Abstract
Some human cancers maintain their telomeres using the alternative lengthening of telomeres (ALT) mechanism; a process thought to involve recombination. Different types of recombinational telomere elongation pathways have been identified in yeasts. In senescing yeast telomerase deletion (ter1-Δ) mutants with very short telomeres, it has been hypothesized that copying a tiny telomeric circle (t-circle) by a rolling circle mechanism is the key event in telomere elongation. In other cases more closely resembling ALT cells, such as the stn1-M1 mutant of Kluyveromyces lactis, the telomeres appear to be continuously unstable and routinely reach very large sizes. By employing two-dimensional gel electrophoresis and electron microscopy, we show that stn1-M1 cells contain abundant double stranded t-circles ranging from ∼100 to 30 000 bp in size. We also observed small single-stranded t-circles, specifically composed of the G-rich telomeric strand and tailed circles resembling rolling circle replication intermediates. The t-circles most likely arose from recombination events that also resulted in telomere truncations. The findings strengthen the possibility that t-circles contribute to telomere maintenance in stn1-M1 and ALT cells.
INTRODUCTION
Telomeres protect chromosome ends from nucleolytic degradation, end-to-end fusion and other processes that could compromise genome integrity (1). They are composed of tandem short DNA repeats and the specialized proteins that bind them (2). Telomeres are normally maintained by the enzyme telomerase that compensates for gradual sequence loss due to incomplete DNA replication by adding telomeric repeats onto chromosome termini. In most human somatic cells, telomerase activity is very low (3,4). This leads to gradual telomere shortening which, in turn, can trigger replicative senescence, a process where a cell with critically short telomeres permanently exits from the cycle of division (5,6). In contrast, the great majority of cancers are able to maintain their telomere lengths indefinitely. In most cases, this occurs because of an up-regulation of telomerase activity (7). However, some cancers maintain their telomere lengths through a telomerase-independent process termed alternative lengthening of telomeres (ALT) (8). The telomeres in ALT cells are highly heterogeneous, often extremely long and appear to be maintained through homologous recombination (9).
Much of what is known about recombinational telomere elongation (RTE) comes from studies in yeast, particularly Saccharomyces cerevisiae and Kluyveromyces lactis. Yeast mutants lacking telomerase undergo growth senescence and most cells eventually die (10,11). The cells that survive senescence are found to have lengthened telomeres through a process dependent upon RAD52 and other genes involved in homologous recombination (12–14). Recombination in and near telomeres is greatly increased when the telomeres become short (15,16). Work in both K. lactis and S. cerevisiae has suggested that RTE lengthens telomeric repeat arrays (Type II RTE) through a ‘roll and spread’ mechanism (15,17–19). According to this model, a small duplex DNA circle consisting of telomeric repeats (t-circle), formed by recombination in cells with critically short telomeres, is used as a template for extending at least one telomere, through a rolling circle copying event. Once one long telomere is formed, other telomeres become extended by copying its sequence. Appreciable evidence supports this model. We have shown that the sequence of a single long telomere is preferentially copied to all other telomeres during survivor formation (15). Cells transformed with telomeric circles (t-circles) routinely acquire telomeres extended by tandem copies of the sequence of the transformed circle (17,18). In addition, t-circles are abundant in at least some types of cells with dysfunctional telomeres including human ALT cells and a K. lactis mutant with altered telomeric repeats (20–22). For recent reviews of RTE and t-circles see references 23 and 24.
It has recently been shown that certain K. lactis mutant cells display a form of RTE that is distinct from the RTE that occurs in ter1-Δ mutants. The stn1-M1 mutant causes an amino acid substitution in Stn1, a protein that forms a complex with Cdc13 and Ten1 in S. cerevisiae. Stn1 was shown to bind G-rich telomeric substrates as well as to provide an essential protective function at the telomeres (25,26). This mutation displays moderate growth defects and leads to the rapid formation of very long and heterogeneous telomeres. Unlike a ter1-Δ mutant that appears to have a telomere-capping defect only when telomeres become very short, the stn1-M1 mutant has a continuous capping defect that is independent of telomere length. Other examples of this type of RTE, now known as Type IIR (runaway) RTE, have been seen in K. lactis telomerase RNA mutants, which generate mutant telomeric repeats (15). The most notable of these appear to be due to alterations in the telomeric repeat that reduce the binding affinity of the double strand telomere-binding protein Rap1 (27). Certain S. cerevisiae cdc13 and stn1 mutants provide similar examples of RTE independent of the length of the telomeres (28–30). The features of yeast Type IIR RTE are particularly similar to those observed in human ALT cells. Learning more about how telomere maintenance occurs in stn1-M1 cells is, therefore, of considerable interest. Whether t-circles contribute to either the formation or the maintenance of the long telomeres of Type IIR RTE or of ALT cells is currently unknown. In this study, we show that a broad range of sizes of t-circles is produced in the stn1-M1 mutant.
