Abstract
Top1 inhibition by camptothecin (CPT) perturbs RNA polymerase II (Pol II) density at promoters and along transcribed genes suggesting an involvement of Top1 in Pol II pausing. Here, we demonstrate that Top1 inhibition favors Pol II escape from a promoter-proximal pausing site of the human HIF-1α gene in living cells. Interestingly, alternative splicing at exon 11 was markedly altered in nascent HIF-1α mRNAs, and chromatin structure was also affected with enhanced histone acetylation and reduced nucleosome density in a manner dependent on cdk activity. Moreover, CPT increases transcription of a novel long RNA (5′aHIF1α), antisense to human HIF-1α mRNA, and a known antisense RNA at the 3′-end of the gene, while decreasing mRNA levels under normoxic and hypoxic conditions. The effects require Top1, but are independent from Top1-induced replicative DNA damage. Chromatin RNA immunoprecipitation results showed that CPT can activate antisense transcription mediated by cyclin-dependent kinase (cdk) activity. Thus, Top1 inhibition can trigger a transcriptional stress, involving antisense transcription and increased chromatin accessibility, which is dependent on cdk activity and deregulated Pol II pausing. A changed balance of antisense transcripts and mRNAs may then lead to altered regulation of HIF-1α activity in human cancer cells.
INTRODUCTION
Human DNA topoisomerase I (Top1) regulate DNA superhelicity during fundamental DNA transactions by transiently cleaving and resealing DNA molecules (1,2). Top1 is the sole target of camptothecin (CPT) and close derivatives, which are anticancer drugs used in standard therapy of human ovarian, small-cell lung, and colorectal cancers (3). CPT interacts reversibly with its target preventing strand relegation and increasing the half-life of Top1-DNA cleavage complexes (Top1cc). A Top1cc can become lethal when it collides with DNA replication forks, leading to irreversible DNA breaks and the activation of S-phase checkpoint, G2 arrest and cell death (3). Although the established cellular effects of CPT are peculiar of DNA damage responses, CPT inhibition of Top1 occurs primarily in actively transcribed regions (2,3). In contrast to the replication-dependent effects, the transcription-dependent effects of Top1cc are not fully understood.
Top1 is enriched in transcribed genomic regions as established by Top1 DNA cleavage sites (1–3) and chromatin immunoprecipitation (ChIP) (4). Top1 has been shown to activate gene transcription, and to bind to general transcription factors at promoters (5–7). A broad and general inhibition of transcription elongation is an immediate effect of CPT in cultured cells (3,8), likely due to the stalling of elongating RNA polymerases by Top1ccs (3,8) and/or by persistent transcription-generated DNA supercoils (9). Nevertheless, several groups have reported more specific effects at transcriptional levels for CPT. For instance, CPT can activate the initiation step of transcription (10) and the expression of selective genes in human cells (11). In yeast, TOP1 gene deletion preferentially up-regulates the expression of telomere-proximal genes by increasing core histone acetylation and chromatin accessibility (12).
Strikingly, CPT-induced Top1ccs have immediate and specific effects on RNA polymerase II (Pol II). CPT triggers a high phosphorylation degree of the largest subunit (Rpb1) of Pol II (4,13,14), showing an effect on a critical step of transcription regulation. Hyperphosphorylation occurs selectively on Ser-5 residues of the conserved heptapeptide repeats of the carboxy-terminal domain (CTD) possibly mediated by Cdk7, component of TFIIH (13). Interestingly, a recent report showed that CPT can disrupt the large inactive P-TEFb complex, thus releasing a free active P-TEFb complex (containing the Cdk9 subunit), which can then contribute to CPT-increased phosphorylation of Pol II (15).
A second immediate effect of CPT on Pol II has been reported by us previously, and can be correlated to the hyperphosphorylation of Rpb1 (4). Short cell treatments with CPT induce a redistribution of chromatin-bound Pol II along transcribed genes in human cancer cells, apparently by enhancing the escape of Pol II from promoter-proximal pausing sites. Remarkably, this early specific CPT effect is independent from replication and replicative DNA damage (4). However, the release of Pol II from pausing into elongation remained to be established.
Thus, to define the molecular mechanisms triggered by Top1ccs at transcribing regions, we have here investigated the CPT effects at transcription and chromatin levels at the hypoxia-inducible factor 1α (HIF-1α) gene locus in human cancer cells. HIF-1α is a transcription factor and a master regulator of the cell response to oxygen deprivation (16,17), and a target of anti-angiogenesis and anticancer agents (17). HIF-1α is subjected to post-translational regulation, as it is degraded at high oxygen tensions by an oxygen-mediated hydroxylation of conserved prolyl and asparaginyl residues, to which the von Hippel-Lindau protein (pVHL) E3 ligase binds targeting HIF-1α to proteasomal degradation (17). We selected to study the HIF-1α gene because previous reports demonstrated that CPT can markedly reduce HIF-1α protein accumulation in hypoxic cells in a manner independent from the hydroxylation pathway and from replicative DNA damage promoted by CPT (18,19).
We here provide evidence that Top1ccs trigger the escape of Pol II from promoter-proximal pause site of the HIF-1α gene, activate a novel antisense transcript at the 5′-end of the HIF-1α locus and induce a more accessible chromatin structure in a manner dependent on cyclin-dependent kinase activity. Thus, Top1 inhibition by CPT can result in a specific transcriptional stress leading to an imbalance of antisense and sense RNA ratios, which may alter the regulation of cancer-relevant factors such as HIF-1α.
MATERIALS AND METHODS
All methods and materials are fully described in the Supplementary Data.
Cell lines and treatments
Human HCT116, HCT116(top1 siRNA) (20), MRCV fibroblast, and Jurkat cells were cultured with standard techniques. Cells were maintained at 37°C in a humidified incubator containing 5% CO2 in air (normoxic conditions, 20% O2). Hypoxia treatments were in a cell chamber with 1% O2 at 37°C. All drug treatments were performed on exponentially growing cells at 37°C. In case of co-treatments, cells were first incubated with aphidicolin (3 μM) or DRB (50 μM) for 15 min, or caffeine (5 mM) for 30 min, and then, CPT was added to the medium for the indicated time.
RNA purification and primer-specific cDNA preparation
Total RNA was purified with the acid phenol method. RNA pellets were resuspended in TE, and DNA was digested with DNAse I. Total RNA (1 µg) was used to prepare cDNA using random primers and SuperScript III (Invitrogen) as suggested by the manufacturer. When specific primers were used for reverse transcription with SuperScript III, conditions for the reaction were as follows: 5 min at 65°C and 50 min at 55°C. Negative controls were cDNAs prepared with no primer during reverse transcription reactions.
Northern blotting
For Northern blot analysis, we used the procedures of (21) with minor modifications. Strand-specific probe was designed to be 40 nucleotides in length and was synthesized by Intregrated DNA Technologies (IDT) with the 3′ StarFire extension (5′-CAG CCC CAA TTC TAA ATA AGC TCT TAG ATT TTC CTC AGC C/NNNNNN/-3′). The probe was then 3′-end 32P-labeled at high specific activity with the IDT StarFire kit (IDT) using the manufacturer's; instructions. Hybridization was performed for 16 h in 7% SDS, 500 mM sodium phosphate (pH 7.0), 1 mM EDTA, 25% formamide at 42°C, and then washed with 2× SSC, 0.5% SDS at 25°C for 20 min.
