Abstract
The RNA-binding protein nuclear factor 90 (NF90) has been implicated in the stabilization, transport and translational control of several target mRNAs. However, a systematic analysis of NF90 target mRNAs has not been performed. Here, we use ribonucleoprotein immunoprecipitation analysis to identify a large subset of NF90-associated mRNAs. Comparison of the 3′-untranslated regions (UTRs) of these mRNAs led to the elucidation of a 25- to 30-nucleotide, RNA signature motif rich in adenines and uracils. Insertion of the AU-rich NF90 motif (‘NF90m’) in the 3′UTR of an EGFP heterologous reporter did not affect the steady-state level of the chimeric EGFP-NF90m mRNA or its cytosolic abundance. Instead, the translation of EGFP-NF90m mRNA was specifically repressed in an NF90-dependent manner, as determined by analysing nascent EGFP translation, the distribution of chimeric mRNAs on polysome gradients and the steady-state levels of expressed EGFP protein. The interaction of endogenous NF90 with target mRNAs was validated after testing both endogenous mRNAs and recombinant biotinylated transcripts containing NF90 motif hits. Further analysis showed that the stability of endogenous NF90 target mRNAs was not significantly influenced by NF90 abundance, while their translation increased when NF90 levels were reduced. In summary, we have identified an AU-rich RNA motif present in NF90 target mRNAs and have obtained evidence that NF90 represses the translation of this subset of mRNAs.
INTRODUCTION
In mammalian cells, gene expression is potently regulated at the post-transcriptional level. RNA-binding factors, including noncoding RNA and RNA-binding proteins (RBPs), influence many post-transcriptional processes, including pre-mRNA splicing and mRNA transport, degradation, storage and translation (1,2). Some RBPs affect one specific post-transcriptional process; for example, tristetraprolin (TTP) and KH-type splicing regulatory protein (KSRP) promote mRNA degradation (3–5). However, most RBPs perform multiple post-transcriptional functions. For example, HuR (human antigen R) stabilizes some target mRNAs but modulates the translation of other target mRNAs (6), AUF1 (AU-binding factor 1) promotes the decay of some mRNAs but can also stabilize and promote the translation of other target transcripts (7–11), the T-cell intracellular antigen-1 (TIA-1) and the TIA-1-related protein (TIAR) are implicated in splicing and translational repression of target transcripts (12–14) and the polypyrimidine tract-binding protein can modulate splicing, stability and translation of target mRNAs (15,16). In general, these RBPs associate with sequences within the 3′-untranslated region (UTR) of the target transcript, but sometimes with the 5′UTR or with the coding region (17,18, reviewed in 19).
One multi-functional RBP is the nuclear factor (NF)90, also named NFAR (nuclear factor associated dsRNA)-1, double-stranded (ds) RNA-binding protein (DRBP76) and interleukin (IL) enhancer-binding factor 3 (ILF3). NF90 was first identified as a 90-kDa protein that interacted with the NFAT (nuclear factor activated in T-cells) DNA site present in the IL-2 promoter (20,21). With two dsRNA-binding domains (DRBDs), NF90 was later shown to interact with the β-glucosidase mRNA (22,23) and viral RNA (24); it was also shown to bind the protein kinase activated by dsRNA (PKR) (25,26). Through alternative splicing, the gene that encodes NF90 also gives rise to NF110; both proteins are ubiquitously expressed and are primarily present in the nucleus, although they can be transported to the cytoplasm under specific conditions, such as during the cell division cycle and in response to damaging agents (27,28). Although NF110 has been investigated in some depth (29), NF90 has been studied in greater detail.
As mentioned earlier, NF90 can bind to the NFAT DNA sequence, and thereby modulates the transcription of IL-2 (20,30,31). However, NF90 potently regulates gene expression through its association with RNA. NF90 was shown to bind the several mRNAs containing AU-rich 3′UTRs; this interaction led to the stabilization of the IL-2 mRNA during T-cell activation, the mitogen-activated protein kinase phosphatase 1 (MKP-1) mRNA in response to oxidative damage, and the vascular endothelial growth factor mRNA in response to hypoxic stress (28,32,33). NF90 also stabilized the mRNAs encoding the cyclin-dependent kinase inhibitor p21Cip1 and the myogenic transcription factor MyoD (34); additionally, NF90 bound the NF90 mRNA and possibly affected its expression (35). In vitro, NF90 associated with the β-glucosidase mRNA and inhibited its translation (22,23). Pfeifer and colleagues (2008) recently showed that NF90 can also function as a general inhibitor of mRNA export to the cytoplasm; while this effect alone could block translation broadly, the authors proposed that NF90 could further prevent translation through its association with polysomes (36).
Our previous studies showing that NF90 selectively associated with the MKP-1 mRNA, together with the specific interaction of NF90 with other mRNAs (28,35), led us to postulate that NF90 may interact preferentially with specific RNA sequences. To test this possibility, we studied the collection of mRNAs that interacted with NF90 by performing ribonucleoprotein immunoprecipitation (RNP IP) analysis followed by microarray identification of bound mRNAs. Comparison of the interacting mRNAs led to the identification of a 25- to 30-nt-long, highly AU-rich motif sequence shared by NF90-associated mRNAs. The addition of the consensus motif to a heterologous GFP reporter revealed that NF90 selectively repressed the translation of a chimeric mRNA bearing this sequence but did not appear to influence the stability of the chimeric mRNA. NF90 was also found to repress the translation of endogenous mRNAs bearing the AU-rich signature sequence, without affecting their half-lives.
MATERIALS AND METHODS
Cell culture and transfection
HeLa cells were maintained in Dulbecco’s modified essential medium (Invitrogen), supplemented with 10% (v/v) bovine calf serum (Hyclone, Logan, UT, USA). For silencing NF90 expression, Oligofectamine (Invitrogen) was used to transfect cells with siRNAs. The NF90 siRNA was GCCCACCUUUGCUUUUUAU; it targetted NF90 at exon 18 and therefore selectively reduced NF90 and did not affect NF110 levels. AllStars negative control siRNA was from Qiagen.
Illumina oligonucleotide microarray analysis
The quality and quantity of RNA in the material obtained after IP reactions using either anti-NF90 or IgG antibodies were assessed using an Agilent 2100 bioanalyzer and the RNA 6000 nanochips. The RNA was used to generate biotin-labeled cRNA using the Illumina TotalPrep RNA Amplification Kit (Ambion; Austin, TX, USA cat # IL1791), which was then hybridized to Illumina's; Sentrix HumanRef-8 Expression BeadChips (Illumina, San Diego, CA, USA), containing 24 000 well-annotated RefSeq transcripts with ∼30-fold redundancy. The arrays were scanned using an Illumina BeadStation 500X Genetic Analysis Systems scanner and the image data extracted using Illumina BeadStudio software, version 1.5. The data were normalized by Z-score transformation and used to calculate differences in signal intensities. Significant values were calculated from three independent experiments, using a two-tailed Z-test and P < 0.01. The complete list of NF90-enriched mRNAs identified on arrays is shown in Supplementary Table S1.
Computational analysis
The top ∼800 human transcripts enriched in NF90 IP served as the experimental dataset (Supplementary Table S1) for the computational identification of the NF90 motif. The transcript sequences (UniGene) were scanned with RepeatMasker (www.repeatmasker.org) to remove repetitive sequences, and complete, high-quality 3′UTR sequences were selected for further analysis. The selected sequences were first divided into 100-base-long subsequences with a 50-base overlap between consecutive sequences and were organized into 50 datasets. Common RNA motifs were then elucidated from each of the 50 random datasets. The top 10 candidate motifs from each random dataset were selected and used to build the stochastic context-free grammar (SCFG) model, which summarizes the folding, pairing and additional secondary structure features. The SCFG model of each candidate motif was then used to search against the experimental 3′UTR dataset as well as the entire human UniGene 3′UTR dataset to obtain the number of hits for each motif. The motif with the highest enrichment in the experimental dataset compared with the entire UniGene dataset was considered to be the top NF90 candidate motif. The enrichment was examined by Fisher’s exact test. The identified RNA motif for NF90 forms a stem-loop.