MATERIALS AND METHODS
Yeast strains
The strain 7B520 (ura3-1 his2-2 trp1) was described previously (31). The stn1-M1 and stn1-M1 ter1-Δ strains used here were also described previously (32). All strains were routinely grown at 30°C.
DNA isolation
Genomic DNA used to generate the telomere restriction fragments separated by one- or two-dimensional (1D or 2D) gel electrophoresis was isolated from 96 (1.5 ml) YPD liquid overnight cultures. Low molecular weight extrachromosomal telomeric DNA for electron microscopy examination was treated with RNA at 37°C for an hour and then isolated by running uncut genomic DNA on 0.8% agarose gels at 90V for 60 min. DNA migrating between 500 and 3500 bp linear markers was excised from the gel and electro-eluted onto 12 000 to 14 000 MWCO Spectra/Por dialysis tubing (Spectrum Laboratories Incorporated, Ranch Dominguez, CA). Electro-eluted DNA was subsequently concentrated using microcon YM-10 spin columns as directed by manufacturer (Amicon Bioseperations, Raleigh, NC). High molecular weight DNA for electron microscopy (EM) examination was attained by spheroplasting and isolating nuclei as described previously (33) with the following modification: the lytic enzyme used was 100 µg/ml Zymolyase 100T (Seikagaku).
Southern and in-gel hybridizations
For 1D gel electrophoresis, EcoRI (NEB Beverly, MA) digested or undigested genomic DNA was separated on a 0.8% SeaKem LE agarose gel (Lonza, Rockland Inc., Rockland, ME). For 2D gel analysis of low molecular weight DNA, uncut, RNase-treated genomic DNA was separated in a 4% NuSieve 3:1 agarose gel initially containing 0.6 µg/ml chloroquine (Lonza, Rockland, ME) as previously described (20). In the above experiments, the gels were blotted onto Hybond N+ membrane and probed with either Klac1-25 G-strand telomeric probe (5′-ACGGATTTGATTAGGTATGTGGTGT-3′) or the Klac-25-1 C-strand telomeric probe (3′-ACACCACATACCTAATCAAATCCGT-5′). All hybridizations were carried out in the presence of 500 mM Na2HPO4 and 7% sodium dodecyl sulfate (SDS) and the washes were done in 100 mM Na2HPO4 and 2% SDS. For 2D gel electrophoresis of high molecular weight DNA, EcoRI-digested DNA was separated along with 50 ng of circularized HindIII λ fragments, as described previously (33). The gel was blotted, probed with 32P-labelled K. lactis telomeric C-strand and then subsequently probed with 32P-labelled HindIII λ fragments.
Electron microscopy
Low molecular weight gel-extracted DNA from stn1-M1 and stn1-M1 ter1-Δ cells was incubated with 20 µg/ml T4 gene 32 protein (gift of Nancy Nossal, NIH, Bethesda, MD) for 5 min in a buffer containing 10 mM HEPES pH 7.5 and 1 mM EDTA. The samples were treated with 0.6% glutaraldehyde on ice for 10 min and chromatographed over a 2.5 ml BioGel A-1.5 M column (Bio-Rad, Hercules, CA). Fractions containing DNA and DNA–protein complexes were prepared for EM by absorption onto negatively charged carbon-coated grids in the presence of spermidine followed by dehydration through a series of gradated ethanol washes, air dying and rotary shadowcasting with tungsten at 1 × 10−6 Torr (33,34). For examination of high molecular weight telomeric DNA, isolated genomic DNA was digested with AluI, HpaII and NlaIII (NEB Beverly, MA), at enzyme concentration of 1 U/mg for 2 h, and then supplemented with an equal amount of each enzyme for an additional 2 h. The telomere restriction fragments were then separated by size exclusion chromatography and the eluted fractions monitored for DNA concentration and telomeric DNA abundance. Telomere-enriched fractions were prepared for EM by surface spreading on a denatured protein film (33,35). Samples were examined on an FEI Tecnai 12 instrument (Hillsboro, OR). Images were captured using a Gatan Ultrascan US4000SP digital camera (Gatan, Pleasanton, CA) and molecule dimensions determined using Gatan Digital Micrograph 3.0 software. Images for publication were captured on sheet film, and digitized using ACT-1 software (Nikon, Tokyo, Japan) and a Nikon SMZ1000 stereoscope. Brightness and contrast were adjusted using Adobe Photoshop (Adobe Systems, San Jose, CA).
