicing through interfering with binding of U2AF65. These studies, combined with our previous investigations demonstrating a role for Hu proteins and TIA-1/TIAR in controlling NF1 exon 23a inclusion, highlight the complex nature of regulation of this important alternative splicing event.
INTRODUCTION
It is now well established that alternative splicing is an important means of gene regulation. This process allows a diverse host of mRNA messages to be generated from a single gene, which is essential given that there are a limited number of genes from which a myriad of functionally distinct protein products must be made. The most recent estimates, which have been obtained using new technologies such as deep sequencing, suggest that as many as 94% of all human genes undergo alternative splicing (1,2). Alternative splicing has been demonstrated to be important in the establishment of tissue specificity as well as in development. This phenomenon is especially robust and diverse in the nervous system, where it is responsible for the modulation of functions such as axon guidance, membrane physiology and synapse formation (3–5).
Alternative splicing is regulated by both cis-acting elements and trans-acting factors. cis-Acting elements are located within flanking introns or within alternative exons themselves, and may act as enhancers or silencers (6,7). Several trans-acting splicing regulators that direct neuron-specific splicing have been identified, including Nova-1/2, Hu proteins, ETR-3, nPTB and Fox-1/2 (8–16). There are a host of other splicing regulators that include SR proteins, hnRNPs, PTB and TIA-1/TIAR which regulate splicing in other tissues (17). With the extensive nature of alternative splicing in the nervous system, it is expected that many more regulatory elements and proteins are involved. Recent studies have shown that there are conserved sequences, which are enriched in the intronic regions that surround alternatively spliced exons (1,18). One of these studies looked at greater than 24 000 alternative splicing events in human tissues and cell lines, and uncovered over 100 enriched 4–7 nucleotide sequences or “words,” many of which are known as binding sites for key splicing regulators such as the CUG-BP and ETR-3 like factors (CELF), Fox 1 and Fox 2, PTB, and muscleblind-like proteins (18). Interestingly, this same study found a large number of enriched sequences for which binding partners have not been identified. Thus, it is clear that there is still a great deal to be learned about trans-acting factors that can regulate alternative splicing.
The CUG-binding protein (CUG-BP) and embryonic lethal abnormal vision type RNA-binding protein-3 (ETR-3) like factors (CELF) are a family of highly conserved RNA-binding proteins which consist of six members. CELF proteins are well-characterized regulators of alternative splicing. In vitro and transfection studies using mutated mini-gene reporters have demonstrated that the CELF proteins bind to UG-rich sequences in the introns flanking alternative exons of their target pre-mRNAs (19–23). The structure of these proteins is conserved with three RNA recognition domains, two of which are separated by a divergent hinge domain. The CELF protein family is subdivided, based on sequence similarities, into two subfamilies. CUG-BP1 and ETR-3 make up one of the subfamilies, and the second subfamily is comprised of CELF members 3–6. The founding member of the CELF family, CUG-BP1, was originally identified in a screen for proteins that could bind to a CUG-repeat probe in an in vitro gel shift assay (24,25). The interest in proteins that could bind to this RNA motif was born out of the knowledge that a CUG trinucleotide expansion is present in the 3′ untranslated region of the DMPK gene of myotonic dystrophy (DM) patients. The second well-characterized CELF protein, ETR-3, was found in a screen for apoptotic factors in the mouse brain and in a screen for factors involved in the development of the embryonic heart (26,27). The members of the second subfamily of CELF proteins were identified based on their sequence homology to CUG-BP1 and ETR-3. CUG-BP1 and ETR-3 are the most comprehensively studied CELF proteins and have widespread distribution with enrichment in the brain, heart and muscle (28–30). CELF3, CELF4 and CELF5 are brain-specific proteins, and CELF6 is enriched in the brain and testes (29,30).
CELF proteins have a myriad of functions in the cell, the best-characterized of which are in the regulation of the alternative splicing of a number of target genes, including cardiac troponin T (cTNT) and the insulin receptor (19,22,23,30–34). These proteins have been demonstrated in both tissue-specific and developmental stage-specific alternative splicing events. The CELF proteins can act as either positive or negative regulators of alternative splicing. For example, CUG-BP1 and CELF6 promote skipping of exon 11 in the insulin receptor pre-mRNA, while all six family members promote inclusion of exon 5 of the cardiac troponin T pre-mRNA (30). Importantly, ETR-3 plays a key role in neuron-specific splicing control, where it acts as either a positive or a negative regulator of two alternative exons (14).
In DM, many CELF protein targets are aberrantly spliced. DM is characterized as a disease of RNA toxicity, in which a CUG trinucleotide expansion in the 3′ untranslated region of the myotonic dystrophy kinase gene leads to an up-regulation of CUG-BP1 and sequestering of another RNA-binding protein, muscleblind-like 1. Several animal models have been generated to abnormally express CUG-BP1. These animals mimic the splicing mis-regulation of CELF protein targets in DM (32,35). Since the CELF proteins' function as splicing regulators is so important in human disease and development, it is imperative to identify additional CELF protein-mediated splicing targets. In recent years, much research effort has been put towards reaching this end, however relatively few new CELF targets have been identified.
Neurofibromatosis type I (NF1) is a common autosomal dominant genetic disorder that affects 1 in 3500 individuals (36). The NF1 gene is highly conserved, and it possesses at least three alternatively spliced exons: 9br, 23a and 48a. NF1 exon 23a is of particular interest, because it lies within the only well-characterized domain of the NF1 protein product, neurofibromin. This domain is referred to as the GTPase activating protein-related domain (GRD), because it shares sequence identity with the mammalian GAP domain and it can complement yeast Ira-1 and Ira-2 mutants (37,38). The NF1 GRD negatively regulates the activities of the oncogene Ras and thus, NF1 is considered a tumor suppressor gene. Interestingly, the inclusion NF1 exon 23a alters Ras-GAP activity. The form of NF1 that does not include exon 23a shows 10-times greater activity in down-regulation of Ras than the form that includes this exon (39,40) The splicing pattern of NF1 exon 23a is highly conserved, with the exon being included in most tissues but skipped in neurons as demonstrated in Figure 1 (40). Mouse models in which NF1 exon 23a is ablated exhibit learning and memory deficits (41). These findings suggest that it is important to have a balance between the two NF1 mRNA species.
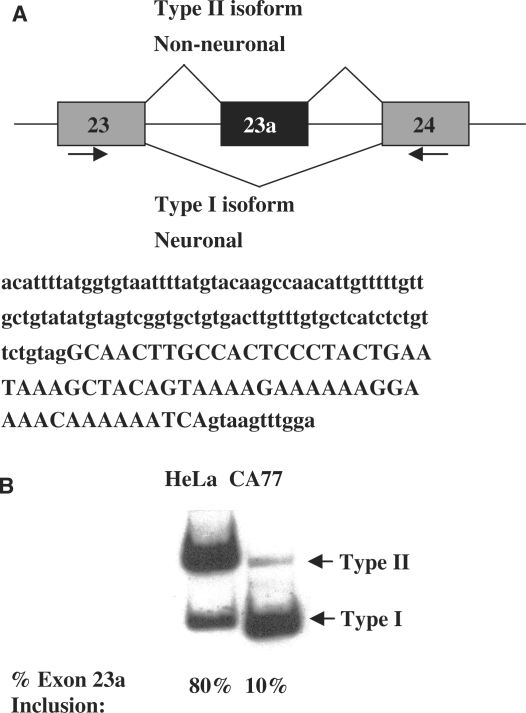
Figure 1.
NF1 Exon 23a alternative inclusion patterns in cell lines. (A) Schematic diagram showing the alternative splicing pattern of NF1 exon 23a. The Type II isoform includes exon 23a and is found in most non-neuronal tissues. The Type I isoform skips exon 23a and is found in neuronal tissues. The sequence of NF1 exon 23a and a portion of its surrounding introns are shown in uppercase and lowercase letters, repectively. (B) Endogenous NF1 splicing patterns in cell model systems. Total RNA was isolated from the CA77 and HeLa cell lines, and endogenous NF1 exon 23a inclusion was analyzed by RT-PCR assay using oligonucleotides indicated by arrows in panel A. The resulting PCR amplification products are either 267 nucleotides for exon 23a inclusion or 204 nucleotides for exon 23a skipping.