The identification of the RNA motif in unaligned sequences was conducted using FOLDALIGN software (37), and the identified motif was modeled by the SCFG algorithm and searched against the transcript dataset using the COVE and COVELS software packages (38). The motif logo was constructed using WebLogo (http://weblogo.berkeley.edu/). RNAplot was used to depict the secondary structure of the representative RNA motifs. The computation was performed using the NIH Biowulf computer farm. Both UniGene and RefSeq datasets were downloaded from NCBI.
Cell fractionation, RNA purification and western blot analysis
Cells were incubated on ice for 5 min in a cytoplasmic lysis (RSB) buffer containing 10 mM Tris–HCl (pH 7.4), 100 mM NaCl, 2.5 mM MgCl2, 40 µg/ml digitonin, 1000 U/ml of RNaseOUT and protease inhibitors, and then centrifuged (2000×g) for 8 min. After removing the supernatant (cytoplasmic fraction), the pellet was resuspended in the 200 μl RSB buffer + 40 µg/ml digitonin (containing 1000 U/ml RNaseOUT and protease inhibitors), kept on ice for 5 min, and centrifuged (2000×g) for 8 min; this step was repeated twice, and then the supernatant (the cytosolic fraction) was removed. The pellet was incubated on ice for 10 min in the presence of 100 µl of the RIPA buffer and was centrifuged (20 800×g). The supernatant (nuclear fraction) was collected for analysis.
For western blot analysis, whole-cell lysates (10 μg) were resolved by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto polyvinylidene difluoride (PVDF) membranes. Primary antibody incubations were performed using antibodies that recognized NF90 (mouse monoclonal anti-DRBP76, BD Transduction Laboratories), GAPDH, EGFP, α-tubulin, HuR, cyclin A, cdc2 (mouse monoclonal, Santa Cruz Biotech.), cyclin I (rabbit polyclonal, SantaCruz Biotech.), eIF4E (rabbit polyclonal, Cell Signaling) and β-actin (mouse monoclonal, Abcam). Following incubation with the appropriate secondary antibodies, the signals were detected with the ECL™ reagent (Amersham Biosciences).
Binding assays: IP and biotin pulldown
IP of endogenous NF90-mRNA complexes was carried out using previously described methods (39–41). Briefly, 20 million HeLa cells per sample were lysed and used for IP [1 h at 4°C in the presence of 30 µg of either an anti-NF90 antibody (mouse monoclonal, BD Transduction Laboratories) or control mouse IgG (Santa Cruz Biotech.]. RNA was isolated using acid phenol-chloroform (Ambion).
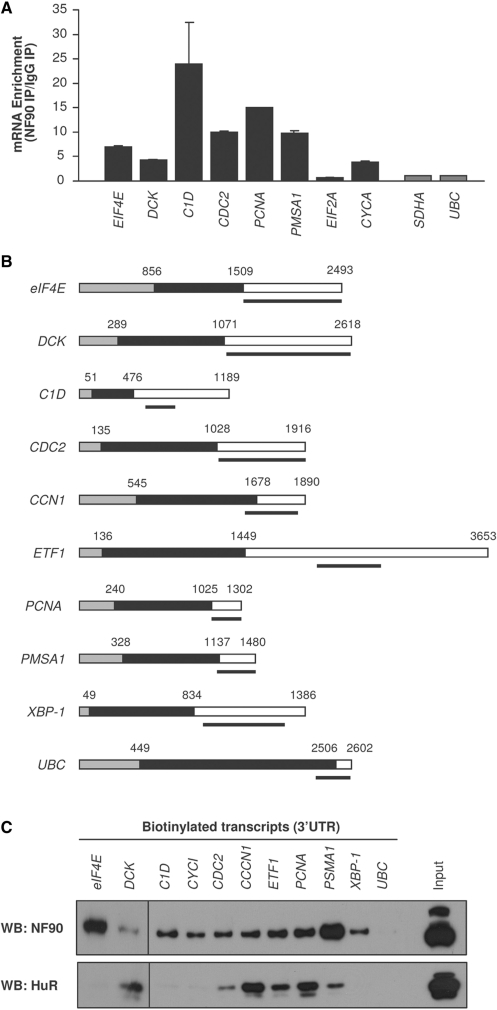
For biotin pulldown assays, PCR fragments containing the T7 RNA polymerase promoter sequence (T7): CCAAGCTTCTAATACGACTCACTATAGGGAGA were used as a templates for in vitro transcription of the 3′UTRs of eIF4E, DCK, C1D, CDC2, CCN1, ETF1, PCNA, PMSA, XBP-1 and UBC using biotinylated CTP. Two µg of biotinylated transcript was incubated with 80 µg of lysate (cytoplasmic or nuclear) for 30 min at room temperature, following which the complexes were isolated with streptavidin-coated magnetic Dynabeads (Dynal) and the pulldown material was analyzed by standard western blotting.
Oligonucleotides used to prepare templates for in vitro biotinylated transcription and pulldown assay
The following primer pairs (forward and reverse, respectively) were used:
(T7) GAAGACACCTTCTGAGTATTCTCA and CAGTTTTGTACACTGTCTTAATATGAA for eIF4E,
(T7) TCTTGCTGAAGACTACAGGCAG and GAGTCATGACAAATAAATAATAAATTTTTATT for DCK,
(T7) GGTCAGTCTTGCAAGTACCATTT and GGGACATAGAGGCAACATGC for C1D,
(T7) CTTTCTGACAAAAAGTTTCCATATG and TCCCAAAGCTAGTAATTTTAGTTAAAT for CDC2,
(T7) AGTGGGTGCAAGCAGACCT and TTCTGGCTCACTCCAAATCA for CCN1,
(T7) CTATTGCTGGGATTGGGAGA and AGGGTTTCAGGCCAACTTTT for ETF1,
(T7) CCCAAGATCGAGGATGAAGA and TGCATTTAGAGTCAAGACCCTTT for PCNA,
(T7) GGCTGATGAACCAATGGAAC and CAGGCGGTGAAACAATTTTA for PMSA1,
(T7) TCAGCCCCTCAGAGAATGAT and TCTGCTATCCTCCAGGCAGT for XBP-1 and
(T7) GGTGCTCCGTCTCAGAGGT and TTGAAAGGAAAGTGCAATGAAA for UBC.
RT followed by real-time, quantitative PCR
Total RNA or RNA isolated from the IPs was reverse transcribed (RT) using random hexamers and subjected to quantitative PCR (qPCR) to assess the abundance of the products using the gene-specific primer pairs listed below. Analysis was performed using the QuantiTect SYBR Green PCR kit (Qiagen) and an ABI Prism 7000 detection system with the ABI Prism 7000 SDS software.
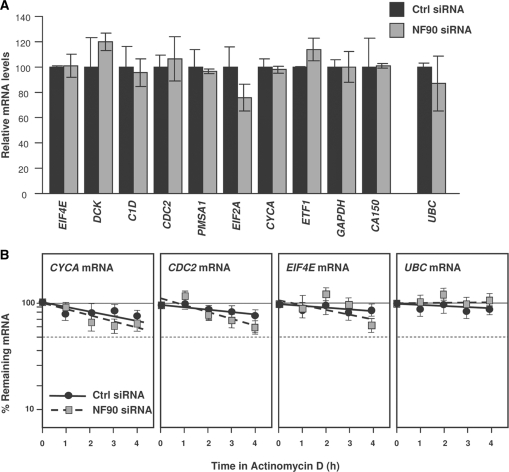
To measure the relative mRNA stabilities in cells with normal NF90 or reduced NF90 levels, cultures were treated with actinomycin D (2 μg/ml) for the times shown. At subsequent times, mRNA levels were measured by RT-qPCR, normalized to 18S rRNA levels, and plotted on a semi-logarithmic scale to calculate the time required for each mRNA to reach one-half of its initial abundance (50%, dashed line).