The HIF-1α mRNA exon 2 was used as probe to determine the mRNA levels. Hybridization was performed for 16 h in 7% SDS, 500 mM sodium phosphate (pH 7.0), 1mM EDTA at 65°C. Final wash was at 65°C for 45 min.
ChIP
The ChIP protocol and measurements of cDNA by qrt-PCR were essentially as reported already (4). The RNA-ChIP (RIP) method was as the ChIP protocol with some modifications. Chromatin was fragmented to an average size of 1000 nucleotides. Precipitated RNA was extensively treated with DNase I, ethanol-precipitated and treated again with DNase I. Then, cDNA was prepared with random primers, and analyzed by qrt-PCR. Negative controls were cDNA prepared with no primers (NP), and non-retrotrascribed samples. At least four dilutions of genomic DNA were run to generate the standard curve. For all specific antibodies, the RNA recovered values were at least 10-fold more enriched than non-immune controls, and background levels set by NP and α-satellite DNA. Moreover, recovered RNA values were at least 100-fold more enriched than non-retrotranscribed samples.
RESULTS
CPT induces a reduction of Pol II at the HIF-1α gene promoter in a Top1 and cdk-dependent manner
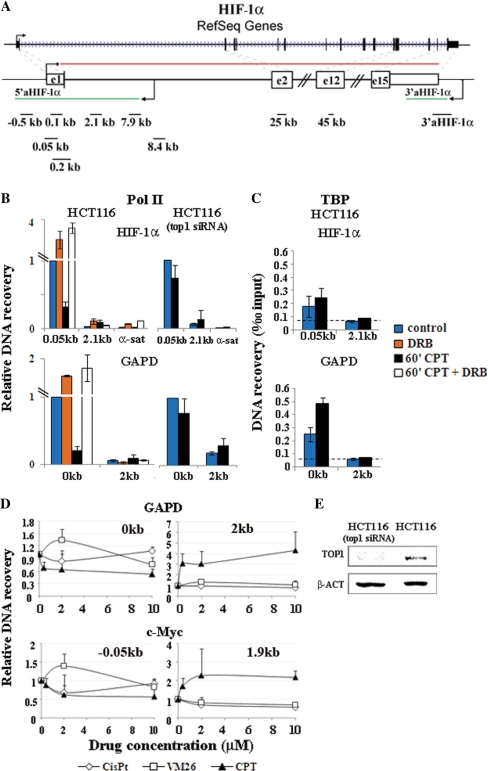
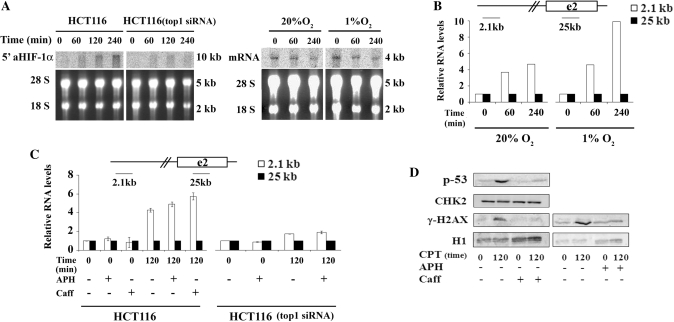
To investigate the escape of Pol II from pausing sites induced by Top1ccs, we first determined the presence of a Pol II pausing site at the promoter of the HIF-1α gene and the specificity of the CPT effect in living cells by ChIP. The human HIF-1α gene is 52.738-kb long and is constituted of 15 exons. The intron size ranges from 0.084 to 4.8 kb, with the exception of the first intron, which is 24.542 kb (16) (Figure 1A). Experiments were carried out with human control HCT116 colon cancer cells and a HCT116 cell line with the Top1 gene silenced by siRNA, which had Top1 contents reduced by ∼5-fold as compared to parent HCT116 cells (20) (Figure 1E). The results showed that Pol II accumulated at higher levels at promoter-proximal sites in control cells (Figure 1B). This is consistent with the polymerase pausing close to the HIF-1α gene promoter. CPT reduced Pol II density at promoters by 80-70% in HCT116 cells, whereas the reduction was much less (25–30%) in HCT116-siTop1 cells (Figure 1B). While CPT abolished the difference in polymerase density between the HIF-1α promoter and an internal site in HCT116 cells, it did not in HCT116-siTop1 cells. Identical results were observed in other transcribed genes such as GAPD (Figure 1B and data not shown). The drug effect is specific for Pol II as the general transcription factor TBP was not affected by CPT treatments (Figure 1C). Thus, Pol II likely pauses at a promoter-proximal site at the HIF-1α gene locus, and Top1 is required for the specific reduction of Pol II density induced by CPT.
Figure 1.
CPT-induced reduction of Pol II density at the HIF-1α gene promoter is dependent on Top1 and Cdk activity. (A) The human HIF-1α gene is indicated with exons (boxes) and introns (lines). Red and green lines show sense and antisense transcripts. Black lines and numbers (distance from mRNA start) indicate the amplicons used in this work. The lower map is not to scale. (B) HCT116 and HCT116(Top1-siRNA) cells were treated for 1 h with CPT (10 µM) and/or DRB (50 µM). DNA recovery was determined at promoter-proximal (0.05 or 0 kb) and -distal regions (2.1 and 2 kb) of HIF-1α and GAPD genes. α-sat, control α-satellite DNA. Values are normalized to promoter-proximal regions in untreated cells, and are means ± SD of at least four determinations from two independent experiments. Mean recovery with non-immune IgG was 0.08. (C) TBP levels at promoters. Values are means ± SD of four determinations from two independent experiments. The dashed line shows recovery levels with non-immune IgG. (D) Effects of CPT, teniposide (VM26) and cis-platin (Cis-Pt) on Pol II density in human MRC5 cells at gene promoters (0 kb), and internal gene regions (2 and 1.9 kb). Values are normalized to untreated cells, and are means ± SD of at least four determinations from two independent experiments. (E) Top1 content of HCT116(Top-siRNA) cells was about 20% of that of HCT116 cells by western blotting. β-Actin (β-ACT) is a loading control.
To further evaluate the specificity of the CPT effects, we investigated two other DNA damaging agents, cisplatin and VM-26, in comparison with CPT. The two agents promote DNA damage that is independent from Top1 (22,23). The data show that Pol II density was decreased even with the lowest CPT dose (0.4 µM) at promoter-proximal pausing sites of the studied genes (Figure 1D), while it increased at sites around 2 kb downstream to the promoters (Figure 1D). In contrast, cisplatin and VM-26 decreased Pol II levels at both promoter and internal gene sites at higher concentrations only (Figure 1D, and data not shown). Thus, the findings further support that the CPT-induced reduction of Pol II accumulation at the promoters of HIF-1α and other genes likely requires Top1ccs.
We then asked whether Cdk activity is needed for the CPT effect on Pol II accumulation at the promoter of the HIF-1α gene. Cdk9 and Cdk7, which phosphorylate Pol II to promote transcription elongation, are the main targets of 5,6-di-chloro-1-β-d-ribofuranosyl-benzimidazole (DRB). We then determined if DRB could abolish the CPT effects. The results showed that CPT-induced redistribution of Pol II along the HIF-1α and other transcribed genes was abolished by DRB (Figure 1B), demonstrating that Cdk activity is needed for the altered distribution of Pol II along the studied genes induced by Top1cc.