Recently, our laboratory established that the RNA-binding proteins, TIA-1, TIAR and the Hu family play important roles in the regulation of NF1 exon 23a inclusion (13). These proteins bind to AU-rich sequences found in the introns flanking the alternative exon, with TIA-1 and TIAR functioning as positive regulatory elements, which promote exon 23a inclusion in non-neuronal tissues. Conversely, Hu proteins bind to AU-rich sequences flanking this exon to promote its skipping in neuronal tissues. Consistent with the notion that regulation of alternative splicing in mammals is complicated, often involving multiple sequence elements and protein factors, our sequence analysis suggests that additional regulatory mechanisms exist that control alternative inclusion of NF1 exon 23a.
The intronic sequence located immediately upstream of exon 23a is highly enriched in UG-rich elements, which have been shown to be the binding sites of the CELF family of RNA-binding proteins. In this report, we present evidence to support that CELF proteins indeed function as regulators for inclusion of NF1 exon 23a. They promote skipping of this exon in neuron-like cells. All of the CELF protein members, except CELF6 strongly promote NF1 exon 23a skipping when individually over-expressed in non-neuronal cells. Additional support for the role of CELF proteins in the regulation of this alternative exon is provided by siRNA knockdown of ETR-3, and the over-expression of a dominant negative CELF protein in neuron-like cells. Furthermore, we show that both endogenous and recombinant CELF proteins bind strongly to an NF1 RNA substrate and block binding of U2AF65, an essential splicing factor that interacts with the polypyrimidine-tract sequence at the 3′ splice site. Finally, we demonstrate that recombinant CELF proteins block NF1 splicing by in vitro splicing assays.
MATERIALS AND METHODS
Plasmids
The human NF1 reporter constructs used in transfection experiments consist of NF1 exon 23a with part of the flanking introns inserted into the first intron of the human metallothionein (HMT) gene (13). The CELF protein expression plasmids for CUG-BP1, ETR-3 and CELF3-6 were gifts from Dr Tom Cooper, Baylor College of Medicine. The plasmid used as template for in vitro transcription of RNA probe 1 was generated by PCR-mediated cloning, while the plasmid used as template for in vitro transcription of RNA probe 2 was generated by oligonucleotide-mediated cloning using oligonucleotides NF1-977 and NF1-978 (Table 1). The cloned plasmids were confirmed by DNA sequencing. The rat ETR-3 cDNA was generated by RT-PCR amplification using RNA isolated from CA77 cells and ETR-3 5′ and ETR-3 3′ oligonucleotides (Table 1). The resulting PCR product was digested with BamHI and XbaI, and ligated into the BamHI–XbaI-digested pcDNA3.1HisC vector (Invitrogen). This clone was confirmed by sequencing.
Table 1.
Oligonucleotides used in this study
| Oligonucleotide | Sequence |
|---|---|
| DS8 | TTGACCATTCACCACATTGGTGTGC |
| HMT3 | ATCTGGGAGCGGGGCTGT |
| NF1 5′ | AAGTTCTTCCATGCCATCATCAG |
| NF1 3′ | ATTCTAGGTGGTGGCTTTTTATCTA |
| Beta Actin 5′ | TGGGCGACGAGGCCCAGAGCA |
| Beta Actin 3′ | GTCAGGTCCCGGCCAGCCAGG |
| CELF3 5′ | GACCGGAAGCTCTTTGTGGGG |
| CELF3 3′ | AGGGTCCGGCTGCTGTGA |
| CELF4 5′ | TTCATTGGGCAGATCCCCCG |
| CELF4 3′ | CAGGCTTCACCTGGATCGGCC |
| CELF5 5′ | GACCGGAAGCTGTTCGTGGG |
| CELF5 3′ | TTGACCACCAGGCTGGAGGA |
| CELF6 5′ | TTCGTGGGGCAGATCCCGCGG |
| CELF6 3′ | GGCAGGGTCTTCTGCTCGTGC |
| NF1-977 | AGCTT GGT GTA ATT TTA TGT ACA AGC CAA CAT TGT TTT TGT TGC TGT ATG TAG TCG GTG CTG TGA CTT GTT TGT GCT CAT CTC TGT TCT GTA G |
| NF1-978 | GATCC CTA CAG AAC AGA GAT GAG CAC AAA CAA GTC ACA GCA CCG ACT ACA TAC AGC AAC AAA AAC AAT GTT GGC TTG TAC ATA AAA TTA CAC C |
| ETR-3 5′ | AAAGGATCCATGAACGGAGCTTTGGATCATTCCGAC |
| ETR-3 3′ | AAATCTAGATCAGTAAGGTTTTGCTGTCGTTTTT |
Cell culture and cell transfection
HeLa cells were maintained according to instructions from the American Type Culture Collection (Manassas, VA). CA77 cells, a cell line derived from rat medullary thyroid carcinoma (a gift from Drs. Alison Hall at Case Western Reserve University, Cleveland, OH and Andrew Russo, University of Iowa, Iowa City, IA), were cultured in DMEM/F-12 (Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen) and 1% pen/strep (Invitrogen). HeLa cells were transfected as described earlier (42). Transfection of CA77 cells was carried out the same as HeLa cells except that Lipofectamine 2000 (Invitrogen) was used, and cells were grown for 72 h instead of 48 h after transfection. Co-transfections were carried out using 1 µg of the NF1 reporter plasmid and 0.5–1 µg of CELF protein plasmid. Other CA77 transfections were carried utilizing the Nucleofector Kit V with the Nucleofector II device (Lonza, formerly Amaxa). These transfections were performed with the standard protocol recommended by Lonza using 1 µg of the HMT-NF1 863/499 reporter and 4 µg of either an empty vector or the CELF dominant negative expression plasmid. Cells were grown for 72 h after the transfection before protein lysates were harvested.
RNA and protein analysis
Procedures for total RNA and protein isolation and RT–PCR analysis were described previously (42). Oligonucleotide pairs DS8 and HMT3, and NF1 5′ and NF1 3′ were used to analyze the NF1 reporter RNA and endogenous NF1 RNA, respectively (13). Low PCR cycle numbers were used to analyze RNA isolated from HeLa (16-20 cycles) or CA77 (19-22 cycles) cells. Endogenous NF1 RNA in all of the cell lines was analyzed using between 22 and 26 PCR cycles. Quantification of exon inclusion was determined by PhosphorImager analysis using a Typhoon Trio (GE Healthcare). The results shown are representative of at least three independent transfections for each experiment. The effect of CELF proteins on RNA processing of the reporter pre-mRNA was calculated as the percentage of NF1 exon 23a inclusion [exon 23a inclusion/(exon 23a inclusion + exon 23a exclusion)]. Western blot analysis from transfected cell lysates was carried out with anti-Xpress antibody (Invitrogen).
siRNA-mediated knockdown of ETR-3
The rat ETR-3 siRNA duplex was synthesized by Dharmacon, and its target sequence is UCGGCAUGAAACGCUUGAA. Co-transfections were performed in CA77 cells using either 200 pmol of PTB siRNA as a negative control, or 200 pmol of ETR-3 siRNA and 1 µg of HMT-NF1 863/499 reporter. Transfections were carried out using Lipofectamine 2000 (Invitrogen) using a previously described protocol (13).
In vitro assays
Pre-mRNAs were transcribed in vitro from plasmid templates using the MEGAscript SP6 kit from Ambion. UV cross-linking reactions were carried out in a volume of 50 µl containing 44% (vol/vol) HeLa cell nuclear extract, 2 mM ATP, 20 mM creatine phosphate, 0.6 mM MgCl2, 1.5% polyethylene glycol, 0.15 mM dithiothreitol, and 5 × 105 c.p.m. of 32P-labeled RNA. Reaction mixtures were incubated at 30°C for 10 min, and heparin was added to a final concentration of 2 µg/µl, followed by UV irradiation (254 nm) at 4°C for 10 min. Reaction mixtures were subsequently treated with 30 µg of RNase A at 37°C for 30 min. Cross-linked polypeptides were immunoprecipitated using monoclonal antibodies against CUG-BP1 (3B1, Invitrogen), ETR-3 (1H2, Santa Cruz Biotechnology, Inc), or U2AF65 (Sigma). Immunoprecipitated proteins were separated on sodium dodecyl sulfate–10% polyacrylamide electrophoresis gels. In vitro splicing reactions were performed as described earlier (44). Recombinant GST, GST-CUG-BP1 and GST-TIAR proteins were prepared from bacteria, and either 0.75 or 1.5 µg of these proteins were used in the assays. Quantification of the percentage of efficiency of lariat formation was obtained using a PhosphorImager. The number of uridine residues present in both the full substrate and the exon 23a-containing lariat were determined from the sequence information. Then the exon 23a-containing lariat data from the PhosphorImager was normalized to the data for the full substrate. After normalization, the percentage of efficiency of lariat formation was determined by the following calculation: [(exon 23a-containing lariat)/(exon 23a-containing lariat + full substrate)].