RESULTS
Detection of t-circles in the stn1-M1 mutant by 2D gel electrophoresis
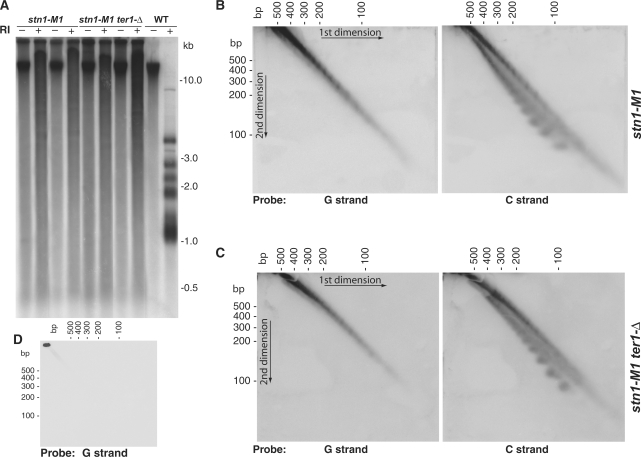
The stn1-M1 mutant generates extremely long telomeres independently of telomerase using Type IIR RTE (Figure 1A) (32). Some of the telomeric hybridization signal seen in uncut genomic DNA from the stn1-M1 and stn1-M1 ter1-Δ cells (the latter deleted for the telomerase RNA gene) appeared throughout the length of the gel. This indicated that some of the smear of telomeric signal, particularly at low molecular weights, likely represented extrachromosomal telomeric sequences. Similar smears largely composed of double- and single-stranded t-circles that hybridized to telomere probes were seen in the long telomere mutant ter1-16T (21). Therefore, we hypothesized that t-circles were being produced in stn1-M1 cells. As a test of this, we electrophoresed uncut genomic DNA from stn1-M1 and stn1-M1 ter1-Δ cells on 4% agarose 2D chloroquine gels to separate low molecular weight t-circles. Filters blotted from these gels were hybridized to oligonucleotide probes matching sequence from either the G-rich or the C-rich strands of K. lactis telomeric DNA (Figure 1B and C). With a G-strand telomeric probe (left panels of Figure 1B and C), we observed similar diagonals of closely spaced diffuse spots in both types of cells. With a C-strand telomeric probe (right panels of Figure 1B and C), we observed the same diagonal plus an additional arc of spots. These results are very similar to the ladders formed from double- and single-stranded t-circles that were observed previously in DNA from ter1-16T cells (21). The ladders of spots represented DNA species containing different integral numbers of the 25 bp K. lactis telomeric repeat. Our results suggested that stn1-M1 and stn1-M1 ter1-Δ cells, like ter1-16T, produce very small double-stranded t-circles as well as very small single-stranded t-circles that were composed specifically of the G-rich strand of telomeric sequence. As expected, wild-type cells produced no observable low molecular weight telomeric spots (Figure 1D).
Figure 1.
Gel analysis of low molecular weight telomeric DNA from stn1-M1 and stn1-M1 ter1-Δ cells. (A) Southern blot of uncut and EcoRI-digested genomic DNA from wild type (strain 7B520), stn1-M1 and stn1-M1 ter1-Δ run on a 1D 0.8% agarose gel and hybridized to a telomeric probe. (B–C) Southern blots of uncut genomic DNA from stn1-M1 and stn1-M1 ter1-Δ run on 2D 4% agarose gels hybridized to either the C-strand or G-strand telomeric probes. (D) Southern blot of uncut genomic DNA from a wild-type control hybridized to G-strand telomeric probe.
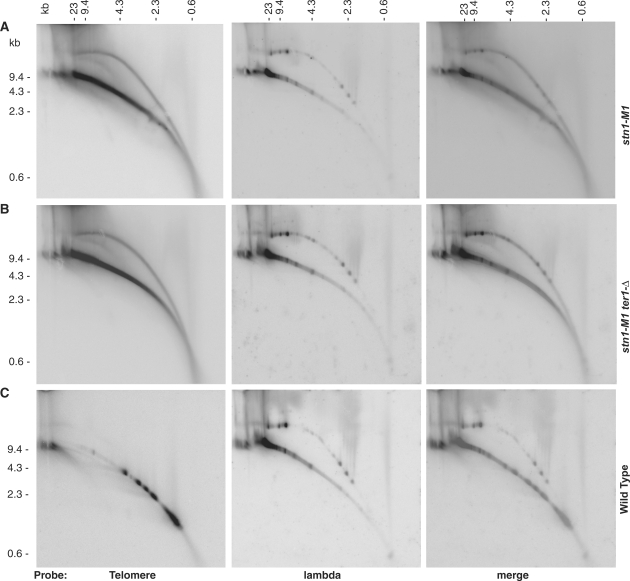
We next separated EcoRI-digested genomic DNA from stn1-M1, stn1-M1 ter1-Δ and a wild-type control on low percentage agarose 2D gels (first dimension in 0.6% agarose gel and 1.1% agarose in second dimension) to test for the possible presence of high molecular weight t-circles. EcoRI does not cleave K. lactis telomeric DNA but does separate the rest of the genomic DNA into a variety of sizes that can be visualized on gels. HindIII fragments of phage λ DNA were ligated into circles and separated along with the EcoRI-digested genomic DNA as a circular DNA control. The results of this experiment showed that a significant proportion of signal produced from a telomeric probe in both stn1-M1 and stn1-M1 ter1-Δ cells was present in an arc migrating with the double-stranded relaxed circle controls (Figure 2A and B). This was not observed with DNA from wild-type cells (Figure 2C). We conclude that stn1-M1 and stn1-M1 ter1-Δ cells contain abundant high molecular weight t-circles.
