Abstract
HuD is a neuronal ELAV-like RNA-binding protein (RBP) involved in nervous system development, regeneration, and learning and memory. This protein stabilizes mRNAs by binding to AU-rich instability elements (AREs) in their 3′ unstranslated regions (3′ UTR). To isolate its in vivo targets, messenger ribonucleoprotein (mRNP) complexes containing HuD were first immunoprecipitated from brain extracts and directly bound mRNAs identified by subsequent GST-HuD pull downs and microarray assays. Using the 3′ UTR sequences of the most enriched targets and the known sequence restrictions of the HuD ARE-binding site, we discovered three novel recognition motifs. Motifs 2 and 3 are U-rich whereas motif 1 is C-rich. In vitro binding assays indicated that HuD binds motif 3 with the highest affinity, followed by motifs 2 and 1, with less affinity. These motifs were found to be over-represented in brain mRNAs that are upregulated in HuD overexpressor mice, supporting the biological function of these sequences. Gene ontology analyses revealed that HuD targets are enriched in signaling pathways involved in neuronal differentiation and that many of these mRNAs encode other RBPs, translation factors and actin-binding proteins. These findings provide further insights into the post-transcriptional mechanisms by which HuD promotes neural development and synaptic plasticity.
INTRODUCTION
mRNA stability is now recognized as a critical post-transcriptional mechanism controlling the expression of a large number of mammalian genes. Transcript stability is dictated by cis-acting elements, mostly localized in the 3′ untranslated region (3′ UTR) and by the activity of trans-acting factors, such as microRNAs (miRNAs) and RNA-binding proteins (RBPs) (1–3). The best characterized cis-acting instability-conferring sequence is the AU-rich element [ARE; (4,5)]. These sequences were originally described in short-lived transcripts encoding cytokines, oncogenes, and growth factors, however, mRNAs for proteins serving a wide variety of functions have been recently identified as containing AREs (6). AREs regulate mRNA decay by interacting with ARE-binding proteins that either trigger mRNA decay [e.g. KH homology splicing regulatory protein (KSRP), tristetraprolin (TTP), AUF1 and butyrate response factor 1 (BRF1)] or stabilization (e.g. Hu proteins) (1,2). In addition to AREs, other elements regulating mRNA stability have been described. For example, a GU-rich element (GRE) that is a target of the CUG-binding protein 1 (CUGBP1/EDEN-BP) was recently identified in the 3′ UTR of many unstable mRNAs (7–9). Furthermore, a few reports have shown the presence of instability sequences in the 5′ UTR (10) and coding region (11); yet, how common these non-3′ UTR localizations are is presently unclear.
Hu proteins are human homologs of Drosophila embryonic lethal abnormal vision (ELAV, 12) and the best-known mRNA stabilizing factors. There are four mammalian ELAV-like/Hu proteins. While HuC and HuD are exclusively expressed in neurons, HuB is found in neurons and gonads, and HuR is ubiquitously expressed in all tissues. Amongst the trans-acting factors that mediate mRNA stabilization, HuD has been shown to play a vital role in neural development and brain physiology, as recently demonstrated by mouse lines either lacking or over-expressing the protein (13–16). HuD knock-out mice show neurogenesis deficits as well as abnormal motor control and development of cranial nerves (13). Over-expression of HuD in transgenic mice leads to alterations in hippocampal physiology and deficits in several spatial and associative learning and memory tasks (16,17). In addition to its role in the central nervous system, HuD has also been associated with axonal re-growth after peripheral nerve injury (18,19). Overall, these studies demonstrate that HuD is required for normal neural development, nerve regeneration and synaptic plasticity (20,21).
At the molecular level HuD binds to several unstable mRNAs and as a result of this interaction the target transcripts are stabilized (22,23). HuD has three RNA-recognition motifs (RRM) and we have previously shown that the first two RRMs of the HuD protein are necessary for binding to GAP-43 mRNA, one of HuD’s best characterized targets (22). However, these domains by themselves are not sufficient to stabilize GAP-43 mRNA in neuronal cells (24) as it also requires the interaction of RRM III with long poly(A) tails (23). Besides GAP-43, other mRNAs such as N-myc, AChE, tau, neuroserpin and MARCKS mRNAs were shown to interact with HuD in vitro and in vivo (2). However, the majority of HuD’s targets in neurons remain to be elucidated.
Several recent studies identified mRNAs associated with mRNPs containing different RBPs using RNA immunoprecipitation (RIP)-Chip assays, a procedure known as ribonomics (25–28). As mRNPs may represent eukaryotic post-transcriptional operons (29), this procedure provides an important tool for defining the biological function of these RNA–protein complexes. HuD was shown to bind other RBPs including the neuronal ELAV-like protein HuB (30), TAP/NXF1, the primary mRNA export receptor (31) and the IGF-2 mRNA-binding protein IMP1a (32). Therefore, in order to determine which of the mRNAs in HuD-containing mRNPs are directly interacting with this RBP, in the present study we combined the original ribonomics protocol with subsequent in vitro pull down assays with GST-HuD. Using this two-step isolation method, nearly 700 new HuD targets were identified. The 3′ UTRs of these targets were then used to discover new HuD-binding motifs and identify the biological and molecular pathways regulated by this protein. As a group, HuD target mRNAs encoded proteins involved in several signaling pathways required for neural differentiation and synaptic remodeling. Interestingly, many of the proteins encoded are also RBPs and translation factors, suggesting that HuD is part of a complex post-transcriptional gene regulatory network.