Oligonucleotides for RT-qPCR-mediated detection of mRNAs in IP materials and in total RNA
Oligonucleotide pairs (forward and reverse, respectively) to detect mature mRNAs were as follows:
CACATGAAGCAGCACGACTT and GGATGTTGCCGTCCTCCTTG for EGFP,
CAGATGGGCACTCTGGTTTT and CTCCCCGTTTGTTTTTCTCA for eIF4E,
AGCAAGGCATTCCTCTTGAA and AACCATTTGGCTGCCTGTAG for DCK,
GCTGTGGATGAGATGCTGAA and TGCCAGCCTTTTTCTTGTCT for C1D,
TTTTCAGAGCTTTGGGCACT and CCATTTTGCCAGAAATTCGT for CDC2,
GGCGTGAACCTCACCAGTAT and TCTCGGCATATACGTGCAAA for PCNA,
GCGTCAGGAGTGTTTGGATT and GACATGGCTCTGCAGTCAAAfor PMSA1,
GCCAAATTGCCCTATCTCAA and CAGAAAAATGGGCAAAGGAA for EIF2A,
AAGAAGCCAGCTGAATCTCAAA and GGTCCAGGTAAACTAATGGCTGAA for CYCA,
TAGCTGGATCCGCTGACTTT and AACAGTACTTGCCCGTGTCC for ETF1,
ACATCAAGAAGGTGGTGAAGCAGG and CCAGCAAGGATACTGAGAGCAAGAG for GAPDH,
GCATCCTGCAGCAGATACAA and CAGCTTCACCGACTTCATCA for MYBBP1a,
CCACCTGCCTTCTCACTAGC and TGGGAAATCTCAAGGACTGG for P2RY2,
CAAGAAGCAGGTCTCGATCC and CCACGTTTCTGGCTCTTCTC for SFRS5,
GGAATGGAGCAGTTGGAAAA and TGACCTGCAGTGTCCCATAA for RAB23,
GTCGATCCAAAAGTGGCAAT and TTCCATGGCAGCTTCTTTCT for TCERG1,
TACATCGACACCACCTGGAA and GCCAGGTTCCTCATCAAAAA for SURF4 and
ATTTGGGTCGCGGTTCTTG and TGCCTTGACATTCTCGATGGT for UBC.
Analysis of translation: polysome gradients and nascent translation assay
For polysome analysis, 48 h after transfection of control siRNA or NF90 siRNA, HeLa cells were incubated with 0.1 mg/ml cycloheximide for 10 min. Cytoplasmic extracts (1 mg each) were prepared and fractionated through a linear sucrose gradient [10–50% (w/v)], as previously reported (28). Ten fractions were collected using a fraction collector (Brandel) and monitored by optical density measurement (A254). The RNA in each fraction was isolated with Trizol LS (Invitrogen). Following RT, qPCR analysis was performed using primer pairs for EGFP, UBC, CYCA and EIF4E.
The levels of nascent (de novo translated) EGFP, GAPDH, Cdc2 and Cyclin A were measured by incubating HeLa cells briefly (20 min) with 1 mCi l-[35S]methionine and l-[35S]cysteine (Easy Tag TMEXPRESS, NEN/Perkin Elmer, Boston, MA, USA) per 60 mm plate, as shown earlier (28). Cells were lysed in RIPA buffer [10 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1% NP-40, 1 mM EDTA, 0.1% SDS and 1 mM DTT] and the IP reactions were carried out in 1 ml TNN buffer [50 mM Tris-HCl (pH 7.5), 250 mM NaCl, 5 mM EDTA, 0.5% NP-40] for 16 h at 4°C using antibodies that recognized EGFP, GAPDH, Cdc2 or Cyclin A (Santa Cruz Biotech.) or using IgG1. Following extensive washes in the TNN buffer, the IP samples were resolved by SDS-PAGE, transferred onto PVDF filters, and visualized and quantified using a PhosphorImager (Molecular Dynamics).
RESULTS
High-throughput identification of NF90 target mRNAs
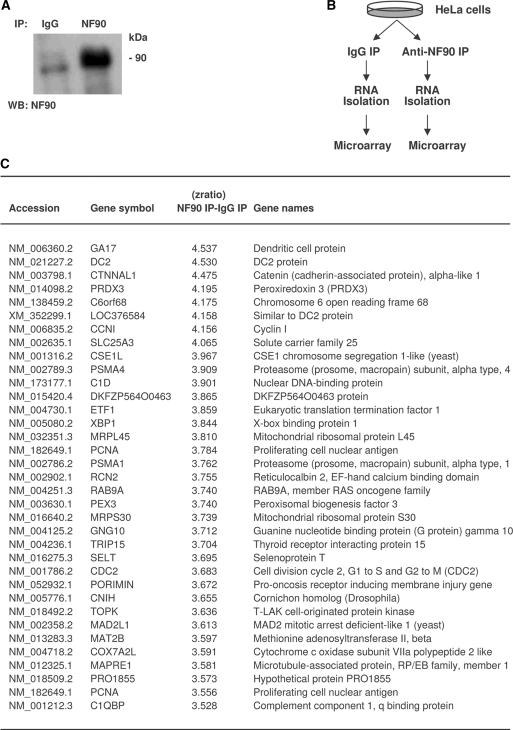
In order to identify the collection of mRNAs associated with NF90, we performed RNP IP analysis using an anti-NF90 antibody, under conditions that preserved the association of NF90 ribonucleoprotein complexes, as previously described (28,35). The RNA in the NF90 IP samples and in the control IgG IP samples (Figure 1A) was isolated and used for reverse transcription and hybridization of Illumina microarrays (‘Materials and Methods‘ section; Figure 1B). A partial list of transcripts that were specifically enriched in NF90 IP relative to IgG IP is shown (Figure 1C); see Supplementary Table S1 for a complete list of mRNAs enriched in NF90 IP samples.
Figure 1.
Identification of NF90 target mRNAs en masse by RNP IP and microarray analysis. (A) Following IP from untreated HeLa cells using either anti-NF90 or IgG antibodies, NF90 was detected by western blot analysis. (B) Schematic of the experimental approach. HeLa cell lysates were subjected to IP with either IgG or anti-NF90 antibodies. The collections of RNAs isolated from each IP reaction were identified using microarrays. (C) Partial table of mRNAs enriched in the NF90 IP samples relative to the control IgG IP samples; relative enrichment is indicated in the form of Z ratios (details in the ’Materials and Methods‘ section).
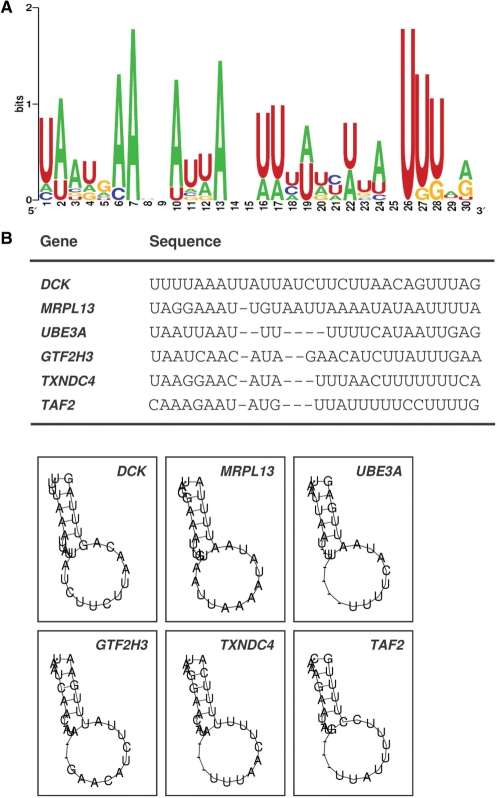
The top ∼800 transcripts specifically isolated in association with NF90 (the experimental dataset) were used for computational analysis to identify the conserved RNA signature motif, based on both primary sequences and secondary structures (‘Materials and Methods’ section). Among the 100 possible candidate motifs identified from the experimental dataset, one motif comprising 25–30 nucleotides showed the highest frequency of hits per kb in the experimental dataset over the entire UniGene transcript sequence database. The logo of this motif, which is a graphic representation of the relative frequency of nucleotides at each position, revealed a bi-partitite sequence, highly A-rich in one half and U-rich in the other half (Figure 1A); together, nucleotides A and U comprised 92% of the signature motif sequence. Depicted in Figure 2B and C are six specific hits of the AU-rich NF90 sequence on specific mRNAs and their predicted folding patterns into hairpin loop structures.