Figure 2.
Two-dimensional gel electrophoresis of high molecular weight telomeric DNA from stn1-M1 and stn1-M1 ter1-Δ cells. EcoRI-digested DNA (8 µg) from stn1-M1 (A), stn1-M1 ter1-Δ (B), and wild type (7B520) (C) cells along with 50 ng of ligated phage λ HindIII fragments were separated on low percentage 2D agarose gels. The main, lower arc in each panel represents linear DNA while the upper arc present in most panels represents open circular form DNA.
Visualization of t-circles by electron microscopy
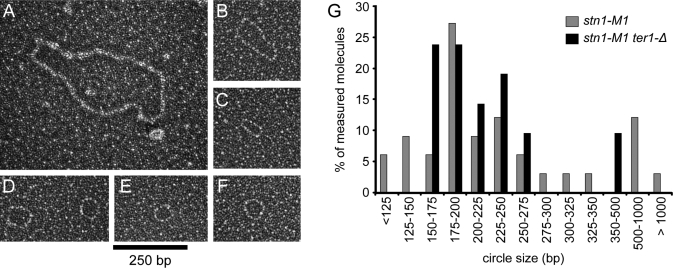
To confirm the presence of small t-circles in stn1-M1, we examined extrachromosomal telomeric DNA from stn1-M1 and stn1-M1 ter1-Δ by EM. Undigested samples of stn1-M1 and stn1-M1 ter1-Δ genomic DNA were separated on a 0.8% agarose gel and DNA running between 500 and 3500 bp (relative to linear DNA markers) was extracted. Once purified, this DNA was incubated with T4 gene 32 single strand DNA-binding protein and visualized by EM (Figure 3A–F). In both the stn1-M1 and stn1-M1 ter1-Δ samples, we observed a high percentage of the DNA molecules to be circular. Scoring DNAs by EM, 19.5% ± 7.8% were circular in the stn1-M1 sample and 30.0% ± 6.8% were circular in the stn1-M1 ter1-Δ sample. Surprisingly, <2% of the circles were single-stranded as judged by gp32 binding. Double-stranded circles lengths from the stn1-M1 and stn1-M1 ter1-Δ samples were measured and the results are shown in Figure 3G. Nearly all of the circles observed were less than 1 kb and most measured between 175 and 300 bp. This is very similar to the size distribution of t-circles observed by EM in DNA isolated from ter1-16T cells using the same protocol (21). The reason why single-stranded circles were much less common in this experiment compared to the 4% agarose 2D gels is not clear.
Figure 3.
Electron microscopy visualization of low molecular weight DNA circles from stn1-M1 and stn1-M1 ter1-Δ cells. (A–F) Electron micrographs of low molecular weight double-stranded t-circles. Estimated circle sizes are 1320, 428, 191, 216 and 258, 187 and 201 bp, respectively, for A–F. Samples were mounted onto thin carbon foils, shadowcasted with tungsten and are shown in negative contrast. Bar is equivalent to 250 bp. (G) Size distribution of observed DNA circles from stn1-M1 and stn1-M1 ter1-Δ. Measured double-stranded circles observed in low molecular weight extrachromosomal DNA isolated from stn1-M1 (n = 33) and stn1-M1 ter1-Δ (n = 21) cells.
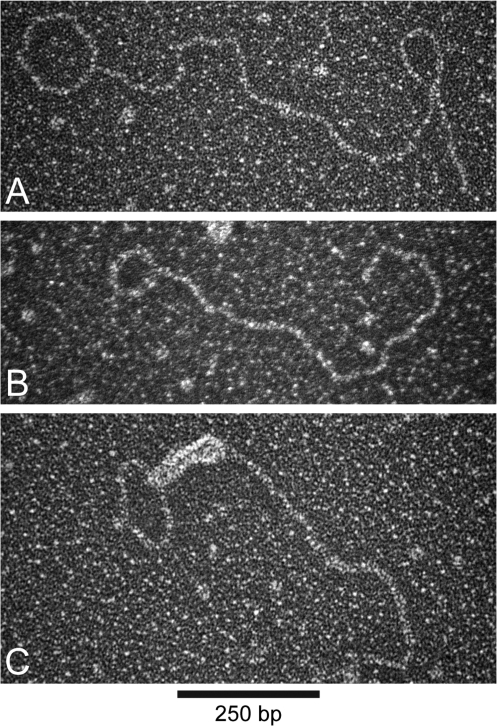
In addition to fully circular DNA molecules, we also visualized a small number of double-stranded DNA circles with tails of varying sizes, examples of which are shown in Figure 4. These structures represented less than 3.5% of the total number of molecules visualized in the stn1-M1 samples. In a few instances, these molecules were bound by T4 gene 32 protein at the base of the tail indicating the presence of a small region of single-stranded DNA at the circle–tail junction (Figure 4C). These molecules conceivably represent intermediates in the formation or processing of t-circles, or circles undergoing rolling-circle replication.