Preparation of nuclear extracts from HeLa and CA77 cells
HeLa cell nuclear extracts were prepared using S3 suspension culture and standard techniques (42). To make nuclear extracts from CA77 cells, 100 100-mm dishes of CA77 monolayer cells were collected and used according to a standard procedure (42).
RESULTS
Endogenous NF1 pre-mRNA is differentially spliced in two cell models
We utilize a cell-based system to study alternative splicing of the NF1 pre-mRNA. NF1 exon 23a is predominantly included in non-neuronal tissues, while it is mostly skipped in neurons [Figure 1A and (44,45)]. In our system, HeLa cells, which are derived from human cervical cancer cells, are used to mimic the non-neuronal splicing pattern while CA77 cells, which are derived from rat medullary thyroid carcinomas and have a number of neuronal features, are used to mimic the neuronal splicing pattern (46). Semi-quantitative RT–PCR analysis, using total RNA isolated from these two cell types and oligonucleotides that anneal to sequences in exons 23 and 24 of NF1, demonstrates that exon 23a is predominantly included in HeLa cells and predominantly skipped in CA77 cells (Figure 1B). These findings validate our choice to use these cell models to study tissue-specific alternative splicing of NF1 exon 23a.
CELF proteins are differentially expressed in the two cell models
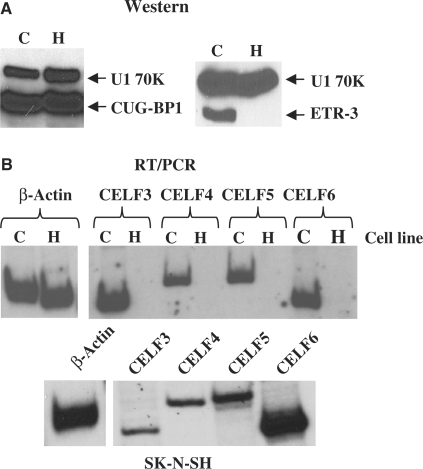
We sought to understand how NF1 exon 23a inclusion is regulated. The intronic sequence upstream of this alternative exon has a series of UG-rich elements (Figure 1A), making it a potential target for regulation by the CELF family of RNA-binding proteins. We hypothesized that CELF proteins could act as negative regulators of NF1 exon 23a inclusion by binding to the UG-rich elements and blocking assembly of spliceosomal components. In order to determine the CELF protein expression profiles in CA77 and HeLa cells, we employed two approaches. First, we carried out a western blot analysis using total protein lysate isolated from the two cell lines and commercially available antibodies against CUG-BP1 or ETR-3. As shown in Figure 2A, CUG-BP1 is present at essentially equivalent levels in the two cell lines. However, ETR-3 is expressed in CA77 cells but undetectable in HeLa cells (Figure 2A). Second, for CELF family members 3–6, for which commercial antibodies are not available, we carried out semi-quantitative RT–PCR using oligonucleotides (Table 1) that were designed in conserved regions between rat and human CELF sequences, which are also unlikely to undergo alternative splicing. We found that CELF3–6 mRNAs are present at a much higher level in total RNA isolates from CA77 than they are in RNAs derived from HeLa cells (Figure 2B). To confirm that our RT–PCR primers worked with human sequences, we performed the RT–PCR using a human neuroblastoma cell line, SK–N–SH, and in this cell line we detected expression of all CELF family mRNAs (Figure 2B, bottom panel). These results demonstrate a correlation between exon 23a skipping and a high level of CELF family member expression and therefore, are consistent with the hypothesis that CELF proteins are potential candidates as negative regulators of NF1 exon 23a inclusion.
Figure 2.
Expression profile of CELF proteins in the two cell models. (A) Total protein was isolated from CA77 (labeled as C) and HeLa (labeled as H) cells and western blot analysis was performed using antibodies against CUG-BP1 or ETR-3 and U1 70K as a loading control. (B) Total RNA was isolated from CA77 and HeLa cells and RT–PCR was performed using oligonucleotides designed to anneal to CELF family members 3–6 in regions conserved between rat and human. PCR amplification products are 201 nucleotides for CELF3, 240 nucleotides for CELF4, 241 nucleotides for CELF5, and 205 nucleotides for CELF6. β-actin was used as a control, and has a PCR amplification product of 376 nucleotides. The human neuroblastoma cell line, SK–N–SH, was used as a control to verify that all of the oligonucleotides worked in both human and rat cells.
Over-expression of CELF proteins in HeLa cells promotes NF1 exon 23a skipping
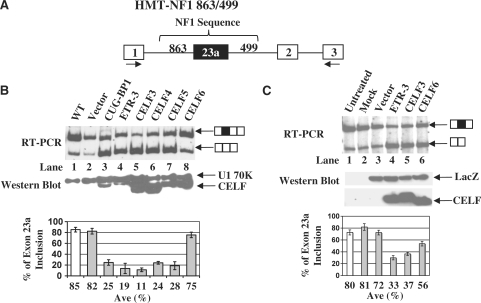
To test the hypothesis that CELF proteins regulate NF1 exon 23a inclusion, we performed co-transfection experiments in which individual human CELF proteins were over-expressed along with an NF1 minigene reporter construct in HeLa cells, the low CELF protein expressing cell line. The minigene reporter construct was generated in previous studies in our laboratory by inserting NF1 sequence containing exon 23a and flanking intronic regions including the presumed CELF regulatory elements into intron 1 of the human metallothionine II gene (Figure 3A) (13). The splicing pattern of NF1 exon 23a was examined by semi-quantitative RT–PCR using oligonucleotides that anneal to exons 1 and 3 of the human metallothionine II gene. The splicing pattern of this reporter pre-mRNA mimics that of the endogenously expressed NF1 pre-mRNA (13). In all cases, except for CELF6, over-expression of CELF proteins strongly promotes skipping of NF1 exon 23a (Figure 3B). CUG-BP1 causes a switch from 85% NF1 exon 23a inclusion in the reporter to 25% inclusion. Likewise, CELF4 and CELF5 decrease inclusion to 24 and 28%, respectively (Figure 3B, lanes 1–3 and 6–7). The most dramatic effects upon NF1 exon 23a inclusion were seen with over-expression of ETR-3, which reduced inclusion to 19% and with CELF3, which reduced the inclusion to 11% (Figure 3B, lanes 1–2 and 4–5). Over-expression of CELF6 with the NF1 reporter does not have a strong effect on the splicing phenotype of exon 23a (Figure 3B, lanes 1, 2 and 8). Western blot analysis performed using protein lysates from transfected cells and an anti-tag antibody (anti-Xpress) shows that CELF family members were over-expressed in HeLa cells (Figure 3B). To further validate our findings, we over-expressed the CELF proteins along with a minigene for a well-studied and verified target of CELF protein-mediated splicing, cardiac troponin T (cTNT). Over-expression of the CELF proteins promoted inclusion of cardiac troponin T exon 5, as expected (data not shown) (29,30).
Figure 3.