MATERIALS AND METHODS
mRNA immunoprecipitation
mRNPs containing HuD were immunoprecipitated using the ribonomics protocol described by Tenenbaum and colleagues (25,26). Briefly, forebrain tissue from the HuD transgenic mice (16) were homogenized using a Dounce homogenizer in Polysome Lysis Buffer (100 mM KCl, 5 mM MgCl2, 10 mM HEPES pH 7.0, 0.5% Nonidet P-40, 1 mM dithiothreitol, 100 U/ml RNAsin, 0.2% vanadyl ribonucleoside complexes, leupeptin and aprotinin). Lysates were cleared by centrifugation at 14 000 × g for 10 min at 4°C and frozen at −80°C until use. Protein G-Sepharose beads (Sigma, St. Louis, MO) were coated with anti myc-monoclonal antibody (9B11 mouse mAb, Cell Signaling Technology, Danvers, MA) or non-immune mouse IgG (Sigma, St. Louis, MO) under constant rocking in NT2 buffer [50 mM Tris (pH 7.4), 150 mM NaCl, 1 mM MgCl2 and 0.05% Nonidet P-40] supplemented with 5% BSA. Brain extracts (100 μl) were immunoprecipitated with 50 μl of coated beads in 850 μl of NT2 buffer supplemented with 8 units of RNAsin, 1 mM vanadyl ribonucleoside complexes, 1 mM DTT and 15 mM EDTA for 2 h at room temperature under constant rocking. After beads were washed six times with ice-cold NT2 buffer, proteins were digested with proteinase K and mRNAs purified by phenol–chloroform extraction and ethanol precipitation.
GST HuD pull down assays
Recombinant GST-HuD protein was prepared from BL21 Escherichia coli transformed with pGEX-2T-HuD plasmid as described previously (23,33). Briefly, BL21 cells were induced with IPTG and lysed by sonication in Fast Break Cell Lysis Reagent (Promega, Madison, WI) supplemented with Halt Protease inhibitor (Pierce, Rockford, IL) and RQ1 DNase (Promega, Madison, WI). After centrifugation for 15 min at 4°C at 27 000 × g, the supernatant was incubated for 1 h at 4°C with MagneGST Glutathione Particles (Promega, Madison, WI). Beads were washed with Binding/Wash buffer (Promega, Madison, WI) and incubated with 600 ng of amplified RNA from the previous ribonomics step for 30 min at 4°C in binding buffer (10 mM HEPES, 3 mM MgCl2, 40 mM KCl, 1 mM DTT, 400 U of RNase out and 5% glycerol). After incubation and washes with binding buffer without glycerol, RNA was extracted as described above.
mRNA amplification and array hybridization
Total RNA (10 ng) from the first and second purification steps was converted to double-stranded cDNA using the GeneChip® Expression 3′-Amplification Two-Cycle Target Labeling and Control Reagents Kit (Affymetrix, Santa Clara, CA) according to manufacturer’s protocols. The resulting cDNA was used for the in vitro synthesis of biotin-labeled cRNA using the GeneChip® IVT Labeling Kit (Affymetrix). cRNA was cleaned using the GeneChip® Sample Cleanup Module and fragmented into 35–200 base pair fragments using a magnesium acetate buffer (Affymetrix). In total, 6.5 µg of labeled cRNA was hybridized to Affymetrix GeneChip® Mouse Genome 430 2.0 for 16 h at 45°C. The arrays were washed and stained according to the manufacturer’s recommendations using the GeneChip Fluidics Station 450 (Affymetrix). Each array was scanned using the GeneChip® Scanner 3000 (Affymetrix) and globally scaled to 150 using the Affymetrix GeneChip® Operating Software (GCOS v1.4).
Microarray analysis
Samples were run in triplicates. For target identification, raw data files from the Affymetrix arrays were normalized to the RNA levels used for cRNA synthesis and log2 of the normalized values were used to calculate the enrichment ratios of HuD IP versus IgG IP and GST-HuD versus GST pull downs. Probes with absent calls in at least two out of three replicates in the HuD IP and the GST-HuD pull down chips but not in the controls (IgG IP and GST pull down) were excluded from the analysis and the average enrichment for each probe was used for statistical analyses. A similar analysis was used to identify mRNAs that were up-regulated in the brains of HuD transgenic overexpressor mice (HuD-Tg). Briefly, mRNAs from three HuD-Tg mice and three control (non-transgenic) littermates were analyzed by Affymetrix 430 2.0 chips and mRNAs that had ‘present’ calls and were significantly upregulated (P < 0.05), as determined using GeneSpring 9.0 (Agilent Technologies, Santa Clara, CA), were used for HuD motif analyses. In addition, we generated a list of all the mRNA expressed in mouse forebrain using transcripts from control forebrains with ‘present’ calls and raw expression values more than 100, which corresponds to twice the background level. This set, which consisted of 17 010 transcripts corresponding to 9757 genes, was used as reference for HuD target analyses (see below).
Sequence analysis
The 3′ UTR, coding region (CR) and 5′ UTR sequences of HuD targets, the genes in the Affymetrix 430 2.0 chip, the mouse forebrain set described above and all the genes available in the Ensembl database were downloaded from Ensembl BioMart, release 49, Mus musculus genes NCBIM37 (http://www.ensembl.org/biomart/index.html). The datasets were made non-redundant based on gene ID using our own Perl scripts. If more than one sequence was available for a gene, the longest was chosen for further analysis. 3′ UTR nucleotide composition and length, presence of each ARE subtype, and U stretches were analyzed using custom scripts written in Perl v5.8.8 and BioPerl 1.5.2 modules (http://www.bioperl.org), which are available upon request. Differences between datasets were analyzed by two by two contingency tables with Chi-square test using the R statistical package version 2.7.1.