Figure 4.
Visualization of tailed circles from stn1-M1 and stn1-M1 ter1-Δ mutant cells. (A–B) Electron micrographs of tailed circle DNA structures. Circular and tail portions of molecules shown in A–B are estimated at 439 and 1361, and 252 and 918 bp, respectively. (C) Electron micrograph of a tailed circle structure with a ds and ss DNA tail. The DNA was incubated with T4 gene 32 protein, gluteraldehyde, crosslinked and prepared for EM as in Figure 3. The loop portion of the molecule shown in C is 400 bp and the total length of the single-stranded and double-stranded tail is equivalent to 796 bp. Bar is equivalent to 250 bp.
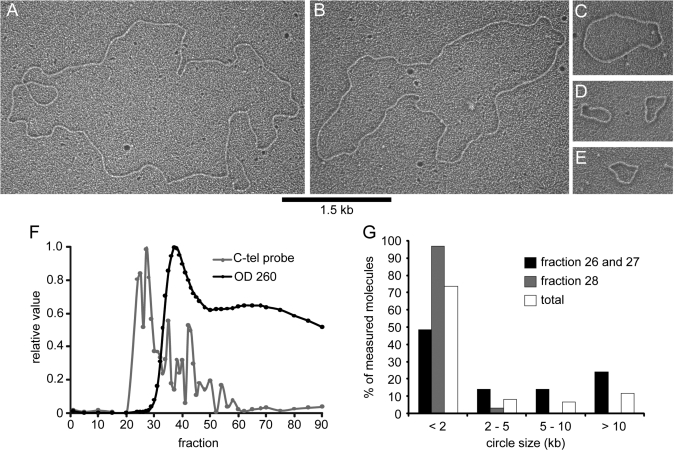
To visualize the large t-circles by EM, high molecular weight telomere restriction fragments from stn1-M1 cells were enriched by gel-exclusion chromatography, as described previously (21,33). Briefly, crude nuclei from stn1-M1 cells were isolated and total genomic DNA was digested with AluI, HpaII and NlaIII restriction enzymes. This reduces the genomic DNA to very small sizes while leaving telomeric repeat tracts intact. The digested genomic DNA was then separated in a long gel-filtration chromatography column, and the eluted fractions were assayed for total and telomeric DNA content (Figure 5F). The fractions highly enriched for telomeric DNA were concentrated and examined by EM (34). In the telomere-enriched fractions, we observed a large number of t-circles ranging in size from 0.3 to 31 kb (Figure 5A–E). The majority of circles were small, with 73% of the circles from the telomeric-enriched fractions measuring less than 3 kb in total length, and 11.5% measuring over 10 kb (n = 61) (Figure 5G). These results are similar to previous experiments in human ALT cells where the bulk of circular molecules were rather small compared to the average telomere size (20).
Figure 5.
Visualization of DNA circles from high molecular weight telomere-enriched DNA from stn1-M1. (A–E) Electron micrographs of DNA circles observed in the telomere-enriched fractions from stn1-M1. Circle lengths for the molecules in A–E, respectively, are estimated at 15.7, 12.1, 3.1, 1.2 and 1.2, and 0.9 kb for fractions A–E, respectively. Note that there is also a ∼0.8 kb linear fragment in the lower right portion of panel A. Samples were prepared by coating the DNA in denatured cytochrome c and are shown in negative contrast. Bar is equivalent to 1.5 kb. (F) Relative DNA abundance and telomeric DNA content of eluted fractions from gel chromatography separation of AluI, HpaII and NlaIII dirgest of stn1-M1 genomic DNA. Molecules shown in A–E are from fractions 26 to 28 (F). Size distribution of the measured circles from fractions 26 to 27 (n = 29) and 28 (n = 32).
DISCUSSION
The results presented here demonstrate that a broad size range of t-circles, from ∼100 to >30 kb, are produced in the K. lactis stn1-M1 mutant that maintains its telomeres using Type IIR RTE. Both small and large t-circles are formed independently of the presence of telomerase in the mutant cells. As telomeric DNA lacks the ability to initiate replication, the t-circles present in stn1-M1 cells must be recent products of recombination rather than heritably replicating episomes. Because stn1-M1 rad52 cells are inviable (32), it has not been possible to directly demonstrate that homologous recombination is required for formation of t-circles in these cells. However, t-circles in other systems, including K. lactis ter1-16T cells, have previously been demonstrated to depend upon Rad52 or other homologous recombination genes (19,21,22,36). The presence of the small t-circles from stn1-M1 cells as a discrete series of spots on 2D gels, as seen previously with t-circles from the ter1-16T mutant (21), strongly favours the idea that these spots are composed of integral numbers of telomeric repeats, as expected for a recombination process dependent upon homology.