Over-expression of CELF proteins in HeLa cells promotes exon 23a skipping. (A) Schematic diagram of the HMT-NF1 863/499 reporter construct. (B) Co-transfection of HeLa cells with the HMT-NF1 reporter (2 µg) and CELF protein expression plasmids (1 µg for CUG-BP, ETR-3, CELF4 and CELF5 and 0.5 µg for CELF3 and CELF6). Total RNA was isolated from transfected cells and semi-quantitative RT–PCR was performed using oligonucleotides indicated in panel A. The percentage of NF1 exon 23a inclusion is displayed in the bar graphs. Error bars indicate standard deviations and n = 3. Total protein was isolated from transfected cells and western blot analysis was carried out using Anti-Xpress antibody. Anti-U1 70K antibody was used as a loading control. (C) Co-transfection of HeLa cells with LacZ (4 µg) and CELF protein expression plasmids (4 µg for ETR-3, CELF3 and CELF6). Total RNA was isolated from transfected cells and semi-quantitative RT–PCR, using oligonucleotides that anneal to NF1 exons 23 and 24, was performed to determine the endogenous NF1 exon 23a inclusion. The percentage of NF1 exon 23a inclusion is displayed in the bar graph, with error bars indicating standard deviations. Total protein was isolated from the transfected cells and western blot analysis was performed using Anti-Xpress antibody to detect either tagged CELF proteins or LacZ which was used as a loading control.
In order to confirm the effects of CELF proteins on endogenous NF1 exon 23a inclusion, we over-expressed representatives of both of the CELF subfamilies and assayed the splicing profile of the endogenously expressed NF1 pre-mRNA using semi-quantitative RT-PCR and oligonucleotides that anneal to NF1 exons 23 and 24. When over-expressed, ETR-3 and CELF3 strongly promote NF1 exon 23a skipping, while over-expression of CELF6 has a more modest effect on the skipping of endogenous NF1 exon 23a (Figure 3C). Western blot analysis, using an anti-tag antibody (anti-Xpress), shows that these proteins were over-expressed at similar levels (Figure 3C). These data indicate that increased levels of CELF family members, with the exception of CELF6, strongly promote skipping of NF1 exon 23a.
Disruption of endogenous CELF proteins promotes NF1 exon 23a inclusion
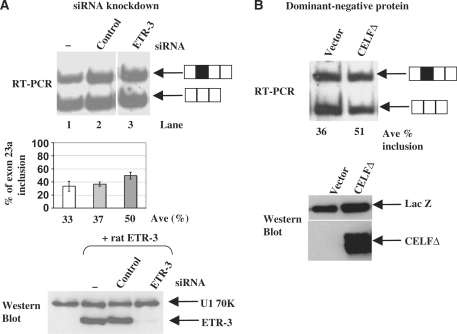
To establish the importance of endogenous CELF proteins in the regulation of NF1 exon 23a inclusion, we disrupted the function of the endogenous proteins in CA77 cells, the high CELF protein-expressing cell line, using two methods. First, we performed a siRNA knockdown experiment. We reduced ETR-3 level by siRNA treatment and examined the effect on exon 23a inclusion from the co-transfected NF1 reporter. Introduction of a control siRNA did not affect the NF1 exon 23a splicing profile, as shown in Figure 4A (top panel, lanes 1–2). When an siRNA duplex targeting rat ETR-3 was introduced in the co-transfection experiments, the exon 23a inclusion was increased from 33% in the wild-type to 50% with 200 pmol ETR-3 siRNA (Figure 4A, top panel, lanes 1 and 3). To confirm that the ETR-3 protein levels were reduced by the siRNA, we over-expressed rat ETR-3 in HeLa cells and simultaneously co-transfected the cells with siRNA targeting ETR-3 in HeLa cells. We found that ETR-3 was knocked down efficiently after being over-expressed in HeLa cells (Figure 4A, bottom panel).
Figure 4.
Reduction of endogenous CELF proteins in CA77 cells promotes exon 23a inclusion. (A) Co-transfection of CA77 cells with the HMT-NF1 reporter (2 µg) in the absence or presence of 200 pmol of control or 200 pmol ETR-3 siRNAs. The percentage of NF1 exon 23a inclusion is displayed in the bar graphs. Western blot analysis was carried out using HeLa cells in which rat ETR-3 and either 200 pmol control siRNA or 200 pmol siRNA targeting ETR-3 were co-transfected. U1 70K is used as a loading control. Lane 1 contains neither ETR-3 nor siRNA. (B) Co-transfection of CA77 cells with the HMT-NF1 863/499 reporter (2 µg) plus vector control plasmid (4 µg) or CELF dominant negative expression plasmid (4 µg). The average percentage of exon 23a inclusion from two independent transfections are indicated below each lane. CA77 cells were co-transfected with HMT-NF1 863/499 reporter (1 µg) plus vector control plasmid (4 µg) or CELF dominant negative expression plasmid (4 µg) using an electroporation apparatus, and the total protein lysate was taken from these cells for western blot analysis. Western blot analysis was carried out using Anti-Xpress antibody to detect the CELF dominant negative and anti-U1 70K antibody as a loading control.
Second, we performed co-transfection experiments using a truncated CELF4 protein, CELFΔ, which was demonstrated to function as a dominant negative protein by interfering with the endogenous CELF proteins in other systems (47). The CELFΔ protein contains the first N-terminal RNA recognition motif (RRM) and a portion of the second N-terminal RRM, a full hinge domain, and a portion of the C-terminal RRM (47). When the CELFΔ protein was over-expressed along with the NF1 reporter in CA77, the high CELF-expressing cell line, exon 23a inclusion was increased from 36 to 51% (Figure 4B). A western blot analysis using protein lysate isolated from transfected CA77 cells confirmed expression of the truncated protein (Figure 4B). Taken together, these results strongly suggest that CELF proteins are negative regulators of NF1 exon 23a inclusion.
CELF proteins bind strongly to a NF1 RNA substrate and block binding of U2AF 65
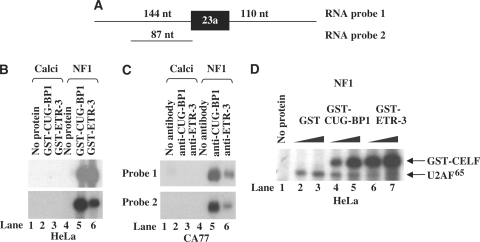
Given that the intronic sequence upstream of NF1 exon 23a is UG-rich, as shown in Figure 1A, and that CELF proteins function as negative regulators of exon 23a inclusion when their levels are manipulated in cell culture, we hypothesized that the CELF proteins bind strongly to the NF1 pre-mRNA. In order to test whether CELF proteins could bind to NF1 RNA, we utilized two substrates to generate 32P-UTP-labeled in vitro RNA transcripts. The first one of these substrates, probe 1, includes 144 nt of NF1 sequence upstream to exon 23a, exon 23a itself, and 110 nt of sequence downstream from the exon (Figure 5A). The second substrate, probe 2, includes only 87 nt of NF1 sequence upstream to exon 23a (Figure 5A). We performed UV crosslinking/immunoprecipitation (IP) assays to assess binding of CUG-BP1 and ETR-3 to the NF1 RNA transcripts. We first performed these assays using HeLa nuclear extract, which contains only CUG-BP1, to which we added recombinant GST-CUG-BP1 or GST-ETR-3. We used a calcitonin in vitro transcribed RNA substrate as a negative control, because this substrate lacks the UG-rich elements (15). We used either anti-CUG-BP1 or anti-ETR-3 antibodies for the immunoprecipitation, and we found that the recombinant CELF proteins could bind strongly to both of the NF1 substrates (Figure 5B, lanes 5–6), but they were unable to bind to the calcitonin RNA substrate (Figure 5B, lanes 2–3). We wanted to assess the ability of the endogenous CELF proteins to bind to the NF1 RNA substrates, so we used nuclear extract isolated from CA77 cells, which contain all of the CELF family members, to perform additional UV crosslinking/IP assays. We used either anti-CUG-BP1 or anti-ETR-3 antibodies to perform these assays, and the calcitonin RNA substrate as a negative control. We found that both endogenous CUG-BP1 and ETR-3 could bind strongly to the NF1 RNA substrates but not to the calcitonin RNA substrate (Figure 5C, lanes 2–3 and 5–6).
Figure 5.