Motif search and analysis
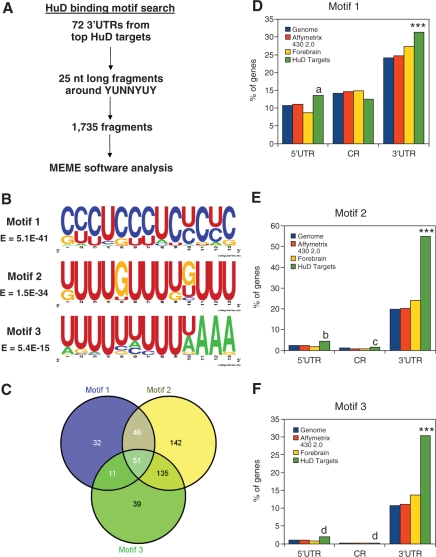
Bioinformatics analyses for HuD-binding motifs considered both the 3′ UTR sequences of the 72 most enriched targets and the restrictions imposed by the three-dimensional structure of the complex between the first two RRMs in HuD and two different Class I and II AREs (34). The 3′ UTR sequences of the 72 most enriched HuD targets downloaded from Ensembl BioMart and 25 nucleotide (nt) long fragments containing the consensus binding sequence YUNNYUY in the middle were extracted. Since the YUNNYUY sequence is not very restrictive, One-thousand seven-hundred thirty-five fragments were obtained. These sequences were used as a training set for motif search using MEME software [http://meme.sdsc.edu/meme; (35,36)]. A diagram of the implemented motif search strategy is shown in Figure 3A. Probability matrices of the new HuD-binding motifs were represented graphically using WebLogo [http://weblogo.berkeley.edu/; (37)]. For analyzing the frequency of each new HuD-binding motif in the different datasets and the location of each motif, we used a Perl script that searches for the following regular expressions (allowing one mismatch): [CG][CT][CT]TC[CT][CT]TC[TC]C[TC]C, [TG]TTTGTTT[TG][GT]TTT, and TTTTTTTTT[TA]AAA, for motifs 1, 2 and 3, respectively. Differences between datasets were analyzed by two by two contingency tables with Chi-square test using the R statistical package version 2.7.1.
Figure 3.
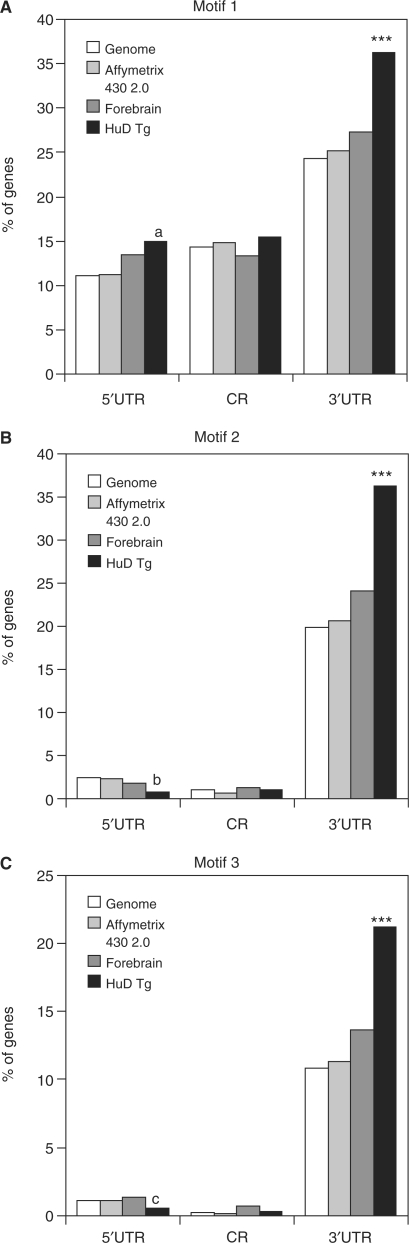
Identification of the HuD-binding motifs. (A) Experimental strategy implemented to identify HuD-binding motifs. (B) WebLogo representations of the three identified HuD-binding sequences. (C) Venn diagram showing the distribution of the three new HuD-binding motifs in the HuD targets. (D) Frequency of motif 1 in the 5′ UTR, coding region (CR), and 3′ UTR of mouse genome (blue bars), Affymetrix 430 2.0 chip genes (red bars), forebrain transcripts (yellow bars) and HuD targets (green bars). (E) Percentage of presence of motif 2 in the same groups as Panel D. (F) Same analysis for motif 3. ***P < 0.0001 of HuD targets versus each of the three reference datasets. (a) P < 0.05 HuD targets versus genome and forebrain, (b) P < 0.05 versus genome, P < 0.01 versus chip and P < 0.0001 versus forebrain, (c) P < 0.05 versus chip and versus forebrain and (d) P < 0.05 versus forebrain.