Figure 2.
Sequence and structure of the predicted AUF1 signature motif identified among NF90-associated transcripts. (A) Probability matrix (graphic logo) of an NF90 signature motif, elucidated from the array-derived experimental dataset, indicating the relative frequency with which each nucleotide is likely to be found at each position within the motif. (B) Specific sequence and secondary structure of six representative examples of the NF90 motif in specific mRNAs; the corresponding gene names are shown.
The NF90 signature motif confers translational repression to target mRNAs
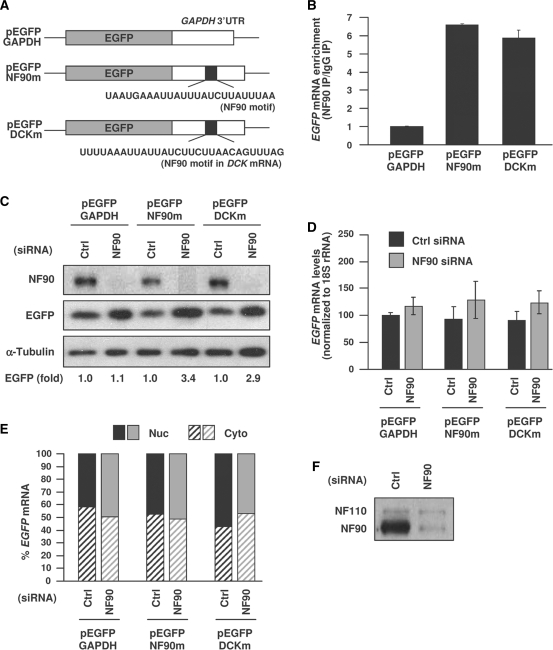
To assess the influence of this signature motif upon the mRNA sequence in which it is found, we studied the consequences of adding the motif to a heterologous reporter. A control plasmid expressing EGFP with a 400 bp segment of the housekeeping GAPDH 3′UTR (pEGFP-GAPDH) was used as the backbone reporter. It was used to generate a reporter construct that contained the ‘ideal’ (most frequent nucleotides at each position) AU-rich NF90 motif, pEGFP-NF90m, as well as another reporter construct that contained one specific motif hit on the DCK mRNA, pEGFP-DCKm (Figure 3A). Following transfection of the plasmids into HeLa cells, NF90 RNP IP analysis showed that both EGFP-NF90m and EGFP-DCKm mRNAs were enriched in NF90 IP samples relative to IgG samples by ∼6-fold greater than the control, EGFP-GAPDH mRNA (Figure 3B), indicating that the two chimeric reporter mRNAs containing the NF90 motif had become NF90 targets.
Figure 3.
Analysis of reporter chimeric RNAs bearing the NF90 signature motif. (A) Schematic of plasmids used in reporter analyses: a control plasmid expressing a chimeric RNA that contained the EGFP CR linked to the GAPDH 3′UTR (pEGFP-GAPDH), a chimeric RNA in which the NF90 ‘ideal’ motif (the top nt position at each site in Figure 2A) was inserted in the middle of the GAPDH 3′UTR (pEGFP-NF90m) and a chimeric RNA in which the specific NF90 motif hit in the DCK mRNA was inserted in the middle of the GAPDH 3′UTR (pEGFP-DCKm). (B) HeLa cells were transfected with plasmids pEGFP-GAPDH, pEGFP-NF90m and pEGFP-DCKm; 24 h later cells were lysed and the association of the expressed chimeric RNAs with NF90 was assessed by RNP IP, followed by RT-qPCR detection of EGFP PCR products. The data are represented as enrichment of each transcript in NF90 IP relative to IgG IP samples. (C) Cells were transfected with control or NF90-directed siRNAs for 24 h and then with the plasmids described in panel (A); an additional 24 h later, EGFP signals were assessed by western blot analysis and EGFP intensities (as well as the intensities of loading control α-tubulin) were measured by densitometry. The relative changes in EGFP signal intensities, normalized to differences in loading control signals, are indicated (fold). (D) In cells that were processed as in (C), the whole-cell abundance of chimeric EGFP mRNAs was measured by RT-qPCR. (E) Following transfection as explained in panel (C), the nuclear and cytosolic fractions were prepared and the abundance of each of the three EGFP chimeric reporters was tested by RT-qPCR analysis; data represent the average from two experiments showing similar results. (F) Representative western blot analysis of NF90 and NF110 in cells transfected with either Ctrl siRNA or NF90 siRNA. The data in (B) and (D) represent the means and S.E.M. from three independent experiments.
Next, an intervention to silence NF90 by transfection with small interfering (si)RNA was used to study the influence of NF90 on the expression of the two NF90 reporter target transcripts. As shown in Figure 3B, NF90 was silenced effectively, reaching a >90% reduction in abundance, when compared to the intensity seen in the control (Ctrl siRNA) transfection group. The presence of NF90m or DCKm lowered slightly the basal EGFP levels (compare the three Ctrl siRNA samples), but NF90 silencing increased EGFP expression in each case: by ∼3.4-fold for the EGFPm reporter and 2.9-fold for the DCKm reporter. This increase did not appear to be due to selective mRNA stabilization when NF90 was silenced, since the levels of EGFP-NF90m mRNA (or the EGFP-DCKm mRNA, not shown) were not significantly different between the Ctrl siRNA group and the NF90 siRNA group (Figure 3D). Likewise, the change in translation did not appear to be due to the NF90-mediated retention of target transcripts in the nucleus, as suggested by earlier studies looking globally at polyadenylated mRNAs (36). As shown in Figure 3E, the relative abundance in the nucleus and cytosol of the control transcript EGFP-GAPDH did not change as a function of NF90, nor did EGFP-NF90m or EGFP-DCKm mRNAs show altered distribution relative to EGFP-GAPDH, regardless of the NF90 status. Evidence that the NF90 siRNA (which was designed to target exon 18, unique to NF90) did not silence the related protein NF110 (which lacks exon 18) is shown in Figure 3F. It is important to note that although the antibody could also recognize NF110, the relative abundance of NF110 was much lower in these cells; it is also important to point out that in cells expressing higher levels of NF110, NF90 siRNA also specifically silenced NF90 and did not affect NF110 abundance (not shown).
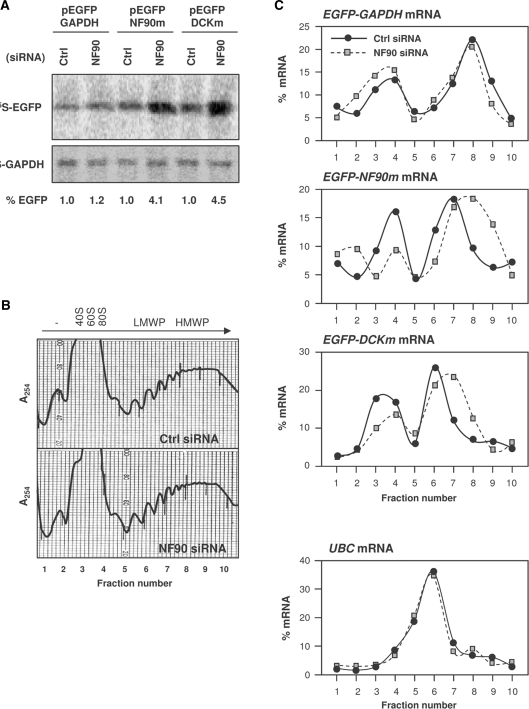
Instead, we postulated that NF90 might specifically lower the translation of these mRNAs. To test this possibility, we first studied the nascent translation of the EGFP reporter vectors. This assay measured the de novo synthesis of the reporter protein (EGFP), expressed from each of the three reporter constructs. Briefly, following siRNA transfections, cells were incubated for a short time period (20 min) with amino acids 35S-Met and 35S-Cys, whereupon lysates were prepared and the abundance of newly synthesized 35S-EGFP was measured by IP using anti-EGFP antibodies. As shown in Figure 4A, nascent EGFP translation in the pEGFP-GADPH transfection group was not influenced significantly after silencing NF90; by contrast, in both pEGFP-NF90m and pEGFP-DCKm transfection groups, nascent EGFP translation from the encoded transcripts (EGFP-NF90m and EGFP-DCKm mRNAs) was significantly upregulated in NF90 siRNA-transfected cells. The effect of NF90 silencing on the increased expression of EGFP was specific, as NF90 silencing did not alter the levels of GAPDH synthesized (35S-GAPDH), nor did it significantly alter the background amino acid incorporation (IgG IP).