Top1cc induces Pol II escape from the promoter-proximal pausing sites
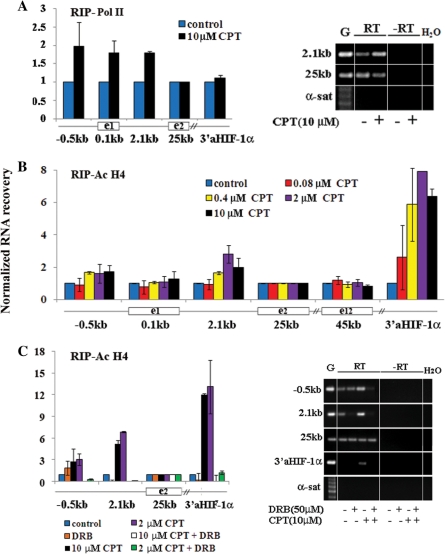
The above data suggest that Top1 may be involved in Pol II pausing regulation and that Top1cc favors Pol II escape. The pausing of elongating Pol II at promoter-proximal sites can be a main mechanism of transcription regulation (24). Interestingly, promoter-proximal pausing can regulate the recruitment of further Pol II at the promoter (25), and the coupling of mRNA maturation to transcript elongation (26). Thus, it is critical to establish whether Top1 inhibition by CPT can enhance Pol II escape from promoter-proximal pausing sites. To this end, we have determined the transcription levels downstream to the pausing site following drug treatments in nascent HIF-1α mRNA. Nascent transcripts were isolated with the RNA ChIP (RIP) method using Abs against acetylated H4 histone or against the N-terminal domain of Pol II large subunit.
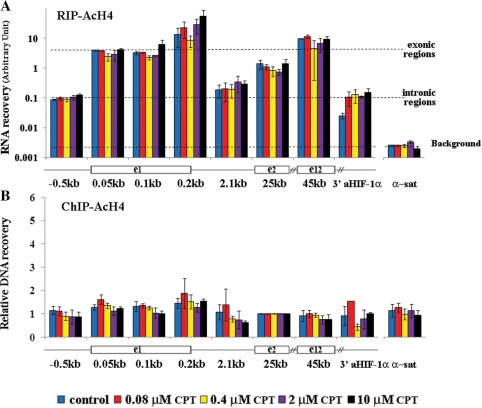
The levels of chromatin-bound RNAs allowed us to make several interesting observations. First, the levels of chromatin-bound RNAs (Figure 2A) were at least 3-log higher in the case of HIF-1α exonic regions than background (as established with samples retro-transcribed without primers) and α-satellite DNA levels in cells untreated and treated with CPT for 1 h. In contrast, HIF-1α gene intronic regions showed intermediate levels of RNA recovery (about 1-log lower than exons, Figure 2A). The observed differences among HIF-1α exons (Figure 2A) have been reported for other genes already (27), and may be due to differences in amplification rates of primer sets. We have then measured the levels of histone H4 acetylation across the entire gene by ChIP, and the results showed that H4 acetylation was not dramatically diverse among the studied fragments and cannot explain the above differences in RNA recoveries (Figure 2B). Thus, as intronic RNAs were lower than exonic RNAs, our observations are consistent with co-transcriptional splicing of HIF-1α mRNA (26). Moreover, we noticed that levels of nascent RNA recovery were not dramatically changed by CPT from 0.08 to 10 µM (Figure 2A), suggesting that Top1ccs do not affect the association of nascent transcripts with chromatin. This is in agreement with the lack of influence on transcription factories by immunofluorescence microscopy (4).
Figure 2.
Levels of chromatin-associated RNAs at the HIF-1α gene locus. (A) HCT116 cells were treated for 1 h with the indicated CPT concentrations. RIP was performed with Abs against anti acetylated histone H4 (Ac-H4). Pelleted RNA was retrotranscribed with random primers, and RNA recovery was determined with qRT-PCR using specific primers corresponding to the indicated regions along the HIF-1α gene, a diagram of which (not in scale) is shown under the graph. The broken lines indicate average recovery of exonic, intronic and centromeric α-satellite (α-sat) DNA regions. The background level was determined with cDNAs retrotranscribed without primers and was similar to α-sat recovery levels. A representative experiment is shown: values are means ± SD of three determinations. (B) Levels of Ac-H4 along the HIF-1α gene after 1-h CPT treatments. ChIP was performed with either anti Ac-H4 or non-immune Abs. Values are normalized to exon 2, and are means ± SD of four to six determinations from two independent experiments.
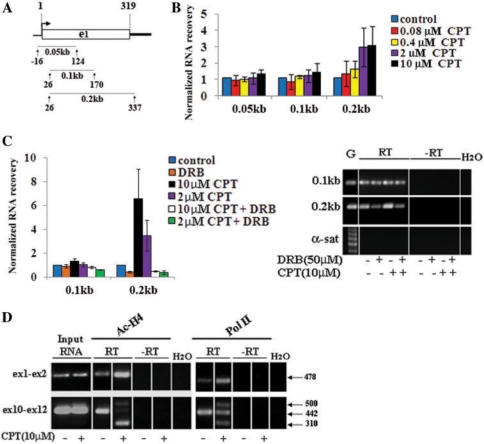
We then determined the transcription levels downstream to the promoter-proximal pausing site of HIF-1α after Top 1 inhibition by CPT. As Pol II is known to pause after 20–50 bases from start (25,26), and the first exon of HIF-1α is 319 bases long, we have compared relative levels of transcription at three overlapping regions spanning exon 1 and the exon 1/intron 1 border (Figure 3A). The findings showed that CPT increased transcription levels at the 3′-end of exon 1 in a dose-dependent manner, while the levels remained unchanged at the 5′-end (Figure 3B). Consistently with the ChIP data on Pol II (Figure 1B), DRB fully abolished the increase of transcription levels induced by Top1ccs (Figure 3C). In contrast, DRB did not affect transcription levels at the 5′-end of exon 1 (Figure 3C), showing that CPT-induced transcription downstream to the pause site was dependent on Cdk9 and/or Cdk7 activity.
Figure 3.
CPT-induced Top1ccs promote Pol II escape from the HIF-1α promoter pause site. (A) Diagram of the HIF-1α exon 1 region, with lines indicating the studied amplicons. Numbers indicate positions from mRNA transcription start. (B) HCT 116 cells were treated for 1 h with CPT, and RIP was performed with anti-acetylated histone H4 Abs (Ac-H4). (C) CPT-induced transcription downstream to the pause site is dependent on Cdk activity. On the right: PCR products of RIP samples. Controls are: genomic DNA (G), no reverse transcription (–RT) and water. α-satellite DNA (α-sat) reactions show no genomic DNA contamination. A representative experiment is reported. (D) Top1cc effects on transcription elongation and alternative splicing of HIF-1α mRNA. Transcription levels at two downstream regions from the promoter pausing site spanning exons 1 and 2, and exons 10 to 12. In the latter case, the observed extra band of 310 bases may correspond to a transcript in which exon 11 has been spliced out. –RT indicates no retrotranscription. In all panels, RNA recovery values, normalized to exon 2 and control samples, are means ± SD of at least four determinations from two independent experiments.