Nitrocellulose filter binding assay
Ribo-oligoribonucleotides (Integrated DNA Technologies, Coralville, IA) were end-labeled with P32gamma-ATP from MP Biomedicals using polynucleotide kinase (New England Biolabs). Labeled oligoribonucleotides were purified using mini Quick Spin Columns (Roche Applied Sciences, Indianapolis, IN) and used immediately for in vitro binding assays. Reaction mixtures (20 µl) contained 50 mM Tris (pH 7.0), 150 mM NaCl, 0.25 mg/ml bovine serum albumin, 0.25 mg/ml tRNA and labeled oligoribonucleotides as indicated. We used 100 ng per reaction of recombinant HuD for motifs 2 and 3, and 600 ng for motif 1. After 20 min incubation at 37°C, the mixtures were diluted 1:6 with wash buffer (20 mM Tris–HCl pH 7.0 and 50 µg/ml tRNA) and filtered through a nitrocellulose membrane using a dot blot apparatus. Membranes were washed twice with wash buffer. To determine the non-specific binding, similar reactions were carried out in parallel with a 100 molar excess of unlabeled oligoribonucleotide. Bound radioactivity was determined using a phosphor-imager (Personal Molecular Imager, BioRad). Serial dilutions of each labeled oligonucleotide were blotted directly onto the membrane and used to measure total radioactivity.
Gene ontology analysis
The Gene Ontology Tree Machine [GOTM, http://bioinfo.vanderbilt.edu/gotm/, (38)] and WebGestalt [http://bioinfo.vanderbilt.edu/webgestalt, (39)] programs were used for these studies. The frequencies and numbers of HuD targets in each gene ontology category were calculated and compared to those in the mouse genome. A hypergeometric distribution test was used to calculate the statistical significance of the observed (HuD targets dataset) over the predicted (genomic) frequency. P-values smaller than 0.01 were considered statistically significant and only gene ontology categories with more than three genes in the HuD targets dataset were considered. Biological pathways associated with HuD targets were identified using the links to the Kyoto Encyclopedia of Genes and Genomes (KEGG) available thorough GOTM and WebGestalt. MetaCore pathway analysis software was also used (GeneGO, Encinitas, CA).
RESULTS
Identification of HuD targets
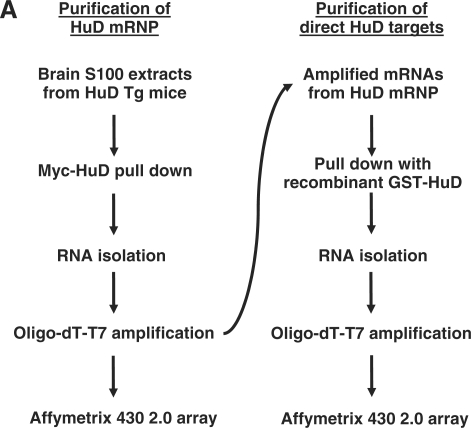
HuD expression is critical for normal brain development and maturation and thus the identification of the repertoire of HuD’s targets is an important first step towards the characterization of the molecular processes regulated by this RBP. In this study, we used two consecutive purification steps to isolate HuD-bound mRNAs (Figure 1). First, neuronal HuD-containing mRNPs were purified from brain extracts of HuD overexpressor mice (14) by immunoprecipitation (IP). Since none of the commercially available HuD antibodies worked well for the IP step, antibodies against the myc-tag present in the HuD transgene were used instead to pull down the protein. Validating the IP step we found that GAP-43 and neuroserpin mRNAs, two of the known targets of HuD, were enriched close to 7- and 70-fold, respectively, in the HuD IP (Supplementary Figure S1). Following mRNA extraction and oligo-dT-T7 directed linear amplification direct targets were subsequently enriched by affinity purification using GST-HuD protein (Figure 1). After each step of purification, mRNAs were isolated, subjected to a two-round linear amplification and hybridized to Affymetrix 430 2.0 arrays.
Figure 1.
Identification of HuD target mRNAs. Experimental strategy implemented for analyzing HuD target mRNAs at genomic scale. The flow chart shows the two sequential steps used for the identification of HuD targets.
Enrichments were calculated by dividing the signal of each probe set in the HuD IP or GST-HuD pull down by the signal of the same probe in the non-immune IgG IP or control GST pull down, respectively. The mean enrichment of mRNAs in the IP step was 2-fold and the use of the GST-HuD step increased this value to 12-fold. As indicated above, one possible explanation for this finding is that many of the mRNAs immunoprecipitated in the first step may be interacting with other proteins in the mRNP. Also, since this is the first time that the RIP-Chip protocol was used for the analyses of Hu protein targets in the brain and Affymetrix arrays were probed with equal amounts of control and HuD IP RNA, the global enrichment in the IP step was lower than that observed for other Hu proteins in cultured cells (25,40). Nevertheless, analyses of the mRNAs enriched in the brain HuD-IP step with those associated with HuR in Jurkat cells (40) resulted in similar enrichment plots for both sets of mRNAs (data not shown). Overall, we found that 5412 probes corresponding to 3529 known genes and expressed sequence tags (ESTs) showed more than 2-fold enrichment after the first step and 1189 probes (1034 genes) had z-scores above 1.5 after the two consecutive isolation steps. Seven hundred probes (673 genes) met both enrichment criteria and were operationally defined as putative HuD targets. Table 1 shows the 70 most enriched mRNAs in the dataset and the complete list of 700 probes is shown in Supplementary Table S1. Although not all the previously reported targets of HuD were included in this dataset, we found that all of them were enriched at least 3-fold after the second purification step and/or contained one of the three RNA-binding motifs described below (Figure 3, Table 3).
Table 1.