We conclude that the t-circle formation in stn1-M1 cells is linked to the chronic telomere-capping defects of this mutant. This defect results not only in highly elongated telomeres but also in abnormal cell and colony growth, large 3′ telomeric overhangs and greatly increased rates of both subtelomeric recombination and telomeric truncation events (32). Unlike the Type II RTE of ter1-Δ mutants, where capping defects occur as a result of telomeres becoming too short, cells undergoing Type IIR RTE have capping defects believed to be independent of telomere length and are results of disruptions in the functioning of telomere proteins. These continuous capping defects presumably underlie the extreme telomere lengths and abundant products of telomeric recombination such as t-circles.
The abundance of t-circles in stn1-M1 cells is compatible with them playing a role in the RTE that occurs in those cells. Although strong circumstantial evidence favours the hypothesis that small t-circles are often involved in the Type II RTE that occurs in yeast ter1-Δ mutants (17,18), there is no evidence to date that t-circles are involved in the Type IIR RTE of stn1-M1 cells or the apparently similar recombinational telomere maintenance of human ALT cells. The proposed role of t-circles in Type II RTE of K. lactis ter1-Δ mutants is in building the first relatively long telomere through a rolling circle copying event in a cell that contains only short telomeres. Once one long telomere is present, t-circles may no longer be necessary, as other telomeres may be lengthened directly by copying the sequence of the long telomere (15). We hypothesize that the most significant possible role for t-circles in Type IIR RTE might be in the initial establishment of the long telomere state (such as would occur in a newly germinated stn1-M1 spore from a STN1/stn1-M1 diploid). There, the formation of extremely long telomeres from the normal length telomeres initially present (∼500 bp in K. lactis) could be accelerated by the rolling circle copying of a t-circle. However, even if not vital to the maintenance of Type IIR RTE, t-circles could play a significant role in telomere elongation due to their high abundance in the cell. It is believed that in K. lactis ter1-Δ mutants, the telomere lengthening from Type II RTE (typically hundreds of bp) is thought to be limited by both the rarity of t-circles in those cells and the poorly processive copying of extremely small (∼100 bp) t-circles. Many, if not most, of the larger t-circles in stn1-M1 cells would likely be capable of producing much greater extensions. It is known, e.g. that 1.6 kb URA3-telomere circles transformed into K. lactis cells routinely produce telomere extensions of >10 000 bp (17,18).
The precise mechanism of t-circle formation in stn1-M1 cells, or in other cells where they have been observed, is currently unknown. A previously proposed hypothesis is that a t-circle can be formed via a t-loop intermediate whereby the 3′-end of the telomere strand invades a more internal region of the same telomere (21,22). In favour of this possibility, t-loop structures have been observed in both human ALT cells and K. lactis ter1-16T cells (20,33). Furthermore, the loop portions of these structures exhibit size distributions similar to those of t-circles observed in the same systems. T-circle formation from t-loops would also be expected to cause a deletion of the telomere involved in the process (37). Among the extrachromosomal structures observed in this study by EM were a small percentage of tailed circles. These likely represent either t-loops or rolling circle intermediates.
Small single-stranded t-circles, specifically composed of the G-rich strand of telomeric sequence, were previously observed in ter1-16T (21) cells where they were approximately as abundant as double-stranded circles, at least for circles <500 bp/nt. Both types of t-circles were absent in ter1-16T rad52 cells, indicating that they are generated by homologous recombination (21). It was suggested that the processing of a t-loop intermediate to form a t-circle often initially produced a transient t-circle that was partially double stranded and partially single stranded. Further processing then could produce more stable t-circles that were either fully double stranded or fully single stranded. The reason why the relative abundance of single-stranded t-circles varied considerably between the EM and 2D gel analysis remains unclear. Variation in the amount lost during purification of yeast DNA is a possible contributing factor. The relatively small sizes of the single-stranded t-circles likely render them vulnerable to being lost during alcohol precipitations or dialysis. In addition, we have observed considerable variation in both the total amount of telomeric signal and the amount of small extrachromosomal telomeric DNA from independent isogenic isolates of stn1-M1 extracted by the same method (J. Xu, S. Iyer, E. Basenko and M. McEachern, unpublished data). It is quite possible that the proportion of single-stranded t-circles may also be naturally variable.
The identification of t-circles in stn1-M1 cells helps further underscore the similarities between the Type IIR RTE of yeast cells and the ALT phenomenon of some human cancers. Learning more about the mechanism of Type IIR RTE in yeast will certainly help to provide insights into how ALT occurs and what mutations underlie it.
FUNDING
National Institutes of Health to M.J.M. (GM61645) and to J.D.G. (GM31819, ES13773) and a summer fellowship from the American Federation for Aging Research to S.I. Funding for open access charge: National Institutes of Health (GM61645).
Conflict of interest statement. None declared.