We also assessed CPT-induced transcription by investigating the matured mRNA of the HIF-1α gene in RIP samples. We found that CPT increased the transcription levels of a region spanning exons 1 and 2 by using Abs against acetylated histone H4 as well as against Pol II (Figure 3D). The increase was not detected in total RNA samples (input RNA in Figure 3D), showing that it was present in the nascent RNA fraction only. As control, we determined the levels of the matured mRNA further downstream, in a region spanning exons 10–12. The findings demonstrated that overall transcript levels were not affected by CPT (Figure 3D), even though the drug markedly altered the splicing at this site as shown by the presence of two additional bands (Figure 3D). Again, the splicing alteration was readily observed in chromatin- and Pol II-associated RNA fractions, but not in input RNAs (Figure 3D).
To evaluate the generality of CPT effects on Pol II escape from pausing, we also determined transcription levels downstream to the c-Myc gene P2 promoter (Figure 4A). The P2 promoter is a well-known elongation pausing site where Pol II density is higher than other gene regions (4). Consistent with above data (Figure 2), chromatin-associated RNA levels were about 10-fold lower in intron 1 than exon 1 (data not shown). Similarly to HIF-1α promoter, 1-h CPT treatments increased transcription downstream to the c-Myc P2 promoter, and DRB fully abolished the CPT effects (Figure 4B). Increased transcription in RIP experiments was detected both with Abs against Ac-H4 histone and against Pol II large subunit (Figure 4B and C). Thus, CPT-induced escape of Pol II was not specific for the HIF-1α promoter, and may rather constitute a specific response to CPT interference with transcription regulation at several genes.
Figure 4.
CPT-induced Top1ccs promote Pol II escape from the c-Myc P2 promoter. (A) Diagram of the studied c-Myc exon 1 region, with lines indicating the studied amplicons. Numbers indicate positions from P2 transcription start. (B) CPT-induced transcription downstream to the P2 pause site is dependent on Cdk activity. HCT116 cells were treated for 1 h with CPT and/or DRB. RIP was performed with Ab against acH4 histone (Ac-H4) or Pol II (Pol II). The gel shows PCR products of RIP samples. Controls are: genomic DNA (G), no reverse transcription (–RT) and water. α-satellite DNA (α-sat) reactions show no genomic DNA contamination. A representative experiment is reported. (C) RIP was performed with Ab against Pol II. All values are means ± S.D. of four determinations from two independent experiments.
Our findings thus demonstrated that Top1cc can specifically increase transcription elongation downstream to the promoters of the HIF-1α and c-Myc genes. Similar CPT-induced increases of 5′-end transcription in RIP experiments were reported for other human genes (28), but the findings were not compared to the levels of Pol II bound to their promoters. Our results also showed marked splicing alterations in nascent mRNAs, demonstrating that Top1cc can affect co-transcriptionally the maturation of HIF-1α mRNA.
Top1cc increases nascent transcripts in HIF-1α intergenic and intron 1 regions in a manner dependent on cyclin-dependent kinase activity
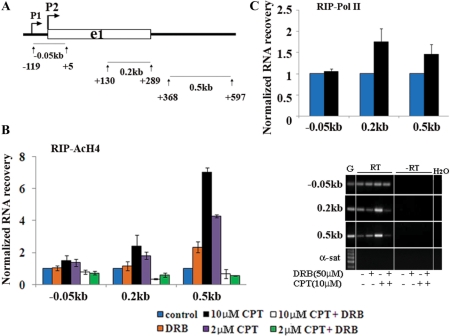
CPT-induced Top1cc can therefore affect the transition of Pol II to the elongation phase by enhancing Pol II escape from the promoter-proximal pausing site. Pol II is known to pause often along transcribed genes (9) and, in particular, Pol II density has a peak at mRNA termination regions suggesting a pausing site (29). Thus, we next determined the levels of nascent RNAs at internal regions and at 5′- and 3′-adjacent regions of the HIF-1α gene to investigate whether Top1cc may also alter Pol II pausing regulation in the body of the gene. First, we noticed that the RNA levels of intergenic regions upstream to the promoter and downstream to the mRNA termination site, were not at background levels, but rather they were very similar to intron 1 levels (Figure 2A). Surprisingly, the recovery of RNAs corresponding to 5′- and 3′-adjacent regions were increased by 1-h treatments with CPT from 0.4 to 10 µM (Figure 5A and B). We also found that CPT increased the levels of RNAs corresponding to the 5′-end of the first intron of HIF-1α gene, whereas the 3′-end of the first intron and other internal gene regions were not affected (data not shown). In addition, DRB fully abolished the increase of nascent RNAs at the HIF-1α gene locus (Figure 5C), showing that the CPT-induced increase of nascent RNA levels was dependent on Cdk activity.
Figure 5.
Levels of specific nascent RNAs are increased by CPT-induced Top1ccs. HCT116 cells were treated for 1 h with CPT and/or DRB as indicated. Immunoprecipitated RNA was retrotranscribed with random primers, and measured with qRT-PCR. Levels of nascent RNAs at HIF-1α gene locus by using Abs anti Pol II (Pol II) are shown in (A) or anti-acetylated histone H4 (Ac-H4) in (B). (C) Activation of nascent RNAs is dependent on cdk activity as shown by RIP experiments performed with anti Ac-H4 Abs. On the right of (A) and (C), a representative gel of PCR products. Samples are: genomic DNA (G), RIP samples (RT). –RT indicates no reverse transcription. α-sat amplicon shows no genomic DNA contamination. In all panels, values are normalized to exon 2 (25 kb) and control samples, and are means ± SD of four to six determinations from two independent experiments.
Observations such as the increased nascent RNAs upstream to the mRNA promoter cannot be easily explained by alterations of Pol II pausing. Moreover, the increase at the 3′-end adjacent region of HIF-1α mRNA was higher than at other regions (Figure 5B), and could be due to CPT-induced alterations of mRNA termination (30). However, we did not detect CPT-induced increased RNA levels at this region when using Abs against Pol II (Figure 5A) suggesting that the nascent RNA was not associated with Pol II. Therefore, perturbation of transcription termination was not the main reason for the increase. As an antisense transcript (3′aHIF-1α) was known to be present at the 3′-end of the HIF-1α gene locus (31), the results suggested that CPT may increase the levels of chromatin-associated 3′aHIF-1α RNA, and that the transcript was not transcribed by Pol II. Thus, the findings raised the unexpected possibility that Top1ccs induced by CPT could increase the transcription of antisense, low-abundance transcripts.