Figure 4.
Influence of the NF90 motif on reporter translation. (A) Nascent EGFP production was monitored following a brief (20-min-long) incubation of HeLa cells with l-[35S]methionine and l-[35S]cysteine after transfection with either Ctrl siRNA or NF90 siRNA, and 24 h later with pEGFP-GAPDH, pEGFP-NF90m or pEGFP-DCKm; 24 h after that, lysates were prepared and subjected to IP using either anti-EGFP or anti-GAPDH antibodies. The incorporation of radiolabeled amino acids into the newly synthesized EGFP and GAPDH proteins was assessed by electrophoresis through 12% SDS-containing polyacrylamide gels and by visualization and quantitation using a PhosphorImager. (B) Representative polysome profiles prepared from cells that were transfected with Ctrl or NF90 siRNAs. The direction of sedimentation (arrow), as well as the components of the translational machinery in each fraction [no ribosome components (–), ribosome subunits (Subunits), monosome (Mono.), LMWP (low molecular weight polysomes) and HMWP (high molecular weight polysomes)] are indicated. (C) Representative distribution of the chimeric EGFP reporter mRNAs (top graphs) as well as the housekeeping UBC mRNA (bottom graph) in Ctrl siRNA and NF90 siRNA populations. The relative distribution of these mRNAs was tested by preparing cytoplasmic lysates from cells treated as in panel (A), fractionating them through sucrose gradients and collecting 10 fractions for analysis. RNA was extracted from each fraction and the levels of EGFP and UBC mRNAs in each fraction from each population were measured by RT-qPCR and plotted as a percentage of the total mRNA in the gradient. The data shown are representative of three independent experiments.
Further confirmation that NF90 selectively repressed the translation of EGRP reporter constructs bearing the NF90 motif was obtained by monitoring the distribution of EGFP-NF90m mRNA on polysome gradients in Ctrl and NF90 siRNA populations. As shown in Figure 4B, there were no major differences between the global polysome profiles seen in Ctrl siRNA- and NF90 siRNA-transfected cells. The levels of specific mRNAs were then measured after isolating RNA from each gradient fraction: –, fractions in which RNA was not associated with ribosome components; 40S, 60S, 80S, small ribosomal subunit, large ribosomal subunit, monosome, respectively; LMWP, low-molecular-weight polysomes; HMWP, high molecular weight polysomes. The RNA in each fraction was then used to perform reverse transcription (RT) and real-time quantitative (q) PCR analysis. This assay showed that the relative distribution of the transcript encoding the housekeeping protein ubiquitin C (UBC mRNA) was comparable between Ctrl and NF90 siRNA groups, in cells that had been transfected with either plasmid pEGFP-GAPDH or plasmid pEGFP-NF90m (bottom graph). However, the chimeric EGFP-GAPDH, EGFP-NF90m and EGFP-DCKm mRNAs showed distinct distribution patterns. Whereas EGFP-GAPDH mRNA distribution essentially overlapped between the Ctrl siRNA and the NF90 siRNA groups (top right graph), the EGFP-NF90m and EGFP-DCKm mRNAs showed a more extensive association with the actively translating polysome fraction (HMWP) in the NF90 siRNA group than in the Ctrl siRNA group (bottom right graph). Together, the results in Figures 3 and 4 indicate that NF90 specifically represses the translation of mRNAs bearing the identified NF90 motif.
NF90 binding to endogenous target mRNAs
Next, we focused our attention on the binding of NF90 to specific target mRNAs. First, using HeLa cells, we validated the interaction of endogenous NF90 with several of the endogenous transcripts identified by microarray analysis (Figure 1). Following RNP IP, we tested the individual enrichment of putative target mRNAs by performing RT followed by qPCR using sequence-specific primer pairs. This analysis showed that all mRNAs were more abundant in NF90 IP samples than in IgG IP samples (except EIF2A mRNA); binding of NF90 to housekeeping transcripts that were not predicted NF90 targets (UBC and SDHA mRNAs) was included to monitor the non-specific binding of NF90 to general cellular mRNAs (Figure 5A). Second, binding of NF90 to endogenous target mRNAs was further assayed by preparing biotinylated transcripts (heavy bar) which spanned sections of the 3′UTR (Figure 5B, white segments) that included NF90 motif hits. Following incubation of the biotinylated RNAs with lysates prepared from HeLa cells, the biotin RNA–protein complexes were pulled down using streptavidin-coated beads and the presence of NF90 in the complexes was detected by western blot analysis. As shown in Figure 5C, all of the biotinylated RNAs associated with NF90, except the negative control, biotinylated UBC. Interestingly, NF110 did not appear to bind the biotinylated transcripts, as it was only detected in the ‘Input’ sample. These interactions appeared to be specific, as probing the biotin pulldown samples with an anti-HuR antibody showed that HuR only associated with a subset of these AU-rich biotinylated sequences.
Figure 5.
RNA-binding assays (array-identified targets). (A) RNP IP analysis of the association of NF90 with mRNAs identified by microarray analysis, using HeLa cells. For the mRNAs shown, the enrichment of the mRNAs in NF90 IP compared with IgG IP is indicated. Differences in sample input were normalized by measuring UBC mRNA. Black bars, array-identified NF90 target mRNAs; gray bars, non-NF90 target mRNAs. Data are the means ±S.E.M. from three independent experiments. (B) Schematic of the biotinylated transcripts prepared for analysis. Gray, 5′UTR; black, CR; white, 3′UTR. (C) Equimolar amounts of the biotinylated transcripts shown in (B) were incubated with HeLa cell lysates. Following pulldown analysis using streptavidin-coated beads, the presence of NF90 and HuR was tested by western blot analysis. A control lysate aliquot was included (Input).
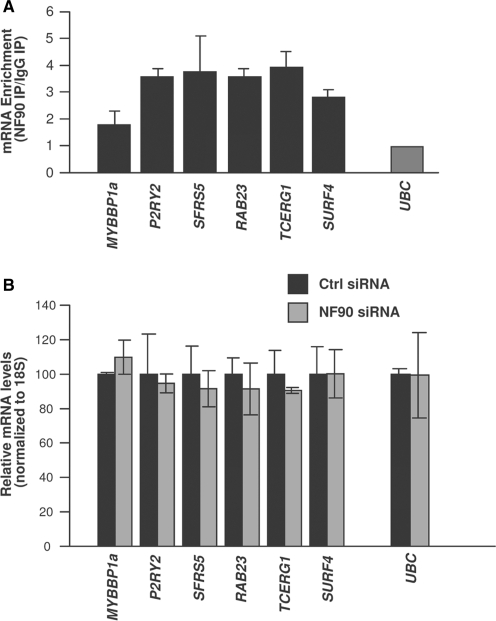
As observed with the chimeric reporter (Figure 3D), NF90 silencing did not significantly increase the steady-state levels of the endogenous target mRNAs (Figure 6A), nor did it affect their half-lives, which were overall high for the mRNAs studied regardless of NF90 abundance (Figure 6B). Based on the presence of at least one hit of the NF90 signature motif, numerous other mRNAs were predicted to be NF90 targets (the complete list of putative target transcripts is provided as Supplementary Table S2). Testing of a subset of these mRNAs verified that these mRNAs did associate with NF90 (Figure 7A), indicating that the presence of the AU-rich signature motif successfully predicted whether an mRNA was a target of NF90. As seen with other targets, NF90 silencing did not alter the mRNA steady-state levels (Figure 7B).