REFERENCES
- 1.Verdun RE, Karlseder J. Replication and protection of telomeres. Nature. 2007;447:924–931. doi: 10.1038/nature05976. [DOI] [PubMed] [Google Scholar]
- 2.Smogorzewska A, de Lange T. Regulation of telomerase by telomeric proteins. Ann. Rev. Biochem. 2004;73:177–208. doi: 10.1146/annurev.biochem.73.071403.160049. [DOI] [PubMed] [Google Scholar]
- 3.Hathcock KS, Chiang YJ, Hodes RJ. In vivo regulation of telomerase activity and telomere length. Immun. Rev. 2005;205:104–113. doi: 10.1111/j.0105-2896.2005.00267.x. [DOI] [PubMed] [Google Scholar]
- 4.Bodnar AG, Ouellette M, Frolkis M, Holt SE, Chiu C-P, Morin GB, Harley CB, Shay JW, Lichtsteiner S, Wright WE. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- 5.Shawi M, Autexier C. Telomerase, senescence and ageing. Mech. Ageing Dev. 2008;129:3–10. doi: 10.1016/j.mad.2007.11.007. [DOI] [PubMed] [Google Scholar]
- 6.Deng Y, Chan SS, Chang S. Telomere dysfunction and tumour suppression: the senescence connection. Nat. Rev. Cancer. 2008;8:450–458. doi: 10.1038/nrc2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harley CB, Kim NW, Prowse KR, Weinrich SL, Hirsch KS, West MD, Bacchetti S, Hirte HW, Counter CM, Greider CW, et al. Telomerase, cell immortality, and cancer. Cold Spring Harbor Symp. Quant. Biol. 1994;59:307–315. doi: 10.1101/sqb.1994.059.01.035. [DOI] [PubMed] [Google Scholar]
- 8.Bryan TM, Marusic L, Bacchetti S, Namba M, Reddel RR. The telomere lengthening mechanism in telomerase-negative immortal human cells does not involve the telomerase RNA subunit. Hum. Mol. Genet. 1997;6:921–926. doi: 10.1093/hmg/6.6.921. [DOI] [PubMed] [Google Scholar]
- 9.Cesare AJ, Reddel RR. Telomere uncapping and alternative lengthening of telomeres. Mech. Ageing Dev. 129:99–108. doi: 10.1016/j.mad.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 10.Lundblad V, Szostak JW. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell. 1989;57:633–643. doi: 10.1016/0092-8674(89)90132-3. [DOI] [PubMed] [Google Scholar]
- 11.McEachern MJ, Blackburn EH. Runaway telomere elongation leads to senescence in yeast. Nature. 1995;376:403–409. doi: 10.1038/376403a0. [DOI] [PubMed] [Google Scholar]
- 12.McEachern MJ, Blackburn EH. Cap-prevented recombination between terminal telomeric repeat arrays (telomere CPR) maintains telomeres in Kluyveromyces lactis lacking telomerase. Genes Dev. 1996;10:1822–1834. doi: 10.1101/gad.10.14.1822. [DOI] [PubMed] [Google Scholar]
- 13.Lundblad V, Blackburn EH. An alternative pathway for yeast telomere maintenance rescues est1-senescence. Cell. 1993;73:347–360. doi: 10.1016/0092-8674(93)90234-h. [DOI] [PubMed] [Google Scholar]
- 14.Chen Q, Ijpma A, Greider CW. Two survivor pathways that allow growth in the absence of telomerase are generated by distinct telomere recombination events. Mol. Cell Biol. 2001;21:1819–1827. doi: 10.1128/MCB.21.5.1819-1827.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Topcu Z, Nickles K, Davis C, McEachern MJ. Abrupt disruption of capping and a single source for recombinationally elongated telomeres in Kluyveromyces lactis. Proc. Natl Acad. Sci. USA. 2005;102:3348–3353. doi: 10.1073/pnas.0408770102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McEachern MJ, Iyer S. Short telomeres in yeast are highly recombinogenic. Mol. Cell. 2001;7:695–704. doi: 10.1016/s1097-2765(01)00215-5. [DOI] [PubMed] [Google Scholar]
- 17.Natarajan S, McEachern MJ. Recombinational telomere elongation promoted by DNA circles. Mol. Cell Biol. 2002;22:4512–4521. doi: 10.1128/MCB.22.13.4512-4521.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Natarajan S, Groff-Vindman C, McEachern MJ. Factors influencing the recombinational expansion and spread of telomeric tandem arrays in Kluyveromyces lactis. Eukaryot. Cell. 2003;2:1115–1127. doi: 10.1128/EC.2.5.1115-1127.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin CY, Chang HH, Wu KJ, Tseng SF, Lin CC, Lin CP, Teng SC. Extrachromosomal telomeric circles contribute to Rad52-, Rad50-, and Polymerase Δ-mediated telomere-telomere recombination in Saccharomyces cerevisiae. Eukaryot. Cell. 2005;4:327–336. doi: 10.1128/EC.4.2.327-336.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cesare AJ, Griffith JD. Telomeric DNA in ALT cells is characterized by free telomeric circles and heterogeneous t-loops. Mol. Cell Biol. 2004;24:9948–9957. doi: 10.1128/MCB.24.22.9948-9957.