Top1 inhibition by CPT increases the levels of a novel antisense RNA at the 5′-end of the HIF-1α gene locus
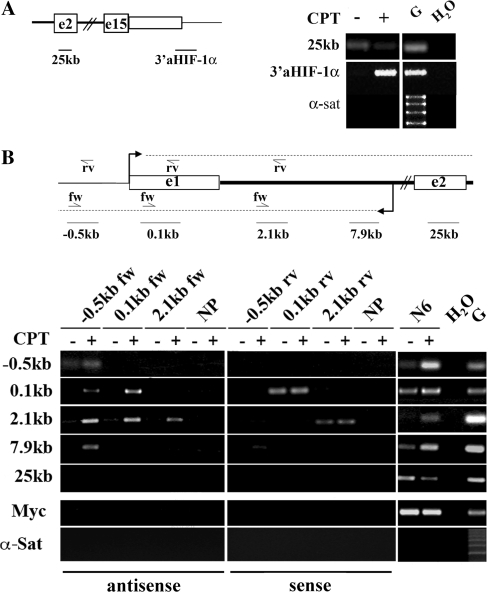
To assess whether CPT induced the transcription of the 3′aHIF-1α antisense RNA, we performed PCR analyses with specific primers in total cellular RNAs following CPT treatments of HCT116 cells. We found that the antisense transcript was indeed increased after 4 h of CPT, while HIF-1α mRNA levels were reduced (Figure 5A), showing that Top1cc induced by CPT can increase the levels of the 3′aHIF-1α antisense RNA. This unexpected finding prompted us to investigate whether CPT could also activate antisense transcription at the 5′-end of the HIF-1α locus. cDNA prepared with strand-specific primers (Figure 5B) demonstrated that antisense transcripts were induced upon 4 h of CPT treatments spanning from an intronic region at about 8 kb from the mRNA start site to a region more than 0.5 kb upstream to the mRNA start (Figure 6B). PCR analyses in the second half of the first introns did not detect antisense transcription (data not shown). Strand-specific cDNAs also showed that the sense pre-mRNA, as detected at the exon 1 and an intron 1 region, was not increased by CPT (Figure 6B). No sense transcription was detected at a region 500 bp upstream to the mRNA promoter (Figure 6B).
Figure 6.
Antisense transcription at the HIF-1α gene locus is increased by Top1cc. Total RNA was extracted from cells untreated (−) or treated with 10 µM CPT for 4 h (+). (A) PCR analysis was performed with primers as indicated in the map. (B) Upper panel: map of the HIF-1α gene region analyzed (not to scale). Dashed lines show the antisense transcript detected at the 5′-end and the mRNA of the HIF-1α gene. Arrows indicate specific primers used for retrotranscription: rv primers target the mRNA, and fw primers target antisense transcripts. Black lines and numbers indicate regions analyzed by PCR. Lower panel: PCR analyses on primer-specific cDNAs from HCT116 cells. Primers used for retrotranscription are indicated on top of the gel, and amplicons used for PCR analyses are indicated on the left of the gel. Negative and positive controls of retrotranscription were no primers (NP) and random primers (N6), respectively. Negative and positive controls of PCR were water (H2O) and genomic DNA (G), respectively. α-satellite DNA (α-sat) shows no genomic DNA contamination.
The antisense RNA was then confirmed with Northern blots by using an oligonucleotide corresponding to the 2.1-kb amplicon of intron 1 (Figure 6B) as strand-specific probe. The novel antisense transcript (5′aHIF1α) is 12 to 10 kb long and increased over 4 h of CPT treatments (Figure 7A). No transcript was detected with an oligonucleotide probe complementary to sense RNAs corresponding to the same intronic region (data not shown). We detected the 5′aHIF1α transcript both under hypoxic and normoxic conditions in a different cell line, the leukemic human Jurkat cells (Figure 7B), while, in agreement with established knowledge (3), CPT decreased the mRNA levels of HIF-1α mRNA under similar conditions (Figure 7A).
Figure 7.
CPT activation of antisense transcription requires Top1 and is independent from replicative DNA damage. (A) Left panel: levels of 5′aHIF1α antisense RNA in cells treated with 10 μM CPT for the indicated time by northern blots. The probe was a 40-nt oligomer corresponding to the 2 kb amplicon and complementary to antisense RNA. Right panel: levels of HIF-1α mRNA in Jurkat cells treated with 10 μM CPT by northern blots of total RNAs with an exon 2 probe. Agarose staining was to check for equal RNA loading and integrity. (B) Increase of the 5′aHIF-1α transcript (2.1 kb amplicon) in Jurkat cells treated with 10 µM CPT in normoxia (20%) or hypoxia (1%). No DNA contamination was detected as established with α-sat. A representative experiment is shown. (C) Total RNA was extracted from untreated cells or cells treated with 10 µM CPT for 120 min with or without aphidicolin (APH) or caffeine (Caff). Left and right panels correspond to HCT116 and HCT116(top1-siRNA) cells, respectively. In all graphs, RNA levels were normalized to mRNA exon 2 (25 kb) and untreated samples, and the values are means ± SD of four determinations from two independent experiments. (D) Western blots of p53 and γ-H2AX after CPT treatments of HCT116 cells with or without Caff or APH. Total cellular proteins (upper two panels) or histones (lower panels) were analyzed by western blots. CHK2 and H1 histone were used as loading controls.
Since collisions between Top1ccs and advancing DNA polymerases can lead to irreversible DNA breaks at replication forks (3), we investigated whether the activation of antisense transcription was dependent on Top1 and/or replicative DNA damage. CPT stimulation of 5′aHIF1α RNA was dependent on Top1, as it was not significantly increased in HCT116(top1 siRNA) cells (Figure 7A). Moreover, the drug effect was independent from replication-mediated DNA breaks and checkpoint activation, as prevention of replicative DNA damage by aphidicolin, a DNA polymerase inhibitor, and of checkpoint activation by caffeine, an inhibitor of mammalian phosphatidylinositol 3-kinase family members (including ATM and ATR), did not affect antisense RNA activation by Top1ccs (Figure 7C and D). Thus, CPT-induced Top1cc increases the cellular levels of low-abundance antisense transcripts (5′aHIF1α and 3′aHIF1α) while reducing the mRNA of the human HIF-1α gene.
Top1 inhibition by CPT induces a more accessible chromatin structure at later time
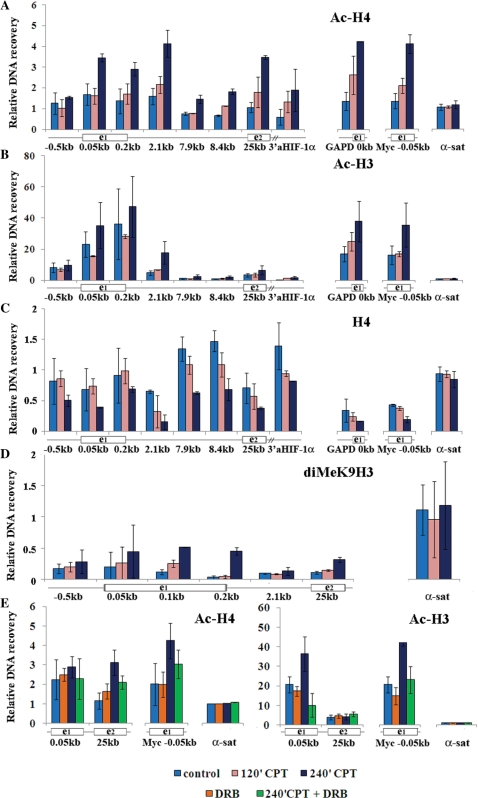
The present findings showed that Top1 inhibition by CPT can interfere with specific transcription regulation mechanisms involving Cdk activity and de-regulated Pol II pausing at promoter-proximal sites leading to the activation of antisense RNAs at the HIF-1α gene locus. As antisense transcripts have been shown to regulate chromatin structure, we next asked whether histone modifications were also affected by Top1cc. As we did not detected any change of histone H4 acetylation after 1 h of drug treatments (Figure 2), we determined core histone acetylation levels at later time points (2 and 4 h) by ChIP in HCT116 cells (Figure 8). We found that histone H4 acetylation at lysines 5, 8, 12 and 16 was increased at promoter and internal gene regions after 4 h, whereas it was substantially unmodified upstream to the HIF-1α mRNA start site, and in satellite DNA (Figure 8A). Interestingly, total H4 histone density had a tendency to be reduced by CPT at transcribed regions of the studied genes (Figure 8C). H3 histone acetylation was also increased, but such increases were restricted to promoter regions (Figure 8B). As antisense transcripts have been suggested to be a trigger for heterochromatin formation and gene repression (32,33), we determined the levels of histone H3K9 dimethylation, a marker of heterochromatin, in cells treated with CPT for 4 h. We found that H3K9 dimethylation levels were very low in the studied HIF-1a gene regions in untreated cells and were not affected by CPT (Figure 8D). Co-treatments with DRB for 4 h suppressed the increase of histone acetylation by CPT (Figure 8E). Thus, the findings showed that CPT also affected histone modifications at the HIF-1α gene locus, but at later time points, promoting a more accessible chromatin structure in a manner dependent on Cdk activity.