Figure 6.
Steady-state levels and stability of array-identified NF90 targets. (A) The steady-state levels of NF90-associated mRNAs identified by microarray analysis was measured by RT-qPCR analysis in HeLa cells expressing normal (Ctrl siRNA) or reduced (NF90 siRNA) NF90 levels. (B) The stability of several NF90 target mRNAs was assessed by incubating HeLa cells (transfected as in panel A) with actinomycin D (2 μg/ml), collecting total RNA at the times shown, measuring the abundance of each mRNA by RT-qPCR, and normalizing it to the abundance of 18S rRNA (also measured by RT-qPCR). UBC mRNA was included as a housekeeping control transcript. Data are the means ±S.E.M. from three independent experiments.
Figure 7.
RNA-binding assays (predicted targets). (A) RNP IP analysis of the association of NF90 with mRNAs predicted to be NF90 targets based on the presence of one or several NF90 hits in their 3′UTRs. The enrichment of the mRNAs in NF90 IP compared with IgG IP is indicated. Differences in sample input were normalized using by measuring UBC mRNA. (B) The steady-state levels of putative NF90 target mRNAs were measured by RT-qPCR analysis in HeLa cells expressing normal (Ctrl siRNA) or reduced (NF90 siRNA) NF90 levels. Data in (A) and (B) are the means ± S.E.M. from three independent experiments.
NF90 silencing promotes the translation of NF90 target mRNAs
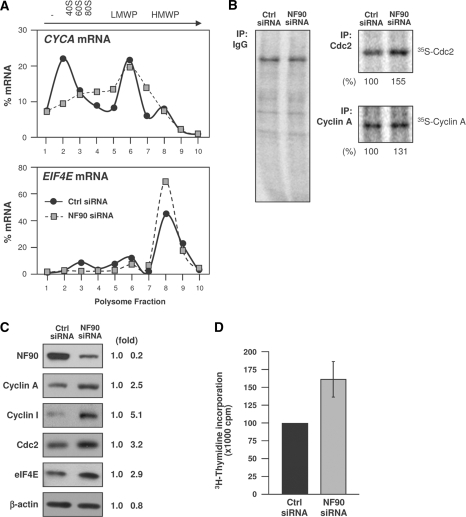
As NF90 did not influence the abundance of endogenous target mRNAs, we examined its consequences on target mRNA translation. First, we examined the association of target mRNAs with polysome gradients. As shown in Figure 8A, the CYCA and EIF4E mRNAs showed proportionately higher levels in the polysomal fractions when NF90 was silenced. Nascent translation analysis showed that translation of the CDC2 and CYCA mRNAs was higher in the NF90 siRNA group, as 35S-Met/Cys incorporation into de novo translated Cdc2 and cyclin A was elevated after silencing NF90 (Figure 8B). Nascent EIF4E could not be tested, as the anti-EIF4E antibody did not precipitate sufficient labeled EIF4E for analysis. Although the increases in polysomal association and nascent translation were relatively modest, NF90 had a marked influence on the steady-state abundance of the encoded proteins. For those tested, NF90 silencing increased their abundance between 2.5- and 5.0-fold (Figure 8C), in keeping with the notion that NF90 functioned as a translational repressor for these target mRNAs. Finally, as several cyclins were higher in NF90-silenced cells, we compared the rate of DNA replication in Ctrl and NF90 siRNA groups. As shown in Figure 8D, NF90 increased DNA biosynthesis moderately in HeLa cells.
Figure 8.
Influence of NF90 on target mRNAs. (A) The relative association of cyclin A and EIF4E mRNAs in cells with normal or reduced NF90 was studied as described in Figure 4. (B) Analysis of nascent translation for Cdc2 and Protein A; new protein synthesis was studied as described in the legend of Figure 4, using anti-cyclin A, anti-Cdc2 and control IgG antibodies. Percent changes in nascent cyclin A and Cdc2 protein levels are indicated. (C) HeLa cells were transfected with Ctrl or NF90 siRNAs; 48 h later, the levels of the proteins shown were assessed by western blot analysis. Signal intensities were quantified by densitometry and represented (fold). (D) By 48 h after transfection with either Ctrl siRNA or NF90 siRNA, the proliferation of HeLa cells was studied by measuring 3H-thymidine incorporation; data are the means ±S.E.M. from three independent experiments.
DISCUSSION
We report the en masse identification of NF90 target mRNAs. Among the 3′UTRs of these mRNAs, we found a shared 25- to 30-nt-long, AU-rich RNA signature motif (Figure 2). Heterologous reporter constructs bearing this motif revealed that NF90 did not change the stability of mRNAs carrying this sequence nor did it change the subcellular concentration of the mRNAs; instead, NF90 prevented the translation of target mRNAs, as determined by studying nascent translation and mRNA distribution on polysome gradients (Figures 3 and 4). The presence of the NF90 signature motif successfully identified endogenous NF90 target transcripts; testing of several endogenous targets further supported a translation inhibitory function for NF90 (Figure 7). In keeping with the proliferative influence of proteins encoded by several NF90 target mRNAs, silencing NF90 moderately enhanced DNA replication (Figure 8).
The discovery that NF90 did not selectively block the transport of target mRNAs (Figure 3E) was unexpected, given earlier findings by Pfeifer and colleagues (36) that NF90 retained mRNAs in the nucleus. However, the Pfeifer study showed that silencing NF90 broadly increased the export of poly(A) and elevated global protein translation. Our results do not reveal an overall increase in translation in NF90 silenced cells, nor do they suggest that target mRNAs bearing the signature motif were selectively retained in the nucleus. The discrepancy between our results and those of Pfeifer et al. could be due to differences in the degree of silencing in both studies, the selective targeting of NF90 in our work and NF90/NF110 in the Pfeifer report, and the times examined after NF90 silencing which were shorter in our study (48 h) than in that of Pfeifer and coworkers (72 h and 96 h). Since NF90 repressed the translation of target mRNAs bearing the NF90 motif, but did not appear to affect their cytosolic abundance, our data suggest that NF90 associates with this subset of target mRNAs after they have reached the cytoplasm. Whether or not the mRNA subset is exported to the cytoplasm by a shared export factor or uses the general mRNA export machinery remains to be studied.
Another question that remains unanswered is the mechanism whereby NF90 represses translation. The fact that the polysomes are smaller suggests that translational initiation is reduced. To explain this effect, it could be hypothesized that binding of NF90 to a target mRNA could block the assembly of components of the translation initiation apparatus on that specific mRNA. Alternatively, NF90 could recruit one or several types of microRNAs onto the target mRNA, thereby repressing translation through the action of RISC (RNA-induced silencing complex), as recently shown for another RBP (42). Yet another mechanism of target mRNA silencing could be linked to the ability of RBPs that bind to double-stranded RNAs (DRBPs) to associate amongst themselves, possibly by forming dsRNA bridges. Thus, it is possible that NF90 recruits onto target mRNAs other DRBPs implicated in gene silencing, such as DICER, PACT and TRBP, which are integral components of the microRNA and RNAi machineries (43,44). It will be interesting to investigate if recombinant purified NF90 binds to the consensus AU-rich signature motif; however, in light of the above-mentioned evidence, the specificity of NF90 is plausibly influenced by proteins that associate with NF90 itself (43,44) or factors (RBPs, microRNAs) that interact directly with the target mRNAs. Therefore, the initial analysis reported here has focused on NF90-mRNA interactions as identified within the cytoplasmic milieu.