Partial list of HuD targets
| Probe set ID | Fold enrichment | Gene symbol | Description | Ensembl |
|---|---|---|---|---|
| 1438554_x_at | 19.43 | Eif4h | Eukaryotic translation initiation factor 4H | ENSMUSG00000040731 |
| 1452833_at | 19.42 | Rapgef2 | Rap guanine nucleotide exchange factor (GEF) 2 | ENSMUSG00000062232 |
| 1424358_at | 19.16 | Ube2e2 | Ubiquitin-conjugating enzyme E2E 2 (UBC4/5 homolog, yeast) | ENSMUSG00000058317 |
| 1434277_a_at | 19.08 | Ypel2 | Yippee-like 2 (Drosophila) | ENSMUSG00000018427 |
| 1416313_at | 18.85 | Mllt11 | Myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 11 | ENSMUSG00000053192 |
| 1434310_at | 18.78 | Bmpr2 | Bone morphogenic protein receptor, type II (serine/threonine kinase) | – |
| 1429839_a_at | 18.63 | Yaf2 | YY1 associated factor 2 | ENSMUSG00000022634 |
| 1435521_at | 18.5 | Msi2 | Musashi homolog 2 (Drosophila) | ENSMUSG00000069769 |
| 1457248_x_at | 18.47 | Hsd17b7 | Hydroxysteroid (17-beta) dehydrogenase 7 | ENSMUSG00000026675 |
| 1435807_at | 18.46 | Cdc42 | Cell division cycle 42 homolog (S. cerevisiae) | ENSMUSG00000006699 |
| 1416767_a_at | 18.45 | RIKEN cDNA 1110003E01 gene | ENSMUSG00000037822 | |
| 1433519_at | 18.43 | Nucks1 | Nuclear casein kinase and cyclin-dependent kinase substrate 1 | – |
| 1450021_at | 18.38 | Ubqln2 | Ubiquilin 2 | ENSMUSG00000050148 |
| 1437457_a_at | 18.38 | Mtpn | Myotrophin | ENSMUSG00000029840 |
| 1416082_at | 18.32 | Rab1 | RAB1, member RAS oncogene family | ENSMUSG00000020149 |
| 1448100_at | 18.31 | RIKEN cDNA 4833439L19 gene | ENSMUSG00000025871 | |
| 1447776_x_at | 18.29 | Rab6 | RAB6, member RAS oncogene family | ENSMUSG00000030704 |
| 1432198_at | 18.29 | RIKEN cDNA A230083H22 gene | ENSMUSG00000039126 | |
| 1448504_a_at | 18.28 | Cbx3 | Chromobox homolog 3 (Drosophila HP1 gamma) | ENSMUSG00000029836 ENSMUSG00000059647 |
| 1415971_at | 18.23 | Marcks | Myristoylated alanine rich protein kinase C substrate | ENSMUSG00000069662 |
| 1426776_at | 18.21 | Wasl | Wiskott–Aldrich syndrome-like (human) | ENSMUSG00000029684 |
| 1433540_x_at | 18.16 | Ppp1cb | Protein phosphatase 1, catalytic subunit, beta isoform | ENSMUSG00000014956 |
| 1426401_at | 18.15 | Ppp3ca | Protein phosphatase 3, catalytic subunit, alpha isoform | ENSMUSG00000028161 |
| 1419112_at | 18.12 | Nlk | Nemo like kinase | ENSMUSG00000017376 |
| 1423895_a_at | 18.11 | Cugbp2 | CUG triplet repeat, RNA-binding protein 2 | ENSMUSG00000002107 |
| 1423220_at | 18.07 | Eif4e | Eukaryotic translation initiation factor 4E | ENSMUSG00000028156 |
| 1458351_s_at | 18.06 | Klhl2 | Kelch-like 2, Mayven (Drosophila) | ENSMUSG00000031605 |
| 1440270_at | 18.05 | Fgf12 | Fibroblast growth factor 12 | ENSMUSG00000022523 |
| 1434082_at | 18.04 | Pctk2 | Serine/threonine-protein kinase PCTAIRE-2 | ENSMUSG00000020015 |
| 1434232_a_at | 18.04 | RIKEN cDNA 2610030H06 gene | ENSMUSG00000073131 | |
| 1428473_at | 18.03 | Ppp3cb | Protein phosphatase 3, catalytic subunit, beta isoform | ENSMUSG00000021816 |
| 1448184_at | 18.03 | Fkbp1a | FK506-binding protein 1a | ENSMUSG00000032966 |
| 1447669_s_at | 18.02 | Gng4 | Guanine nucleotide binding protein (G protein), gamma 4 subunit | ENSMUSG00000021303 |
| 1428970_at | 18.01 | Nat13 | N-acetyltransferase 13 | ENSMUSG00000022698 |
| 1437585_x_at | 17.99 | Zfp161 | Zinc finger protein 161 | ENSMUSG00000049672 |
| 1419246_s_at | 17.99 | Rab14 | RAB14, member RAS oncogene family | ENSMUSG00000026878 |
| 1438007_at | 17.94 | Expressed sequence AI851790 | ENSMUSG00000044071 | |
| 1424852_at | 17.87 | Mef2c | Myocyte enhancer factor 2C | – |
| 1436452_x_at | 17.82 | Tmed2 | Transmembrane emp24 domain trafficking protein 2 | ENSMUSG00000029390 ENSMUSG00000074460 |
| 1428416_at | 17.79 | RIKEN cDNA 3110050N22 gene | ENSMUSG00000043542 | |
| 1417377_at | 17.78 | Cadm1 | Cell adhesion molecule 1 | ENSMUSG00000032076 |
| 1433521_at | 17.75 | Ankrd13c | Ankyrin repeat domain 13c | ENSMUSG00000039988 |
| 1433751_at | 17.73 | Slc39a10 | Solute carrier family 39 (zinc transporter), member 10 | ENSMUSG00000025986 |