Figure 8.
Late effects of CPT on chromatin structure. (A) and (B) HCT116 cells were treated with 10 µM CPT for 2 and 4 h. ChIP was with Abs against acetyl -K5,-K8,-K12 and K16 of histone H4 or acetyl –K9 and –K14 of histone H3. Histone acetylation levels were determined along the HIF-1α gene, GAPD promoter (GAPD 0 kb) and c-Myc P2 promoter (Myc −0.05 kb). Mean DNA recovery with non-immune IgG was 0.41. Values are means ± SD of four to eight determinations from two to four independent experiments. (C) Levels of total histone H4 bound to chromatin. (D) Levels of dimetil -K9 of histone H3 after CPT treatments. A representative ChIP experiment is shown, and values are means ± SD of two to three determinations. Mean DNA recovery with non-immune IgG is 0.59 (E) HCT116 cells were treated with CPT and/or DRB for 4 h. Values are means ± SD of four to six determinations of two independent experiments. In all panels, the values are normalized to the recoveries of α-sat DNA, as the internal standard, and to that of total histone H4 (shown in C).
DISCUSSION
Even though CPT is commonly considered an efficient inhibitor of transcription elongation, the molecular mechanism has not been established fully. Here, we have demonstrated that Top1cc has unexpected transcriptional consequences: (i) increased Pol II escape from promoter-proximal pausing sites of HIF-1α and c-Myc; (ii) increase in non-coding antisense RNAs at the HIF-1α locus; (iii) increase in chromatin accessibility; and (iv) marked alteration of alternative splicing of HIF-1α mRNA. The events require Top1 and are independent from replication and replicative DNA damage. Interestingly, they are not equal in time, as, for instance, (i) and (iv) occur earlier while (iii) occurs later during drug treatments. Remarkably, our findings also showed that inhibition of Cdk9 and Cdk7 activity by DRB can suppress the CPT-induced Pol II escape from promoter-proximal pausing sites, activation of antisense transcription, and increased chromatin accessibility. Even though we show evidence that CPT may affect several genes, the generality of the drug effects will definitively be addressed with a genome-wide approach.
Previous reports showed that CPT promotes the complete hyperphosphorylation of the Rpb1 of Pol II in both primary and transformed cancer cells likely through Cdk7 and/or Cdk9 activity (4,13–15). Thus, we propose that Top1cc can increase the activity of Cdks that phosphorylate Rpb1 promoting transcription elongation at promoter-proximal pausing sites (Figure 9). Hyperphosphorylated Pol II is not competent for recruitment to promoters leading to a reduction of Pol II density at promoter regions of HIF-1α, c-Myc (Figure 1) and other genes (4). Interestingly, Pol II pausing at promoters has recently been shown to favor the recruitment of further Pol II at promoters (25), therefore Top1cc-induced Pol II escape from the pause site may interfere with this mechanism. Reduction of Pol II loading at the promoter will eventually lead to decreased gene transcription.
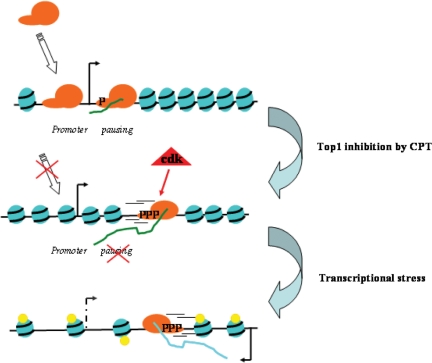
Figure 9.
A model of the molecular mechanism triggered by Top1ccs induced by CPT at transcribed genes. Arrows indicate transcription start sites and direction. The blue spheres are nucleosomes and small yellow spheres are acetylation marks. The orange object is Pol II. Green and light blue lines are sense and antisense transcripts, respectively.
A kinetic analysis of Pol II transcription at a gene-array locus showed that transcription can be inefficient and that Pol II often pauses during elongation (9). Interestingly, while leaving active the entire population of Pol IIs, CPT increased the intragenic pausing efficiency but not the pause time (9), resulting in a reduction of the elongation rate to a one-fourth of the normal rate. CPT does not completely block transcription at the studied gene array, in contrast to other inhibitors such as DRB (9). This is consistent with our data showing that (i) CPT does not destroy transcription factories under conditions in which it destroys replication factories (4,34); (ii) 1-h CPT treatments do not reduce the overall levels of chromatin-associated RNAs along the HIF-1α gene locus (Figure 2); and (iii) CPT readily alters alternative splicing at exon 10–12 site of HIF-1α, which implies that transcription elongation is not abolished by CPT.
A striking finding of the present report is indeed the demonstration that CPT-induced Top1ccs can affect alternative splicing of HIF-1α mRNA co-transcriptionally (Figure 3D). Recently, P-TEFb has been proposed to play a critical role in coupling transcription elongation with alternative splicing (35). Moreover, UV-induced DNA damage has been shown to alter alternative splicing at several genes in human cells by increasing the phosphorylation of Rpb1 of Pol II likely mediated by P-TEFb (36). Two major mechanisms, not mutually exclusive, may allow the coupling of transcription and alternative splicing: (i) recruitment of splicing factors to the transcribing Pol II; and (ii) kinetic coupling involving Pol II elongation rate (37). It has been shown that UV-induced hyperphosphorylation of Pol II may cause a lower elongation rate of Pol II, which may then affect alternative splicing (36). As Top1cc induced by CPT readily triggers Rpb1 hyperphosphorylation (4,13–15) and co-transcriptional alterations of HIF-1α alternative splicing, our findings suggest that Top1ccs may influence the elongation rate of Pol II with a similar mechanism as proposed for UV-induced DNA damage (36).