CELF proteins bind to the NF1 pre-mRNA and block the binding of U2AF65. (A) In vitro RNA substrates: Probe 1 contains NF1 exon 23a and 144 nucleotides of the upstream and 110 nucleotides of the downstream intronic sequences and Probe 2 contains only the 87 nucleotides of intronic sequence upstream to NF1 exon 23a. (B) The in vitro transcribed RNAs (represented in the diagram) were incubated with HeLa nuclear extract to which GST-CUG-BP1 or GST-ETR-3 (3 µg) were added. Anti-CUG-BP1 or anti-ETR-3 antibodies were used in these IP experiments. Binding of CELF proteins to the NF1 RNA substrates was detected by a UV crosslinking/IP assay. (C) Binding of endogenous CELF proteins to the NF1 RNA substrates was detected by a UV-crosslinking/IP assay using the same in vitro transcribed RNAs and CA77 nuclear extract. Anti-CUG-BP1 or anti-ETR-3 antibodies were used in these experiments. (D) Binding of U2AF65 to RNA probe 1 was analyzed by UV crosslinking/IP using HeLa nuclear extract supplemented with increasing amounts of either GST-CUG-BP1 (1.5–3.0 µg) or GST-ETR-3 (1.5–3.0 µg) protein. U2AF65 antibody was used in the immunoprecipitation.
The UG-rich sequences are located immediately upstream of the 3′ splice site of exon 23a. Therefore, we predicted that the CELF proteins most likely interfere with binding of critical splicesome components at or around the 3′ splice site. To test this possibility, we examined binding of a splicing factor by UV crosslinking/IP analysis using HeLa nuclear extract and exogenously provided recombinant GST-CUG-BP1 or GST-ETR-3. We used an antibody against U2AF65, an essential splicing factor that binds to the polypyrimidine-tract of the 3′ splice site, for the IP assays. We found that increasing amounts of the recombinant CELF proteins resulted in a decrease in the levels of U2AF65 binding (Figure 5D, compare lanes 2 and 7). These results allude to the mechanism of CELF protein regulation of NF1 exon 23a, by showing that they can block binding of U2AF65 to the NF1 RNA substrate, and thus prevent the exon from being included.
CELF proteins block splicing of NF1 exon 23a
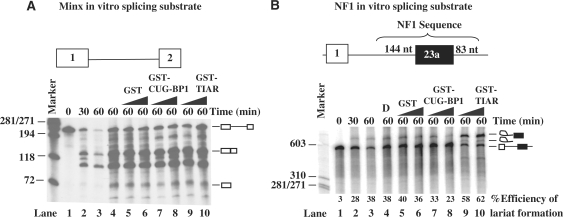
To provide definitive evidence that CELF proteins block splicing of NF1 exon 23a directly, we carried out an in vitro splicing analysis using an RNA substrate shown in Figure 6A. This substrate was generated from the reporter used in earlier cell transfection experiments (Figure 6B) and contains 144 nt of NF1 sequence upstream of exon 23a, exon 23a, and 83 nt of NF1 sequence downstream of NF1 exon 23a. We then analyzed splicing of this substrate in HeLa nuclear extract under standard splicing conditions. We also utilized a strong, classical splicing substrate, Minx, as shown in Figure 6A. Compared to the splicing of Minx RNA, the splicing efficiency of the NF1 substrate is very low (Figure 6B). Only the lariat intermediates can be detected in a time-dependent manner, as shown in Figure 6B. We decided to test the effect of CELF proteins on splicing of the NF1 substrate by monitoring the lariat production, as described in a previous study (48). Addition of increasing amounts of GST-CELF proteins resulted in a decrease in the percentage of efficiency of lariat formation from about 38 to 23%, indicative of blocked splicing (Figure 6B, lanes 3–6 and 7–8). Importantly, addition of GST-TIAR, a protein that was previously identified as a positive regulator of NF1 exon 23a splicing, increased lariat formation, from 38 to 58–62%, consistent with the positive role played by TIA-1/TIAR in splicing of NF1 exon 23a (Figure 6B, lanes 3–6 and 9–10). As expected, neither the recombinant CELF proteins nor the recombinant TIAR protein affected the splicing of the Minx control (Figure 6A). These experiments demonstrate that CELF proteins block splicing of the intron upstream of NF1 exon 23a.
Figure 6.
CELF proteins block the splicing of NF1 exon 23a. (A) In vitro splicing assays were carried out using an in vitro-transcribed Minx RNA containing exons 1 and 2 and an intron in between (represented in the diagram) and HeLa nuclear extract. A splicing time course is shown in lanes 1–3. CELF proteins were added in increasing amounts. (B) In vitro splicing assays were carried out using an in vitro-transcribed RNA containing NF1 exon 23a and both upstream and downstream intronic sequence (represented in the diagram) and HeLa nuclear extract. A splicing time course is shown in lanes 1–3. The ability of CELF proteins to block the splicing of NF1 exon 23a was tested using increasing amounts of GST (0.75–1.5 µg), GST-CUG-BP1 (0.75–1.5 µg), or GST-TIAR (0.75–1.5 µg) and the splicing reaction was carried out for 60 min. Roeder D (labeled as D) with no protein was used as a negative control. The percentage of efficiency of lariat formation is given for the NF1 splicing assay.
DISCUSSION
NF1 is a novel target of CELF-mediated splicing regulation
CELF proteins are important regulators of alternative splicing (16). Several mouse models have been generated that have abnormal expression of CUG-BP1 in the heart (32,35). These animals were made to model the human disease, DM, and they show significant heart defects and interestingly, many of the defects can be attributed to aberrant splicing of a number of well-characterized CELF protein splicing targets (32,35). To fully understand the function of CELF proteins and their roles in human disease, it is important to identify all of the splicing targets of these proteins. Recent work has identified at least 19 mRNA splicing targets that are sensitive to changes in levels of CELF proteins during heart development (49).
Here, we report the identification of a novel target of the CELF proteins, exon 23a of the NF1 pre-mRNA. We show that in cell transfection experiments, changes of CELF protein level by either over-expression or siRNA knockdown leads to a decrease or increase of exon 23a inclusion, respectively. These results strongly argue that CELF proteins promote skipping of this exon. Importantly, NF1 plays an important role in the development of the heart, in addition to the nervous system, since animal models in which NF1 has been deleted do not survive past mid-gestation (50,51) due to heart development related abnormalities. Cardiac dysfunction in mouse models in which CELF activity is elevated or repressed have demonstrated the importance of CELF-mediated alternative splicing in the heart. Also of interest is the finding that there is a change in NF1 Type I (exon 23a skipped) and NF1 Type II (exon 23a included) isoform distribution in the heart during early embryonic development in mice (44). The NF1 splicing changes occur early in heart development between days 11 and 13 (44). NF1 exon 23a was not identified in the aforementioned splicing microarray study, and this is most likely because the study examined splicing events that occurred from embryonic day 14 to adulthood which is clearly after the NF1 splicing patterns have changed (44,49).
These findings hint at the possibility of CELF protein involvement in regulation of the NF1 exon 23a, since both CUG-BP1 and ETR-3 are robustly expressed during early in the heart and then the levels of these proteins decline (31).
NF1 exon 23a is under complex combinatorial control
Tissue-specific alternative splicing is complex and a myriad of factors are often necessary to provide additional layers of regulation for a particular splicing event. One example of a splicing event which is regulated by several different factors is the cell-type specific regulation of the mutually exclusive exons IIIb and IIIc of the Fibroblast Growth Factor Receptor 2 (FGFR2) for which several different auxiliary cis-acting elements and their binding partners have been identified (52–55). Another interesting example occurs in the immune system and involves the regulation of exon 4 of the CD45 gene which can be regulated by hnRNPL, PSF and other factors (56).
Like many other alternative exons, NF1 exon 23a is tightly regulated. The NF1 sequence is highly conserved among mice, rats and humans, and the tissue-specific splicing phenotype is conserved among these species as well (13,57). These facts imply that this alternative splicing event must be especially important. A few previous studies have been carried out in attempts to understand the importance of this regulated splicing event. A mouse model in which NF1 exon 23a has been deleted shows learning disabilities, which are also an important phenotypic characteristic of some NF1 disease patients (41).