Although we previously identified MKP-1 mRNA as a target of NF90, MKP-1 mRNA unexpectedly lacks any predicted hits of the AU-rich NF90 motif described here. Following treatment with H2O2, NF90 abundance increased in the cytoplasm, its binding to MKP-1 mRNA was enhanced and it repressed translation of MKP-1 mRNA but stabilized it (28). It is possible that the subset of target mRNAs whose stability increases by NF90 share a different signature motif. It is also possible that stress treatments (oxidative or otherwise) change the affinity of NF90 for a target RNA sequence. Perhaps phosphorylation of NF90 by kinases [including known kinases such as PKR (25)] may affect its affinity for target transcripts, as shown for HuR (45). Accordingly, it will be interesting to identify a signature motif in NF90 target mRNAs after exposure to a stimulus that alters the subset of NF90-bound mRNAs. For TIAR, a C-rich signature motif was found in target mRNAs under unstimulated conditions; after exposure to ultraviolet light, the signature motif was instead highly AU-rich (41). The signature motif for TIA-1 was also AU-rich (13). Thus, it is possible that under conditions of stress, TIAR or TIA-1, both translational repressors, competes with NF90 for binding to shared target mRNAs. Although such a possibility remains to be tested systematically, at least one example supports this mode of action. NF90 binds with strong preference at a distal, ∼250-base long segment of the MKP-1 3′UTR, while TIA-1 binds with moderate strength throughout the MKP-1 3′UTR (28). In this instance, the association of NF90 to MKP-1 mRNA increased after treatment with H2O2, while binding of TIA-1 to MKP-1 mRNA decreased, suggesting that for MKP-1 mRNA, NF90 and TIA-1 may bind competitively.
HuR can also function as a translational repressor, although its signature motif is mainly U-rich (40). A systematic comparison, validation and analysis of competition and/or cooperation between HuR and NF90 on target-shared mRNAs is also beyond the scope of this study. However, HuR was also shown to interact with the MKP-1 mRNA in a modestly cooperative fashion with NF90, since silencing NF90 slightly lowered the interaction of HuR with MKP-1 mRNA and vice versa (28). Like TIA-1, HuR did not bind exactly to the same MKP-1 3′UTR segment as NF90 did (28); this finding led us to postulate that HuR and NF90 may bind cooperatively by interacting with the MKP-1 3′UTR at the same time. As HuR, TIAR and TIA-1 likely do not bind to the very same location on the shared mRNAs, the relative distance, sequence and dynamics of association between NF90 and these RBPs likely vary among the mRNAs. Therefore, the functional consequences of their association are probably determined by each specific mRNA and need to be analyzed on a case-by-case basis.
In conclusion, this study provides a first systematic analysis of NF90-RNA interactions. Given NF90’s ability to bind DNA and RNA and its influence on mRNA transport, stability and translation, testing its functions individually appears to be most informative. The AU-rich signature element described in this report identifies a particular subset of NF90 target mRNAs subject to translational repression by NF90. Many other RBPs, such as TTP, AUF1, BRF1, KSRP and HuR, also have broad affinity for AU-rich RNA. Therefore, complexes containing NF90 and a target mRNA bearing this motif are likely subject to rich post-transcriptional processes involving other RBPs, which may compete or cooperate with NF90 in determining the post-transcriptional fate of the target transcripts. Further knowledge of the post-translational modification of NF90 and its influence on the metabolism of target mRNAs will give critical insight into the function of this pivotal regulator of gene expression.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Supported in its entirety by the NIA-IRP, National Institute of Health. Funding for open access charge: National Institute on Aging-Intramural Research Program, National Institutes of Health.
Supplementary Material
REFERENCES
- 1.Moore MJ. From birth to death: the complex lives of eukaryotic mRNAs. Science. 2005;309:1514–1518. doi: 10.1126/science.1111443. [DOI] [PubMed] [Google Scholar]
- 2.Glisovic T, Bachorik JL, Yong J, Dreyfuss G. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 2008;582:1977–1986. doi: 10.1016/j.febslet.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carballo E, Lai WS, Blackshear PJ. Feedback inhibition of macrophage tumor necrosis factor-a production by tristetraprolin. Science. 1998;281:1001–1005. doi: 10.1126/science.281.5379.1001. [DOI] [PubMed] [Google Scholar]
- 4.Chen CY, Gherzi R, Ong SE, Chan EL, Raijmakers R, Pruijn GJ, Stoecklin G, Moroni C, Mann M, Karin M. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell. 2001;107:451–464. doi: 10.1016/s0092-8674(01)00578-5. [DOI] [PubMed] [Google Scholar]
- 5.Briata P, Forcales SV, Ponassi M, Corte G, Chen CY, Karin M, Puri PL, Gherzi R. p38-dependent phosphorylation of the mRNA decay-promoting factor KSRP controls the stability of select myogenic transcripts. Mol. Cell. 2005;20:891–903. doi: 10.1016/j.molcel.2005.10.021. [DOI] [PubMed] [Google Scholar]
- 6.Hinman MN, Lou H. Diverse molecular functions of Hu proteins. Cell Mol. Life Sci. 2008;65:3168–3181. doi: 10.1007/s00018-008-8252-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang W, Wagner BJ, Ehrenman K, Schaefer AW, De-Maria CT, Crater D, DeHaven K, Long L, Brewer G. Purification, characterization, and cDNA cloning of an AU-rich element RNA-binding protein, AUF1. Mol. Cell Biol. 1993;13:7652–7665. doi: 10.1128/mcb.13.12.7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liao B, Hu Y, Brewer G. Competitive binding of AUF1 and TIAR to MYC mRNA controls its translation. Nat. Struct. Mol. Biol. 2007;14:511–518. doi: 10.1038/nsmb1249. [DOI] [PubMed] [Google Scholar]
- 9.Mazan-Mamczarz K, Kuwano Y, Zhan M, White EJ, Martindale JL, Lal A, Gorospe M. Identification of a signature motif in target mRNAs of RNA-binding protein AUF1. Nucleic Acids Res. 2009;37:204–214. doi: 10.1093/nar/gkn929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sarkar B, Xi Q, He C, Schneider RJ. Selective degradation of AU-rich mRNAs promoted by the p37 AUF1 protein isoform. Mol. Cell Biol. 2003;23:6685–6693. doi: 10.1128/MCB.23.18.6685-6693.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raineri I, Wegmueller D, Gross B, Certa U, Moroni C. Roles of AUF1 isoforms, HuR and BRF1 in ARE-dependent mRNA turnover studied by RNA interference. Nucleic Acids Res. 2004;32:1279–1288. doi: 10.1093/nar/gkh282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Piecyk M, Wax S, Beck AR, Kedersha N, Gupta M, Maritim B, Chen S, Gueydan C, Kruys V, Streuli M, et al. TIA-1 is a translational silencer that selectively regulates the expression of TNF-α. EMBO J. 2000;19:4154–4163. doi: 10.1093/emboj/19.15.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.López de Silanes I, Galbán S, Martindale JL, Yang X, Mazan-Mamczarz K, Indig FE, Falco G, Zhan M, Gorospe M. Identification and functional outcome of mRNAs associated with RNA-binding protein TIA-1. Mol. Cell Biol. 2005;25:9520–9531. doi: 10.1128/MCB.25.21.9520-9531.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lal A, Abdelmohsen K, Pullmann R, Kawai T, Yang X, Galban S, Brewer G, Gorospe M. Posttranscriptional derepression of GADD45a by genotoxic stress. Mol. Cell. 2006;22:117–128. doi: 10.1016/j.molcel.2006.03.016. [DOI] [PubMed] [Google Scholar]
- 15.Sawicka K, Bushell M, Spriggs KA, Willis AE. Polypyrimidine-tract-binding protein: a multifunctional RNA-binding protein. Biochem. Soc. Trans. 2008;36:641–647. doi: 10.1042/BST0360641. [DOI] [PubMed] [Google Scholar]