Since increased Pol II escape and reduced elongation rate are earlier CPT effects, both of which may likely be dependent on Cdk activation and Rpb1 hyperphosphorylation (Figures 3 and 4) (4,9), we propose that a sustained CPT interference with Pol II regulation may then lead to a more general disturbance of transcription regulation (transcriptional stress) involving impaired chromatin structure and activation/de-repression of antisense transcription (Figure 9).
CPT-induced Pol II escape, cdk activation and transcriptional stress can constitute a response to drug-promoted Top1-mediated DNA breaks or to changed duplex superhelicity due to inhibition of Top1 activity. Interestingly, recent studies have shown that, when CPT freezes a Top1cc, the enzyme catalytic cycle is hindered with a more pronounced effect on the removal of positive supercoils (38). Consistently, the drug action results in the inhibition of enzyme catalytic activity leading to a marked torsional stress of the DNA template in living cells (38,39). Upon siRNA knockdown of Top1 no effect on Pol II density was detected (Figure 1), however Top1 is not completely eliminated from the studied cells. Moreover, these cells are adapted to reduced Top1, therefore the enzyme activity may be replaced by Top2 (2)
Local DNA torsional stress can significantly regulate gene expression in mammalian cells. Localized DNA superhelicity can regulate the transcription of the human c-Myc gene (40), and recently, the widespread divergent transcription at protein-coding gene promoters has been proposed to be determined by increased negative supercoils generated by active transcription of mRNAs (41). Therefore, Top1 inhibition by CPT may promote a supercoiling imbalance locally at promoters, which may then interfere with the regulation of Pol II pausing at promoter-proximal sites. The precise mechanism remains to be established, however one attractive hypothesis is that Cdk activity is coupled to torsional DNA tension and/or nucleosome regulation. Thus, in addition to the well known effects on DNA replication and DNA damage checkpoints, CPT can affect transcription regulation leading to alterations of gene expression patterns, which may be relevant for cancer therapy, particularly at low drug concentrations.
A main finding of the present report is that CPT can lead to an impaired balance of cellular antisense and sense transcripts which may influence the cancer-related HIF-1 pathway. HIF-1 is a heterodimer constituted by HIF-1α or HIF-2α subunits, and a constitutively-expressed HIF-1β subunit. The HIF-α subunits are regulated at high oxygen tensions by the VHL pathway (17). Even though the mechanism of CPT-induced reduction of HIF-1α protein levels (18,19) remains to be defined, our findings show that the human HIF-1α gene locus is complex as at least two non-coding RNAs are present in the antisense orientation relative to the mRNA. An attractive hypothesis is that they may have a role in novel mechanisms of transcriptional and/or post-transcriptional regulation of HIF-1α activity (42,43), with an impact for cancer prognosis and the development of new therapy for human tumors.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Associazione Italiana per la Ricerca sul Cancro [IG 4494 to G.C.]. Ministero dell’Università e della Ricerca (PRIN program) [to G.C.]. University of Bologna PhD Program in Functional Biology of Molecular and Cellular Systems [to L.B. and D.B.]. Funding for open access charge: Associazione Italiana per la Ricerca sul Cancro, Milan, Italy [IG 4494 to G.C.].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank V. Scarlato (University of Bologna) for helpful discussions.
REFERENCES
- 1.Champoux JJ. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 2.Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell Biol. 2002;3:430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 3.Pommier Y. Topoisomerase I inhibitors: camptothecins and beyond. Nat. Rev. Cancer. 2006;6:789–802. doi: 10.1038/nrc1977. [DOI] [PubMed] [Google Scholar]
- 4.Khobta A, Ferri F, Lotito L, Montecucco A, Rossi R, Capranico G. Early effects of topoisomerase I inhibition on RNA polymerase II along transcribed genes in human cells. J. Mol. Biol. 2006;357:127–138. doi: 10.1016/j.jmb.2005.12.069. [DOI] [PubMed] [Google Scholar]
- 5.Merino A, Madden KR, Lane WS, Champoux JJ, Reinberg D. DNA topoisomerase I is involved in both repression and activation of transcription. Nature. 1993;365:227–232. doi: 10.1038/365227a0. [DOI] [PubMed] [Google Scholar]
- 6.Kretzschmar M, Meisterernst M, Roeder RG. Identification of human DNA topoisomerase I as a cofactor for activator-dependent transcription by RNA polymerase II. Proc. Natl Acad. Sci. USA. 1993;90:11508–11512. doi: 10.1073/pnas.90.24.11508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shykind BM, Kim J, Stewart L, Champoux JJ, Sharp PA. Topoisomerase I enhances TFIID-TFIIA complex assembly during activation of transcription. Genes Dev. 1997;11:397–407. doi: 10.1101/gad.11.3.397. [DOI] [PubMed] [Google Scholar]
- 8.Wu J, Liu LF. Processing of topoisomerase I cleavable complexes into DNA damage by transcription. Nucleic Acids Res. 1997;25:4181–4186. doi: 10.1093/nar/25.21.4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Darzacq X, Shav-Tal Y, de Turris V, Brody Y, Shenoy SM, Phair RD, Singer RH. In vivo dynamics of RNA polymerase II transcription. Nat. Struct. Mol. Biol. 2007;14:796–806. doi: 10.1038/nsmb1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ljungman M, Hanawalt PC. The anti-cancer drug camptothecin inhibits elongation but stimulates initiation of RNA polymerase II transcription. Carcinogenesis. 1996;17:31–35. doi: 10.1093/carcin/17.1.31. [DOI] [PubMed] [Google Scholar]
- 11.Collins I, Weber A, Levens D. Transcriptional consequences of topoisomerase inhibition. Mol. Cell Biol. 2001;21:8437–8451. doi: 10.1128/MCB.21.24.8437-8451.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lotito L, Russo A, Chillemi G, Bueno S, Cavalieri D, Capranico G. Global transcription regulation by DNA topoisomerase I in exponentially growing Saccharomyces cerevisiae cells: activation of telomere-proximal genes by TOP1 deletion. J. Mol. Biol. 2008;377:311–322. doi: 10.1016/j.jmb.2008.01.037. [DOI] [PubMed] [Google Scholar]
- 13.Sordet O, Larochelle S, Nicolas E, Stevens EV, Zhang C, Shokat KM, Fisher RP, Pommier Y. Hyperphosphorylation of RNA polymerase II in response to topoisomerase I cleavage complexes and its association with transcription- and BRCA1-dependent degradation of topoisomerase I. J. Mol. Biol. 2008;381:540–549. doi: 10.1016/j.jmb.2008.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Desai SD, Zhang H, Rodriguez-Bauman A, Yang JM, Wu X, Gounder MK, Rubin EH, Liu LF. Transcription-dependent degradation of topoisomerase I-DNA covalent complexes. Mol. Cell Biol. 2003;23:2341–2350. doi: 10.1128/MCB.23.7.2341-2350.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Amente S, Gargano B, Napolitano G, Lania L, Majello B. Camptothecin releases P-TEFb from the inactive 7SK snRNP complex. Cell Cycle. 2009;8:1249–1255. doi: 10.4161/cc.8.8.8286. [DOI] [PubMed] [Google Scholar]