| 1424215_at | 17.72 | Fundc1 | FUN14 domain containing 1 | ENSMUSG00000025040 |
| 1434620_s_at | 17.72 | RIKEN cDNA 2610024E20 gene | ENSMUSG00000036501 | |
| 1421323_a_at | 17.71 | G3bp2 | GTPase activating protein (SH3 domain) binding protein 2 | ENSMUSG00000029405 |
| 1423309_at | 17.7 | Tgoln1 | Trans-golgi network protein | ENSMUSG00000056429 |
| 1426864_a_at | 17.7 | Ncam1 | Neural cell adhesion molecule 1 | ENSMUSG00000039542 |
| 1424683_at | 17.67 | RIKEN cDNA 1810015C04 gene | ENSMUSG00000022270 | |
| 1418067_at | 17.65 | Cfl2 | Cofilin 2, muscle | ENSMUSG00000062929 |
| 1437801_at | 17.62 | Morf4l1 | Mortality factor 4 like 1 | ENSMUSG00000062270 |
| 1428537_at | 17.62 | Csnk1a1 | Casein kinase 1, alpha 1 | ENSMUSG00000024576 |
| 1419971_s_at | 17.61 | Slc35a5 | Solute carrier family 35, member A5 | ENSMUSG00000022664 |
| 1417410_s_at | 17.61 | Prkci | Protein kinase C, iota | ENSMUSG00000037643 |
| 1417411_at | 17.61 | Nap1l5 | Nucleosome assembly protein 1-like 5 | ENSMUSG00000029804 |
| 1418436_at | 17.6 | Stx7 | Syntaxin 7 | ENSMUSG00000019998 |
| 1422748_at | 17.58 | Zeb2 | Zinc finger E-box binding homeobox 2 | ENSMUSG00000026872 |
| 1434820_s_at | 17.57 | Pkig | Protein kinase inhibitor, gamma | ENSMUSG00000035268 |
| 1415911_at | 17.57 | Impact | Imprinted and ancient | ENSMUSG00000024423 |
| 1454976_at | 17.55 | Sod2 | Superoxide dismutase 2, mitochondrial | ENSMUSG00000006818 |
| 1433986_at | 17.52 | cDNA sequence BC024659 | – | |
| 1437016_x_at | 17.46 | Rap2c | RAP2C, member of RAS oncogene family | ENSMUSG00000050029 |
| 1434106_at | 17.46 | Epm2aip1 | EPM2A (laforin) interacting protein 1 | ENSMUSG00000046785 |
| 1416008_at | 17.45 | Satb1 | Special AT-rich sequence binding protein 1 | ENSMUSG00000023927 |
| 1456177_x_at | 17.44 | Zfp706 | Zinc finger protein 706 | ENSMUSG00000062397 |
| 1416501_at | 17.44 | Pdpk1 | 3-phosphoinositide dependent protein kinase-1 | ENSMUSG00000024122 |
| 1423684_at | 17.44 | Hnrpk | Heterogeneous nuclear ribonucleoprotein K | ENSMUSG00000021546 |
| 1426924_at | 17.4 | Rc3h2 | Ring finger and CCCH-type zinc finger domains 2 | ENSMUSG00000075376 |
| 1437288_at | 17.38 | Impad1 | Inositol monophosphatase domain containing 1 | ENSMUSG00000066324 |
| 1429579_at | 17.37 | RIKEN cDNA 6330407I18 gene | – |
The top 70 most enriched mRNAs of the 700 putative HuD targets. The complete dataset is shown in Supplementary Table S1.
Table 3.
Predicted location of HuD-binding motifs in selected known and new HuD targets
| mRNA | Motif 1 | Motif 2 | Motif 3 |
|---|---|---|---|
| Known HuD targets | |||
| GAP-43 | 128 332 | – | – |
| MARCKS | 518 522 526 530 534 538 542 546 | 151 158 275 276 277 278 279 | 111 559 560 |
| Neuroserpin | 1801 1803 1805 1807 1809 1811 1813 1815 1817 1819 1821 1823 1825 1827 1829 1831 1833 1835 1837 1839 1841 1843 | 2073 | 615 |
| N-myc | – | – | 244 245 369 |
| p21 waf1 | 133 1211 | – | – |
| tau | – | – | – |
| VEGF | 939 1261 | – | 397 |
| p27 5UTR | 225 233 306 | 436 | – |
| p27 3UTR | – | 878 879 880 881 882 883 884 885 886 887 888 889 | 475 597 891 |
| New HuD targets | |||
| eIF4e | – | 563 657 | – |
| HuR | – | 1190 1199 | – |
| HuB | – | 456 | – |
| Fkbp1a | 976 | – | – |
| PPP1cb | 985 | – | – |
| PPP3ca | – | 15 | – |
| CaMkinIIα | 1475 1483 1485 1487 1491 1497 1512 3032 3034 3036 3038 3040 3042 3044 3046 3048 3050 3052 3054 3056 3058 3060 3062 3064 3066 3068 3070 | 3294 3298 3302 3306 3312 | – |
| Homer1 | – | – | 754 |
| Rab1 | 891 1247 1249 | – | – |
| N-CAM1 | 75 77 79 81 83 | 744 1108 1109 1317 1334 1351 1352 1353 1354 1355 1356 1357 1358 1359 2833 2837 | 2705 |
| Musashi 2 | 274 276 280 | 253 416 421 422 426 456 461 1558 1563 1570 1575 1764 1765 2258 2263 2760 3632 3638 3889 4248 4253 | 1141 1580 1767 1768 1871 1872 2955 2956 3642 4082 4083 |
The predicted location of each motif in the 3′ UTR of selected HuD targets. As described in the ‘Materials and Methods’ section, Pearl scripts were used to search for the location of each motif, allowing one mismatch in each consensus sequence.