Up until recently, it was not known how NF1 exon 23a inclusion is regulated. Recent work in our laboratory has demonstrated that NF1 exon 23a inclusion is also repressed by the Hu family of RNA-binding proteins, of which three of the four family members are neuron-specific (13,58). We previously demonstrated that Hu proteins block the function of TIA-1/TIAR, most likely through competitive binding, leading to decreased binding of U1 and U6 snRNP interaction with the 5′ splice site downstream of exon 23a (13). Hu proteins also decrease binding of U2AF to the 3′ splice site upstream of exon 23a.
Given that this phenomenon is so complex, it is probable that other positive and negative factors that regulate NF1 exon 23a remain to be identified. An interesting future direction is to determine whether the CELF and Hu proteins function together or independently of one another to achieve their functions as splicing regulators of NF1 exon 23a. Both CUG-BP1 and ETR-3 are robustly expressed during the early development of the embryonic heart, and one Hu protein, HuR, is present. The relative expression levels of HuR, throughout heart development, are not known. It would be interesting to better understand HuR expression in the heart, as this could give us some hints into whether the CELF and Hu proteins can regulated NF1 exon 23a independently.
Redundant and non-redundant functions of CELF proteins
Our results of over-expression studies in HeLa cells show that the different CELF family members have different capabilities in regulating NF1 exon 23a inclusion. Five of the six members of this protein family exhibit strong activity in repressing inclusion of NF1 exon 23a, while CELF6 shows modest regulatory activity in both the mini-gene reporter assays and in the assays in which the endogenous NF1 was considered. This finding is very interesting since the CELF family members are very similar in sequence and CELF6 is about 40% identical to CUG-BP1 and ETR-3 (16). A previous study suggested a possibility that CELF6 binds to other sequence elements in addition to CUG or UG-rich sequences (30). However, the functional significance of this difference remains unclear. It is possible that CELF6 requires a different sequence to operate optimally, and thus it does not function well in our system. In addition, the cellular localization of CELF6 is likely to play a significant role in its ability to regulate splicing. Three of the six CELF proteins, CUG-BP1, ETR-3 and CELF4, have been shown to localize in both the nuclear and cytoplasmic subcellular compartments, suggesting that they might have shuttling activities (28,29,59,60), and it is likely that CELF6 is similar. CELF6 has a nuclear localization signal, but computational prediction methods suggest that it might spend most of its time in the cytoplasm and only shuttle to the nucleus in response to cellular needs (30).
The redundant function of the five CELF members is also illustrated by the siRNA knockdown experiment in CA77 cells. A fairly modest change was observed when ETR-3 was knocked down in CA77 cells. The most plausible explanation for this finding is that knocking down one CELF member is not sufficient to significantly impact the NF1 exon 23a inclusion, since all of the family members are present in CA77 cells as shown in our expression data (Figure 2).
When a CELF dominant negative protein (CELFΔ), as described by Charlet et al. (34), is co-transfected with the NF1 minigene into CA77 cells, a fairly modest increase in NF1 exon 23a inclusion is observed. Previous studies, using both fibroblasts and primary cardiomyocyte cultures, have shown that the CELFΔ protein must be present in excess to the endogenous CELF proteins in order to have an effect on splicing of CELF protein targets (34,61). It is therefore not surprising that we do not see a more dramatic effect on NF1 exon 23a inclusion since there are six different CELF proteins in the CA77 cells. Additionally, there are four Hu proteins present in CA77, which are known negative regulators of this system whose functions are not expected to be hindered by the over-expression of the CELF dominant negative protein.
Mechanism of CELF protein-mediated splicing regulation of NF1 exon 23a
It is clear, based on our in vitro experiments (Figures 5 and 6) that CELF proteins can block splicing of NF1 exon 23a. Specifically, U2AF65 is blocked by the addition of increasing amounts of either recombinant GST-CUG-BP1 or GST-ETR-3 as the U2AF65 band decreases significantly. The mechanism by which the CELF proteins perform these functions is under investigation. There are several potential ways in which the CELF proteins could block U2AF65 binding, and thus block splicing. First of all, it is possible that CELF proteins compete for binding of very specific sites on the pre-mRNA, which prevent the binding of critical spliceosomal machinery components. In the context of U2AF65, this is plausible since the polypyrimidine-tract sequence upstream of the 3′ splice site of exon 23a is a very weak binding site for U2AF65, and since the polypyrimidine-tract is adjacent to and potentially part of several UG-rich elements (Figure 1A). Indeed this mechanism is used by polypyrimidine tract binding protein as demonstrated by others (62,63). A second way in which CELF proteins might work is by binding to other factors in a complex, which can crowd the splicing machinery by steric hindrance. A third way in which CELF proteins could accomplish blocking splicing is by binding to specific UG-rich sites and causing conformational changes in the structure of the mRNA, which then prevent the binding of other necessary splicing factors. Although this mechanism has not been described for the CELF proteins in other systems, thus far, it is a mechanism that is used by other factors such as PTB. In that case, PTB binds to sequences both upstream and downstream of an exon to loop out the exon and promote its skipping (64). Furthermore, recent work has demonstrated the importance of RNA secondary structure and splicing control by showing that the muscleblind-like (MBNL) family of RNA-binding proteins and U2AF65 regulate the cardiac troponin T exon 5 system by binding to different portions of a stem loop structure that is formed encompassing the polypryimidine tract and the MBNL binding sites (65).
In summary, the study reported here identifies NF1 exon 23a as a novel splicing target for the CELF family of proteins and reveals an additional layer of regulation of this alternative exon. These studies highlight the complexity and versatility of tissue-specific alternative splicing regulation.
FUNDING
National Science Foundation (MCB-0237685 to H.L.); the National Institutes of Health (NS-049103 to H.L., T32 HD-007104-32 to V.B., T32GM008613 to M.H.); and the American Heart Association (0815373D to V.B., 0415086B to H.Z, and 0865420D to A.L.). Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank the following individuals for providing reagents: Dr Thomas Cooper at Baylor College of Medicine for providing the CELF expression plasmids, Dr Alison Hall at Case Western Reserve University and Dr Andrew Russo at the University of Iowa for providing the CA77 cell line. We thank members of the Lou lab for helpful discussions and critical reading of the manuscript.