- 16.Galbán S, Kuwano Y, Pullmann R, Jr, Martindale JL, Kim HH, Lal A, Abdelmohsen K, Yang X, Dang X, Liu JO, et al. RNA-binding proteins HuR and PTB promote the translation of hypoxia-inducible factor 1 alpha. Mol. Cell Biol. 2008;28:93–107. doi: 10.1128/MCB.00973-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mitchell P, Tollervey D. mRNA stability in eukaryotes. Curr. Opin. Genet. Dev. 2000;10:193–198. doi: 10.1016/s0959-437x(00)00063-0. [DOI] [PubMed] [Google Scholar]
- 18.Orphanides G, Reinberg D. A unified theory of gene expression. Cell. 2002;108:439–451. doi: 10.1016/s0092-8674(02)00655-4. [DOI] [PubMed] [Google Scholar]
- 19.Abdelmohsen K, Kuwano Y, Kim HH, Gorospe M. Posttranscriptional gene regulation by RNA-binding proteins during oxidative stress: implications for cellular senescence. Biol. Chem. 2008;389:243–255. doi: 10.1515/BC.2008.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Corthesy B, Kao PN. Purification by DNA affinity chromatography of two polypeptides that contact the NF-AT DNA binding site in the interleukin 2 promoter. J. Biol. Chem. 1994;269:20682–20690. [PubMed] [Google Scholar]
- 21.Kao PN, Chen L, Brock G, Ng J, Kenny J, Smith AJ, Corthésy B, et al. Cloning and expression of cyclosporin A- and FK506-sensitive nuclear factor of activated T-cells: NF45 and NF90. J. Biol. Chem. 1994;269:20691–20699. [PubMed] [Google Scholar]
- 22.Xu YH, Grabowski GA. Molecular cloning and characterization of a translational inhibitory protein that binds to coding sequences of human acid beta-glucosidase and other mRNAs. Mol. Genet. Metab. 1999;68:441–454. doi: 10.1006/mgme.1999.2934. [DOI] [PubMed] [Google Scholar]
- 23.Xu YH, Busald C, Grabowski GA. Reconstitution of TCP80/NF90 translation inhibition activity in insect cells. Mol. Genet. Metab. 2000;70:106–115. doi: 10.1006/mgme.2000.3010. [DOI] [PubMed] [Google Scholar]
- 24.Liao HJ, Kobayashi R, Mathews MB. Activities of adenovirus virus-associated RNAs: purification and characterization of RNA binding proteins. Proc. Natl. Acad. Sci. USA. 1998;95:8514–8519. doi: 10.1073/pnas.95.15.8514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patel RC, Vestal DJ, Xu Z, Bandyopadhyay S, Guo W, Erme SM, Williams BR, Sen GC, et al. DRBP76, a doublestranded RNA-binding nuclear protein, is phosphorylated by the interferon-induced protein kinase, PKR. J. Biol. Chem. 1999;274:20432–20437. doi: 10.1074/jbc.274.29.20432. [DOI] [PubMed] [Google Scholar]
- 26.Saunders LR, Perkins DJ, Balachandran S, Michaels R, Ford R, Mayeda A, Barber GN, et al. Characterization of two evolutionarily conserved, alternatively spliced nuclear phosphoproteins, NFAR-1 and -2, that function in mRNA processing and interact with the doublestranded RNA-dependent protein kinase, PKR. J. Biol. Chem. 2001;276:32300–32312. doi: 10.1074/jbc.M104207200. [DOI] [PubMed] [Google Scholar]
- 27.Parrott AM, Walsh MR, Reichman TW, Mathews MB. RNA binding and phosphorylation determine the intracellular distribution of nuclear factors 90 and 110. J. Mol. Biol. 2005;348:281–293. doi: 10.1016/j.jmb.2005.02.047. [DOI] [PubMed] [Google Scholar]
- 28.Kuwano Y, Kim HH, Abdelmohsen K, Pullmann R, Jr, Martindale JL, Yang X, Gorospe M. MKP-1 mRNA stabilization and translational control by RNA-binding proteins HuR and NF90. Mol. Cell Biol. 2008;28:4562–4575. doi: 10.1128/MCB.00165-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reichman TW, Parrott AM, Fierro-Monti I, Caron DJ, Kao PN, Lee CG, Li H, Mathews MB, et al. Selective regulation of gene expression by nuclear factor 110, a member of the NF90 family of double-stranded RNA-binding proteins. J. Mol. Biol. 2003;332:85–98. doi: 10.1016/s0022-2836(03)00885-4. [DOI] [PubMed] [Google Scholar]
- 30.Shi L, Qiu D, Zhao G, Corthesy B, Lees-Miller S, Reeves WH, Kao PN, et al. Dynamic binding of Ku80, Ku70 and NF90 to the IL-2 promoter in vivo in activated T-cells. Nucleic Acids Res. 2007;35:2302–2310. doi: 10.1093/nar/gkm117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi L, Godfrey WR, Lin J, Zhao G, Kao PN. NF90 regulates inducible IL-2 gene expression in T cells. J. Exp. Med. 2007;204:971–977. doi: 10.1084/jem.20052078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shim J, Lim H, J RY, Karin M. Nuclear export of NF90 is required for interleukin-2 mRNA stabilization. Mol. Cell. 2002;10:1331–1334. doi: 10.1016/s1097-2765(02)00730-x. [DOI] [PubMed] [Google Scholar]
- 33.Vumbaca F, Phoenix KN, Rodriguez-Pinto D, Han DK, Claffey KP. Double-stranded RNA-binding protein regulates vascular endothelial growth factor mRNA stability, translation, and breast cancer angiogenesis. Mol. Cell Biol. 2008;28:772–783. doi: 10.1128/MCB.02078-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shi L, Zhao G, Qiu D, Godfrey WR, Vogel H, Rando TA, Hu H, Kao PN. NF90 regulates cell cycle exit and terminal myogenic differentiation by direct binding to the 3′-untranslated region of MyoD and p21WAF1/CIP1 mRNAs. J. Biol. Chem. 2005;280:18981–18989. doi: 10.1074/jbc.M411034200. [DOI] [PubMed] [Google Scholar]
- 35.Pullmann R, Jr, Kim HH, Abdelmohsen K, Lal A, Martindale JL, Yang X, Gorospe M, et al. Analysis of turnover and translation regulatory RNA-binding protein expression through binding to cognate mRNAs. Mol. Cell Biol. 2007;27:6265–6278. doi: 10.1128/MCB.00500-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pfeifer I, Elsby R, Fernandez M, Faria PA, Nussenzveig DR, Lossos IS, Fontoura BM, Martin WD, Barber GN. NFAR-1 and -2 modulate translation and are required for efficient host defense. Proc. Natl. Acad. Sci. USA. 2006;105:4173–4178. doi: 10.1073/pnas.0711222105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gorodkin J, Heyer LJ, Stormo GD. Finding the most significant common sequence and structure motifs in a set of RNA sequences. Nucleic Acids Res. 1997;25:3724–3732. doi: 10.1093/nar/25.18.3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eddy SR, Durbin R. RNA sequence analysis using covariance models. Nucleic Acids Res. 1994;22:2079–2088. doi: 10.1093/nar/22.11.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tenenbaum SA, Lager PJ, Carson CC, Keene JD. Ribonomics: identifying mRNA subsets in mRNP complexes using antibodies to RNA-binding proteins and genomic arrays. Methods. 2002;26:191–198. doi: 10.1016/S1046-2023(02)00022-1. [DOI] [PubMed] [Google Scholar]
- 40.Lopez de Silanes I, Zhan M, Lal A, Yang X, Gorospe M. Identification of a target RNA motif for RNA-binding protein HuR. Proc. Natl. Acad. Sci USA. 2004;101:2987–2992. doi: 10.1073/pnas.0306453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim HS, Kuwano Y, Zhan M, Pullmann R, Jr, Mazan-Mamczarz K, Li H, Kedersha N, Anderson P, Wilce MC, et al. Elucidation of a C-rich signature motif in target mRNAs of RNA-binding protein TIAR. Mol. Cell Biol. 2007;27:6806–6817. doi: 10.1128/MCB.01036-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim HH, Kuwano Y, Srikantan S, Lee EK, Martindale JL, Gorospe M. HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev. 2009;23:1743–1748. doi: 10.1101/gad.1812509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R, et al. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee Y, Hur I, Park SY, Kim YK, Suh MR, Kim VN. The role of PACT in the RNA silencing pathway. EMBO J. 2006;25:522–532. doi: 10.1038/sj.emboj.7600942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abdelmohsen K, Pullmann R, Jr, Lal A, Kim HH, Galban S, Yang X, Blethrow JD, Walker M, Shubert J, Gillespie DA, et al. Phosphorylation of HuR by Chk2 regulates SIRT1 expression. Mol. Cell. 2007;25:543–557. doi: 10.1016/j.molcel.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.