3′ UTR sequence characteristics of HuD targets
The 5′ UTR, coding region (CR), and 3′ UTR sequences of all the mRNAs in the HuD target set, mouse genome, and Affymetrix 430 2.0 chip were extracted from Ensembl BioMart. Since ∼50% of the genome is expressed in the brain, we generated a set of forebrain-expressed mRNAs (see ‘Materials and Methods’ section) and used this set of transcripts to compare with HuD targets as well. As shown in Table 2, the average 3′ UTR length in HuD target mRNAs is surprisingly longer than in then genome, the Affymetrix chip and the forebrain set (HuD targets: 1909 nt versus 1121 in mouse genome, 1142 in the chip and 1325 in forebrain transcripts). Also, we found that the forebrain set had slightly longer 3′ UTRs and slightly shorter 5′ UTRs than the genome and chip sets. The increased 3′ UTR length of forebrain mRNAs relative to the entire mouse transcriptome is consistent with a previous report showing a positive correlation between the 3′ UTR length of mRNAs and their brain-specific expression (41) and with the observations that some brain-expressed mRNAs have longer 3′ UTRs than their counterparts in other tissues (42–44). Regarding the relative abundance of all four nucleotides in the sequence, we found that the 3′ UTRs in the HuD target dataset are slightly more U-rich than the three reference datasets. No difference between the Mus musculus genome and the genes present in the Affymetrix chip were observed, demonstrating that the Affymetrix 430 2.0 chip contains an unbiased representation of all mouse genes. Furthermore, aside for small differences in the lengths of the 5′ and 3′ UTRs, the properties of forebrain transcripts did not differ significantly from those of the other two reference datasets (Table 2, Figure 2).
Table 2.
Characteristics of the datasets used in the present studies
| 5′ UTR |
CR |
3′ UTR |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genome | Chip | Forebrain | HuD targets | Genome | Chip | Forebrain | HuD targets | Genome | Chip | Forebrain | HuD Targets | |
| Number of sequences | 12 397 | 11 036 | 8200 | 421 | 23 679 | 19 029 | 8688 | 608 | 17 918 | 16 359 | 8363 | 572 |
| Length | 380 | 367 | 274 | 359 | 1469 | 1605 | 1739 | 1593 | 1121 | 1142 | 1325 | 1,909 |
| A | 20.48 | 19.92 | 17.97 | 18.3 | 25.52 | 25.5 | 25.97 | 27.63 | 26.6 | 26.45 | 26.39 | 27.86 |
| U | 20.12 | 19.7 | 17.93 | 18.85 | 22.8 | 21.95 | 21.66 | 22.68 | 28.76 | 28.74 | 29.28 | 32.95 |
| C | 29.23 | 29.67 | 30.77 | 30.9 | 25.73 | 25.97 | 25.52 | 23.92 | 22.55 | 22.65 | 22.27 | 19.11 |
| G | 30.18 | 30.71 | 33.34 | 31.95 | 25.95 | 26.58 | 26.85 | 25.77 | 22.08 | 22.16 | 22.06 | 20.08 |
3′ UTR sequences were retrieved from Ensembl BioMart and datasets were made non-redundant by Gene ID. The datasets were analyzed by scripts written in Perl v5.8.8 and using BioPerl modules.
Figure 2.
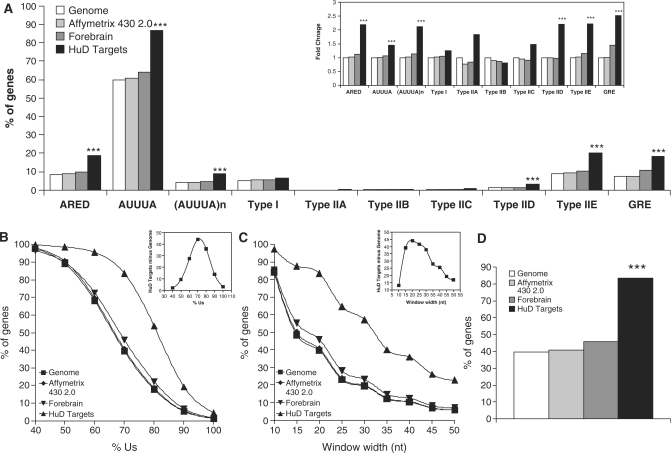
ARE frequency and U content in HuD target genes. (A) The presence of the different AREs was analyzed in 3′ UTR sequences of the entire mouse genome (white bars), the genes present in the Affymetrix 430 2.0 chip (light gray bars), a set of forebrain expressed transcripts (dark gray bars) and in the HuD target dataset (black bars). Inset shows the same data normalized to the frequency in the genome. (B) Analysis of the percentage of Us in 20 nt long stretches in the 3′ UTRs of the mouse genome, Affymetrix 430 2.0 chip, and HuD targets. Inset shows the difference between HuD targets and mouse genome. (C) Percentage of genes with segments of 70% Us as a function of stretch length. Inset shows the difference between HuD targets and mouse genome. (D) Frequency of genes with 20 nt long fragments in the 3′ UTR containing at least 70% Us in all the datasets described in panel A. ***P < 0.0001 after a Chi-square test.