REFERENCES
- 1.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 2.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li Q, Lee JA, Black DL. Neuronal regulation of alternative pre-mRNA splicing. Nat. Rev. Neurosci. 2007;8:819–831. doi: 10.1038/nrn2237. [DOI] [PubMed] [Google Scholar]
- 4.Ule J, Ule A, Spencer J, Williams A, Hu JS, Cline M, Wang H, Clark T, Fraser C, Ruggiu M, et al. Nova regulates brain-specific splicing to shape the synapse. Nat. Genet. 2005;37:844–852. doi: 10.1038/ng1610. [DOI] [PubMed] [Google Scholar]
- 5.Lipscombe D. Neuronal proteins custom designed by alternative splicing. Curr. Opin. Neurobiol. 2005;15:358–363. doi: 10.1016/j.conb.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 6.Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 7.Matlin AJ, Clark F, Smith CW. Understanding alternative splicing: towards a cellular code. Nat. Rev. Mol. Cell Biol. 2005;6:386–398. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- 8.Underwood JG, Boutz PL, Dougherty JD, Stoilov P, Black DL. Homologues of the Caenorhabditis elegans Fox-1 protein are neuronal splicing regulators in mammals. Mol. Cell Biol. 2005;25:10005–10016. doi: 10.1128/MCB.25.22.10005-10016.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ule J, Jensen KB, Ruggiu M, Mele A, Ule A, Darnell RB. CLIP identifies Nova-regulated RNA networks in the brain. Science. 2003;302:1212–1215. doi: 10.1126/science.1090095. [DOI] [PubMed] [Google Scholar]
- 10.Ule J, Stefani G, Mele A, Ruggiu M, Wang X, Taneri B, Gaasterland T, Blencowe BJ, Darnell RB. An RNA map predicting Nova-dependent splicing regulation. Nature. 2006;444:580–586. doi: 10.1038/nature05304. [DOI] [PubMed] [Google Scholar]
- 11.Zhu H, Hasman RA, Barron VA, Luo G, Lou H. A nuclear function of Hu proteins as neuron-specific alternative RNA processing regulators. Mol. Biol. Cell. 2006;17:5105–5114. doi: 10.1091/mbc.E06-02-0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hinman MN, Lou H. Diverse molecular functions of Hu proteins. Cell Mol. Life Sci. 2008;65:3168–3181. doi: 10.1007/s00018-008-8252-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu H, Hinman MN, Hasman RA, Mehta P, Lou H. Regulation of neuron-specific alternative splicing of neurofibromatosis type 1 pre-mRNA. Mol. Cell Biol. 2008;28:1240–1251. doi: 10.1128/MCB.01509-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang W, Liu H, Han K, Grabowski PJ. Region-specific alternative splicing in the nervous system: implications for regulation by the RNA-binding protein NAPOR. RNA. 2002;8:671–685. doi: 10.1017/s1355838202027036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou HL, Baraniak AP, Lou H. Role for Fox-1/Fox-2 in mediating the neuronal pathway of calcitonin/calcitonin gene-related peptide alternative RNA processing. Mol. Cell Biol. 2007;27:830–841. doi: 10.1128/MCB.01015-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barreau C, Paillard L, Mereau A, Osborne HB. Mammalian CELF/Bruno-like RNA-binding proteins: molecular characteristics and biological functions. Biochimie. 2006;88:515–525. doi: 10.1016/j.biochi.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 17.Hanamura A, Caceres JF, Mayeda A, Franza BR, Jr, Krainer AR. Regulated tissue-specific expression of antagonistic pre-mRNA splicing factors. RNA. 1998;4:430–444. [PMC free article] [PubMed] [Google Scholar]
- 18.Castle JC, Zhang C, Shah JK, Kulkarni AV, Kalsotra A, Cooper TA, Johnson JM. Expression of 24,426 human alternative splicing events and predicted cis regulation in 48 tissues and cell lines. Nat. Genet. 2008;40:1416–1425. doi: 10.1038/ng.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Savkur RS, Philips AV, Cooper TA. Aberrant regulation of insulin receptor alternative splicing is associated with insulin resistance in myotonic dystrophy. Nat. Genet. 2001;29:40–47. doi: 10.1038/ng704. [DOI] [PubMed] [Google Scholar]
- 20.Takahashi N, Sasagawa N, Suzuki K, Ishiura S. The CUG-binding protein binds specifically to UG dinucleotide repeats in a yeast three-hybrid system. Biochem. Biophys. Res. Commun. 2000;277:518–523. doi: 10.1006/bbrc.2000.3694. [DOI] [PubMed] [Google Scholar]
- 21.Marquis J, Paillard L, Audic Y, Cosson B, Danos O, Le Bec C, Osborne HB. CUG-BP1/CELF1 requires UGU-rich sequences for high-affinity binding. Biochem. J. 2006;400:291–301. doi: 10.1042/BJ20060490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Faustino NA, Cooper TA. Identification of putative new splicing targets for ETR-3 using sequences identified by systematic evolution of ligands by exponential enrichment. Mol. Cell Biol. 2005;25:879–887. doi: 10.1128/MCB.25.3.879-887.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Philips AV, Timchenko LT, Cooper TA. Disruption of splicing regulated by a CUG-binding protein in myotonic dystrophy. Science. 1998;280:737–741. doi: 10.1126/science.280.5364.737. [DOI] [PubMed] [Google Scholar]
- 24.Timchenko LT, Miller JW, Timchenko NA, DeVore DR, Datar KV, Lin L, Roberts R, Caskey CT, Swanson MS. Identification of a (CUG)n triplet repeat RNA-binding protein and its expression in myotonic dystrophy. Nucleic Acids Res. 1996;24:4407–4414. doi: 10.1093/nar/24.22.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Timchenko LT, Timchenko NA, Caskey CT, Roberts R. Novel proteins with binding specificity for DNA CTG repeats and RNA CUG repeats: implications for myotonic dystrophy. Hum. Mol. Genet. 1996;5:115–121. doi: 10.1093/hmg/5.1.115. [DOI] [PubMed] [Google Scholar]
- 26.Choi DK, Ito T, Tsukahara F, Hirai M, Sakaki Y. Developmentally-regulated expression of mNapor encoding an apoptosis-induced ELAV-type RNA binding protein. Gene. 1999;237:135–142. doi: 10.1016/s0378-1119(99)00312-1. [DOI] [PubMed] [Google Scholar]
- 27.Hwang DM, Hwang WS, Liew CC. Single pass sequencing of a unidirectional human fetal heart cDNA library to discover novel genes of the cardiovascular system. J. Mol. Cell Cardiol. 1994;26:1329–1333. doi: 10.1006/jmcc.1994.1151. [DOI] [PubMed] [Google Scholar]
- 28.Good PJ, Chen Q, Warner SJ, Herring DC. A family of human RNA-binding proteins related to the Drosophila bruno translational regulator. J. Biol. Chem. 2000;275:28583–28592. doi: 10.1074/jbc.M003083200. [DOI] [PubMed] [Google Scholar]
- 29.Ladd AN, Charlet N, Cooper TA. The CELF family of RNA binding proteins is implicated in cell-specific and developmentally regulated alternative splicing. Mol. Cell Biol. 2001;21:1285–1296. doi: 10.1128/MCB.21.4.1285-1296.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ladd AN, Nguyen NH, Malhotra K, Cooper TA. CELF6, a member of the CELF family of RNA-binding proteins, regulates muscle-specific splicing enhancer-dependent alternative splicing. J. Biol. Chem. 2004;279:17756–17764. doi: 10.1074/jbc.M310687200. [DOI] [PubMed] [Google Scholar]
- 31.Ladd AN, Stenberg MG, Swanson MS, Cooper TA. Dynamic balance between activation and repression regulates pre-mRNA alternative splicing during heart development. Dev. Dyn. 2005;233:783–793. doi: 10.1002/dvdy.20382. [DOI] [PubMed] [Google Scholar]
- 32.Ho TH, Bundman D, Armstrong DL, Cooper TA. Transgenic mice expressing CUG-BP1 reproduce splicing mis-regulation observed in myotonic dystrophy. Hum. Mol. Genet. 2005;14:1539–1547. doi: 10.1093/hmg/ddi162. [DOI] [PubMed] [Google Scholar]
- 33.Cooper TA. Muscle-specific splicing of a heterologous exon mediated by a single muscle-specific splicing enhancer from the cardiac troponin T gene. Mol. Cell Biol. 1998;18:4519–4525. doi: 10.1128/mcb.18.8.4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Charlet BN, Savkur RS, Singh G, Philips AV, Grice EA, Cooper TA. Loss of the muscle-specific chloride channel in type 1 myotonic dystrophy due to misregulated alternative splicing. Mol. Cell. 2002;10:45–53. doi: 10.1016/s1097-2765(02)00572-5. [DOI] [PubMed] [Google Scholar]