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Groff-Vindman C, Cesare AJ, Natarajan S, Griffith JD, McEachern MJ. Recombination at long mutant telomeres produces tiny single- and double-stranded telomeric circles. Mol. Cell Biol. 2005;25:4406–4412. doi: 10.1128/MCB.25.11.4406-4412.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang RC, Smogorzewska A, de Lange T. Homologous recombination generates T-loop-sized deletions at human telomeres. Cell. 2004;119:355–368. doi: 10.1016/j.cell.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 23.McEachern MJ, Haber JE. Break-induced replication and recombinational telomere elongation in yeast. Ann. Rev. Biochem. 2006;75:111–135. doi: 10.1146/annurev.biochem.74.082803.133234. [DOI] [PubMed] [Google Scholar]
- 24.Tomaska L, Nozek J, Kramara J, Griffith JD. Telomeric circles: universal players in telomere maintenance? Nat. Struct. Mol. Biol. 2009;16:1010–1015. doi: 10.1038/nsmb.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao H, Cervantes RB, Mandell EK, Otero JH, Lundblad V. RPA-like proteins mediate yeast telomere function. Nat. Struct. Mol. Biol. 2007;14:208–214. doi: 10.1038/nsmb1205. [DOI] [PubMed] [Google Scholar]
- 26.Grandin N, Reed SI, Charbonneau M. Stn1, a new Saccharomyces cerevisiae protein, is implicated in telomere size regulation in association with Cdc13. Genes Dev. 1997;11:512–527. doi: 10.1101/gad.11.4.512. [DOI] [PubMed] [Google Scholar]
- 27.Bechard LH, Butuner BD, Peterson GJ, McRae W, Topcu Z, McEachern MJ. Mutant telomeric repeats in yeast can disrupt the negative regulation of recombination-mediated telomere maintenance and create an alternative lengthening of telomeres-like phenotype. Mol. Cell Biol. 2009;29:626–639. doi: 10.1128/MCB.00423-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grandin N, Charbonneau M. The Rad51 pathway of telomerase-independent maintenance of telomeres can amplify TG1-3 sequences in yku and cdc13 mutants of Saccharomyces cerevisiae. Mol. Cell Biol. 2003;23:3721–3734. doi: 10.1128/MCB.23.11.3721-3734.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grandin N, Damon C, Charbonneau M. Cdc13 prevents telomere uncapping and Rad50-dependent homologous recombination. EMBO J. 2001;20:6127–6139. doi: 10.1093/emboj/20.21.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petreaca RC, Chiu HC, Nugent CI. The role of Stn1p in Saccharomyces cerevisiae telomere capping can be separated from its interaction with Cdc13p. Genetics. 2007;177:1459–1474. doi: 10.1534/genetics.107.078840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wray LV, Jr, Witte MM, Dickson RC, Riley MI. Characterization of a positive regulatory gene, LAC9, that controls induction of the lactose-galactose regulon of Kluyveromyces lactis: structural and functional relationships to GAL4 of Saccharomyces cerevisiae. Mol. Cell Biol. 1987;7:1111–1121. doi: 10.1128/mcb.7.3.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iyer S, Chadha AD, McEachern MJ. A mutation in the STN1 gene triggers an alternative lengthening of telomere-like runaway recombinational telomere elongation and rapid deletion in yeast. Mol. Cell Biol. 2005;25:8064–8073. doi: 10.1128/MCB.25.18.8064-8073.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cesare AJ, Groff-Vindman C, Compton SA, McEachern MJ, Griffith JD. Telomere loops and homologous recombination-dependent telomeric circles in a Kluyveromyces lactis telomere mutant strain. Mol. Cell Biol. 2008;28:20–29. doi: 10.1128/MCB.01122-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Griffith JD, Christiansen G. Electron microscope visualization of chromatin and other DNA-protein complexes. Ann. Rev. Biophys. Bioeng. 1978;7:19–35. doi: 10.1146/annurev.bb.07.060178.000315. [DOI] [PubMed] [Google Scholar]
- 35.Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T. Mammalian telomeres end in a large duplex loop. Cell. 1999;97:503–514. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- 36.Compton SA, Choi J-H, Cesare AJ, Ozgur S, Griffith JD. Xrcc3 and Nbs1 are required for the production of extrachromosomal telomeric circles in human alternative lengthening of telomere cells. Cancer Res. 2007;67:1513–1519. doi: 10.1158/0008-5472.CAN-06-3672. [DOI] [PubMed] [Google Scholar]
- 37.Bhattacharyya MK, Lustig AJ. Telomere dynamics in genome stability. Trends Biochem. Sci. 2006;31:114–122. doi: 10.1016/j.tibs.2005.12.001. [DOI] [PubMed] [Google Scholar]