- 16.Iyer NV, Kotch LE, Agani F, Leung SW, Laughner E, Wenger RH, Gassmann M, Gearhart JD, Lawler AM, Yu AY, et al. Cellular and developmental control of O2 homeostasis by hypoxia-inducible factor 1 alpha. Genes Dev. 1998;12:149–162. doi: 10.1101/gad.12.2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Semenza GL. Targeting HIF-1 for cancer therapy. Nat. Rev. Cancer. 2003;3:721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 18.Rapisarda A, Uranchimeg B, Sordet O, Pommier Y, Shoemaker RH, Melillo G. Topoisomerase I-mediated inhibition of hypoxia-inducible factor 1: mechanism and therapeutic implications. Cancer Res. 2004;64:1475–1482. doi: 10.1158/0008-5472.can-03-3139. [DOI] [PubMed] [Google Scholar]
- 19.Rapisarda A, Zalek J, Hollingshead M, Braunschweig T, Uranchimeg B, Bonomi CA, Borgel SD, Carter JP, Hewitt SM, Shoemaker RH, et al. Schedule-dependent inhibition of hypoxia-inducible factor-1alpha protein accumulation, angiogenesis, and tumor growth by topotecan in U251-HRE glioblastoma xenografts. Cancer Res. 2004;64:6845–6848. doi: 10.1158/0008-5472.CAN-04-2116. [DOI] [PubMed] [Google Scholar]
- 20.Miao ZH, Player A, Shankavaram U, Wang YH, Zimonjic DB, Lorenzi PL, Liao ZY, Liu H, Shimura T, Zhang HL, et al. Nonclassic functions of human topoisomerase I: genome-wide and pharmacologic analyses. Cancer Res. 2007;67:8752–8761. doi: 10.1158/0008-5472.CAN-06-4554. [DOI] [PubMed] [Google Scholar]
- 21.Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 22.Zorbas H, Keppler BK. Cisplatin damage: are DNA repair proteins saviors or traitors to the cell? Chembiochem. 2005;6:1157–1166. doi: 10.1002/cbic.200400427. [DOI] [PubMed] [Google Scholar]
- 23.Capranico G, Binaschi M, Borgnetto ME, Zunino F, Palumbo M. A protein-mediated mechanism for the DNA sequence-specific action of topoisomerase II poisons. Trends Pharmacol. Sci. 1997;18:323–329. doi: 10.1016/s0165-6147(97)01095-x. [DOI] [PubMed] [Google Scholar]
- 24.Core LJ, Lis JT. Transcription regulation through promoter-proximal pausing of RNA polymerase II. Science. 2008;319:1791–1792. doi: 10.1126/science.1150843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gilchrist DA, Nechaev S, Lee C, Ghosh SK, Collins JB, Li L, Gilmour DS, Adelman K. NELF-mediated stalling of Pol II can enhance gene expression by blocking promoter-proximal nucleosome assembly. Genes Dev. 2008;22:1921–1933. doi: 10.1101/gad.1643208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sims RJ, III, Belotserkovskaya R, Reinberg D. Elongation by RNA polymerase II: the short and long of it. Genes Dev. 2004;18:2437–2468. doi: 10.1101/gad.1235904. [DOI] [PubMed] [Google Scholar]
- 27.Gilbert C, Kristjuhan A, Winkler GS, Svejstrup JQ. Elongator interactions with nascent mRNA revealed by RNA immunoprecipitation. Mol. Cell. 2004;14:457–464. doi: 10.1016/s1097-2765(04)00239-4. [DOI] [PubMed] [Google Scholar]
- 28.Listerman I, Sapra AK, Neugebauer KM. Cotranscriptional coupling of splicing factor recruitment and precursor messenger RNA splicing in mammalian cells. Nat. Struct. Mol. Biol. 2006;13:815–822. doi: 10.1038/nsmb1135. [DOI] [PubMed] [Google Scholar]
- 29.Glover-Cutter K, Kim S, Espinosa J, Bentley DL. RNA polymerase II pauses and associates with pre-mRNA processing factors at both ends of genes. Nat. Struct. Mol. Biol. 2008;15:71–78. doi: 10.1038/nsmb1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maniatis T, Reed R. An extensive network of coupling among gene expression machines. Nature. 2002;416:499–506. doi: 10.1038/416499a. [DOI] [PubMed] [Google Scholar]
- 31.Thrash-Bingham CA, Tartof KD. aHIF: a natural antisense transcript overexpressed in human renal cancer and during hypoxia. J. Natl Cancer Inst. 1999;91:143–151. doi: 10.1093/jnci/91.2.143. [DOI] [PubMed] [Google Scholar]
- 32.Grewal SI, Moazed D. Heterochromatin and epigenetic control of gene expression. Science. 2003;301:798–802. doi: 10.1126/science.1086887. [DOI] [PubMed] [Google Scholar]
- 33.Yu W, Gius D, Onyango P, Muldoon-Jacobs K, Karp J, Feinberg AP, Cui H. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202–206. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Capranico G, Ferri F, Fogli MV, Russo A, Lotito L, Baranello L. The effects of camptothecin on RNA polymerase II transcription: roles of DNA topoisomerase I. Biochimie. 2007;89:482–489. doi: 10.1016/j.biochi.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 35.Barboric M, Lenasi T, Chen H, Johansen EB, Guo S, Peterlin BM. 7SK snRNP/P-TEFb couples transcription elongation with alternative splicing and is essential for vertebrate development. Proc. Natl Acad. Sci. USA. 2009;106:7798–7803. doi: 10.1073/pnas.0903188106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Munoz MJ, Perez Santangelo MS, Paronetto MP, de la Mata M, Pelisch F, Boireau S, Glover-Cutter K, Ben-Dov C, Blaustein M, Lozano JJ, et al. DNA damage regulates alternative splicing through inhibition of RNA polymerase II elongation. Cell. 2009;137:708–720. doi: 10.1016/j.cell.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 37.Kornblihtt AR. Coupling transcription and alternative splicing. Adv. Exp. Med. Biol. 2007;623:175–189. doi: 10.1007/978-0-387-77374-2_11. [DOI] [PubMed] [Google Scholar]
- 38.Koster DA, Palle K, Bot ES, Bjornsti MA, Dekker NH. Antitumour drugs impede DNA uncoiling by topoisomerase I. Nature. 2007;448:213–217. doi: 10.1038/nature05938. [DOI] [PubMed] [Google Scholar]
- 39.Duann P, Sun M, Lin CT, Zhang H, Liu LF. Plasmid linking number change induced by topoisomerase I-mediated DNA damage. Nucleic Acids Res. 1999;27:2905–2911. doi: 10.1093/nar/27.14.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kouzine F, Sanford S, Elisha-Feil Z, Levens D. The functional response of upstream DNA to dynamic supercoiling in vivo. Nat. Struct. Mol. Biol. 2008;15:146–154. doi: 10.1038/nsmb.1372. [DOI] [PubMed] [Google Scholar]
- 41.Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, Young RA, Sharp PA. Divergent transcription from active promoters. Science. 2008;322:1849–1851. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lapidot M, Pilpel Y. Genome-wide natural antisense transcription: coupling its regulation to its different regulatory mechanisms. EMBO Rep. 2006;7:1216–1222. doi: 10.1038/sj.embor.7400857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kapranov P, Willingham AT, Gingeras TR. Genome-wide transcription and the implications for genomic organization. Nat. Rev. Genet. 2007;8:413–423. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.