- 35.Wang GS, Kearney DL, De Biasi M, Taffet G, Cooper TA. Elevation of RNA-binding protein CUGBP1 is an early event in an inducible heart-specific mouse model of myotonic dystrophy. J. Clin. Invest. 2007;117:2802–2811. doi: 10.1172/JCI32308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bernards A. Neurofibromatosis type 1 and Ras-mediated signaling: filling in the GAPs. Biochim. Biophys. Acta. 1995;1242:43–59. doi: 10.1016/0304-419x(95)00003-x. [DOI] [PubMed] [Google Scholar]
- 37.Ballester R, Marchuk D, Boguski M, Saulino A, Letcher R, Wigler M, Collins F. The NF1 locus encodes a protein functionally related to mammalian GAP and yeast IRA proteins. Cell. 1990;63:851–859. doi: 10.1016/0092-8674(90)90151-4. [DOI] [PubMed] [Google Scholar]
- 38.Gil R, Seeling JM. Characterization of Saccharomyces cerevisiae strains expressing ira1 mutant alleles modeled after disease-causing mutations in NF1. Mol. Cell Biochem. 1999;202:109–118. doi: 10.1023/a:1007058427880. [DOI] [PubMed] [Google Scholar]
- 39.Yunoue S, Tokuo H, Fukunaga K, Feng L, Ozawa T, Nishi T, Kikuchi A, Hattori S, Kuratsu J, Saya H, et al. Neurofibromatosis type I tumor suppressor neurofibromin regulates neuronal differentiation via its GTPase-activating protein function toward Ras. J. Biol. Chem. 2003;278:26958–26969. doi: 10.1074/jbc.M209413200. [DOI] [PubMed] [Google Scholar]
- 40.Andersen LB, Ballester R, Marchuk DA, Chang E, Gutmann DH, Saulino AM, Camonis J, Wigler M, Collins FS. A conserved alternative splice in the von Recklinghausen neurofibromatosis (NF1) gene produces two neurofibromin isoforms, both of which have GTPase-activating protein activity. Mol. Cell Biol. 1993;13:487–495. doi: 10.1128/mcb.13.1.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Costa RM, Federov NB, Kogan JH, Murphy GG, Stern J, Ohno M, Kucherlapati R, Jacks T, Silva AJ. Mechanism for the learning deficits in a mouse model of neurofibromatosis type 1. Nature. 2002;415:526–530. doi: 10.1038/nature711. [DOI] [PubMed] [Google Scholar]
- 42.Zhu H, Hasman RA, Young KM, Kedersha NL, Lou H. U1 snRNP-dependent function of TIAR in the regulation of alternative RNA processing of the human calcitonin/CGRP pre-mRNA. Mol. Cell Biol. 2003;23:5959–5971. doi: 10.1128/MCB.23.17.5959-5971.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zeng C, Berget SM. Participation of the C-terminal domain of RNA polymerase II in exon definition during pre-mRNA splicing. Mol. Cell Biol. 2000;20:8290–8301. doi: 10.1128/mcb.20.21.8290-8301.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huynh DP, Nechiporuk T, Pulst SM. Differential expression and tissue distribution of type I and type II neurofibromins during mouse fetal development. Dev. Biol. 1994;161:538–551. doi: 10.1006/dbio.1994.1052. [DOI] [PubMed] [Google Scholar]
- 45.Gutmann DH, Zhang Y, Hirbe A. Developmental regulation of a neuron-specific neurofibromatosis 1 isoform. Ann. Neurol. 1999;46:777–782. doi: 10.1002/1531-8249(199911)46:5<777::aid-ana15>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 46.Russo AF, Lanigan TM, Sullivan BE. Neuronal properties of a thyroid C-cell line: partial repression by dexamethasone and retinoic acid. Mol. Endocrinol. 1992;6:207–218. doi: 10.1210/mend.6.2.1569964. [DOI] [PubMed] [Google Scholar]
- 47.Charlet BN, Logan P, Singh G, Cooper TA. Dynamic antagonism between ETR-3 and PTB regulates cell type-specific alternative splicing. Mol. Cell. 2002;9:649–658. doi: 10.1016/s1097-2765(02)00479-3. [DOI] [PubMed] [Google Scholar]
- 48.Grabowski PJ, Padgett RA, Sharp PA. Messenger RNA splicing in vitro: an excised intervening sequence and a potential intermediate. Cell. 1984;37:415–427. doi: 10.1016/0092-8674(84)90372-6. [DOI] [PubMed] [Google Scholar]
- 49.Kalsotra A, Xiao X, Ward AJ, Castle JC, Johnson JM, Burge CB, Cooper TA. A postnatal switch of CELF and MBNL proteins reprograms alternative splicing in the developing heart. Proc. Natl Acad. Sci. USA. 2008;105:20333–20338. doi: 10.1073/pnas.0809045105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jacks T, Shih TS, Schmitt EM, Bronson RT, Bernards A, Weinberg RA. Tumour predisposition in mice heterozygous for a targeted mutation in Nf1. Nat. Genet. 1994;7:353–361. doi: 10.1038/ng0794-353. [DOI] [PubMed] [Google Scholar]
- 51.Brannan CI, Perkins AS, Vogel KS, Ratner N, Nordlund ML, Reid SW, Buchberg AM, Jenkins NA, Parada LF, Copeland NG. Targeted disruption of the neurofibromatosis type-1 gene leads to developmental abnormalities in heart and various neural crest-derived tissues. Genes Dev. 1994;8:1019–1029. doi: 10.1101/gad.8.9.1019. [DOI] [PubMed] [Google Scholar]
- 52.Mauger DM, Lin C, Garcia-Blanco MA. hnRNP H and hnRNP F complex with Fox2 to silence fibroblast growth factor receptor 2 exon IIIc. Mol. Cell Biol. 2008;28:5403–5419. doi: 10.1128/MCB.00739-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Warzecha CC, Sato TK, Nabet B, Hogenesch JB, Carstens RP. ESRP1 and ESRP2 are epithelial cell-type-specific regulators of FGFR2 splicing. Mol. Cell. 2009;33:591–601. doi: 10.1016/j.molcel.2009.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hovhannisyan RH, Carstens RP. Heterogeneous ribonucleoprotein m is a splicing regulatory protein that can enhance or silence splicing of alternatively spliced exons. J. Biol. Chem. 2007;282:36265–36274. doi: 10.1074/jbc.M704188200. [DOI] [PubMed] [Google Scholar]
- 55.Newman EA, Muh SJ, Hovhannisyan RH, Warzecha CC, Jones RB, McKeehan WL, Carstens RP. Identification of RNA-binding proteins that regulate FGFR2 splicing through the use of sensitive and specific dual color fluorescence minigene assays. RNA. 2006;12:1129–1141. doi: 10.1261/rna.34906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Melton AA, Jackson J, Wang J, Lynch KW. Combinatorial control of signal-induced exon repression by hnRNPL and PSF. Mol. Cell Biol. 2007;27:6972–6984. doi: 10.1128/MCB.00419-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bernards A, Snijders AJ, Hannigan GE, Murthy AE, Gusella JF. Mouse neurofibromatosis type 1 cDNA sequence reveals high degree of conservation of both coding and non-coding mRNA segments. Hum. Mol. Genet. 1993;2:645–650. doi: 10.1093/hmg/2.6.645. [DOI] [PubMed] [Google Scholar]
- 58.Okano HJ, Darnell RB. A hierarchy of Hu RNA binding proteins in developing and adult neurons. J. Neurosci. 1997;17:3024–3037. doi: 10.1523/JNEUROSCI.17-09-03024.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Timchenko NA, Cai ZJ, Welm AL, Reddy S, Ashizawa T, Timchenko LT. RNA CUG repeats sequester CUGBP1 and alter protein levels and activity of CUGBP1. J. Biol. Chem. 2001;276:7820–7826. doi: 10.1074/jbc.M005960200. [DOI] [PubMed] [Google Scholar]
- 60.Mukhopadhyay D, Jung J, Murmu N, Houchen CW, Dieckgraefe BK, Anant S. CUGBP2 plays a critical role in apoptosis of breast cancer cells in response to genotoxic injury. Ann. NY Acad. Sci. 2003;1010:504–509. doi: 10.1196/annals.1299.093. [DOI] [PubMed] [Google Scholar]
- 61.Ladd AN, Taffet G, Hartley C, Kearney DL, Cooper TA. Cardiac tissue-specific repression of CELF activity disrupts alternative splicing and causes cardiomyopathy. Mol. Cell Biol. 2005;25:6267–6278. doi: 10.1128/MCB.25.14.6267-6278.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang L, Liu W, Grabowski PJ. Coordinate repression of a trio of neuron-specific splicing events by the splicing regulator PTB. RNA. 1999;5:117–130. doi: 10.1017/s1355838299981530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lin CH, Patton JG. Regulation of alternative 3′ splice site selection by constitutive splicing factors. RNA. 1995;1:234–245. [PMC free article] [PubMed] [Google Scholar]
- 64.Oberstrass FC, Auweter SD, Erat M, Hargous Y, Henning A, Wenter P, Reymond L, Amir-Ahmady B, Pitsch S, Black DL, et al. Structure of PTB bound to RNA: specific binding and implications for splicing regulation. Science. 2005;309:2054–2057. doi: 10.1126/science.1114066. [DOI] [PubMed] [Google Scholar]
- 65.Warf MB, Diegel JV, von Hippel PH, Berglund JA. The protein factors MBNL1 and U2AF65 bind alternative RNA structures to regulate splicing. Proc. Natl Acad. Sci. USA. 2009;106:9203–9208. doi: 10.1073/pnas.0900342106. [DOI] [PMC free article] [PubMed] [Google Scholar]