Subsequent analyses examined the frequency of known mRNA instability elements in the 3′ UTRs of mRNAs in three reference transcripts sets and the HuD target dataset. As shown in Figure 2A, HuD target mRNAs contained significantly higher frequencies of these motifs. Because some of the motifs were very rare, to easily visualize differences the data were normalized to the frequency in the genome (Figure 2A, inset). Of the known instability-conferring sequences, the ARED motif, defined by the following regular expression [AU][AU][AU]U(AUUUA)UUU[AU] with 1 mismatch allowed in the pentamer flanking regions (6) showed an approximate 2-fold enrichment in the HuD target dataset. However, this motif was present in only 20% of the targets. We then examined the presence of the AUUUA pentamer and found a modest but statistically significant increase in the frequency of this sequence in the HuD target dataset. However, given that this motif is present at a high frequency in the genome (∼60% of the genes) and a single AUUUA motif by itself does not constitute an ARE, this sequence is an unlikely candidate for a HuD-recognition motif. The search for ARE motifs was then restricted to overlapping AUUUAs [(AUUUA)n] or AUUUA motifs embedded in a U-rich region (Type I ARE) defined arbitrarily as a 20 nt long stretch with at least 60% U. Overlapping AUUUA pentamers were present at high levels in the HuD target dataset relative to the three reference datasets, but they only accounted for ∼10% of the HuD targets. In contrast, type I AREs were not enriched in this set. Analysis of the type II ARE sub-categories proposed by Wilusz and colleagues (1) revealed that that most of these sub-categories were barely represented in the genome. However, sub-categories IID and IIE were more frequent in the HuD target 3′ UTRs. Finally, we searched for the recently reported G-rich element [GRE; (9)] and found that the frequency of GREs was increased among HuD targets. Taking together all the known instability motifs, only 42.48% of all HuD targets could be explained by these sequences suggesting that other elements are involved in HuD target recognition.
The common feature of the previously analyzed ARE sequences is a high U content. Therefore, we performed a detailed analysis of the U content in the 3′ UTR of HuD targets. Figure 2B shows the frequency of 20 nt long stretches with different percentages of U. As the percentage of U increases, the fraction of genes containing these sequences diminishes, however, the HuD target dataset shows a higher number of mRNAs with high U content. When the difference between percentage of genes in the HuD targets and the Mus musculus genome was plotted versus percentage of U (Figure 2B, inset), the curve showed a peak at 70% U, suggesting that this percentage is needed for HuD binding. Figure 2C presents a similar analysis, but with a fixed percentage of U set at 70 and variation in the length of the window. The results show an expected decrease in the percentage of genes meeting the criteria as the window gets longer, with the maximum discrimination between HuD targets and the mouse genome at a window length of 20 nts (Figure 2C, inset). Analysis of the frequencies of mRNAs having 20 nt long stretches with 70% U in the 3′ UTR (Figure 2D) revealed that 80% of the HuD targets have this characteristic, a value that was highly significant when compared to the genome, chip and forebrain datasets.
Discovery of new HuD-binding motifs
Because ARE and GRE sequences only explained a fraction of the motifs present in HuD targets and the 20 nt and 70% U criteria is not sequence-specific, subsequent studies searched for new HuD-binding motifs. As shown in Figure 3A, the 3′ UTR sequences of the 72 most enriched HuD targets were used to extract 25 nt long fragments containing the sequence YUNNYUY in the middle. This sequence reflects the minimal binding motif that was derived from modeling studies of the crystal structure of the complex of RRMs I and II in HuD with class I and II AREs (34). Using this filter to remove physiologically irrelevant sequences 1735 fragments were obtained, and this new dataset was used as a training set to search for new HuD-binding motifs. Using a multiple expectation maximization algorithm implemented in the MEME software (35,36), we found three motifs with statistically significant E-values, which are represented in Figure 3B by WebLogo graphics (37). Supplementary Table 2 shows the probability matrix for each individual motif. The first motif is pyrimidine-rich with a preponderance of C and a lower U content. This motif is analogous to a C-rich instability element that is present in α-globin and other mRNAs and that is recognized by the poly(C)-binding protein (45,46). The second motif is U-rich with some interspersed Gs, and very similar to the GRE motif (9) and the binding motif of sex-lethal, another homolog of Hu proteins in Drosophila melanogaster (47). The last motif is also U-rich with several As and thus is comparable to the classical type I and II AREs.
If these motifs are indeed recognized by HuD, mRNAs carrying these motifs should be present at higher frequency in the HuD target dataset than in the three reference datasets (mouse genome, Affymetrix chip and forebrain transcripts). As shown in Figure 3D the frequency of motif 1 is increased in the 5′ UTR and the 3′ UTR of HuD targets but not in the coding region. Motifs 2 and 3 showed a very similar pattern with highly significant increases in the 3′ UTR and a slight increase in the 5′ UTR and coding regions (Figures 3E and F). Analysis of the distribution of the three motifs (Figure 3C) showed that ∼80% (456 out of 572) of the target mRNAs have at least one of the motifs, ∼10% have all three motifs in their 3′ UTR and 33% have two motifs. Subsequent analyses mapped the localization of the three motifs in a set of known and novel HuD targets. As shown in Table 3, with the exception of tau, one of the mRNAs identified as a target of HuD in a previous study (32), all the known HuD targets contain at least one of the motifs. Some of the targets such as Musashi 2, N-CAM1, Neuroserpin and CaMkinIIα mRNAs have a large number of motifs while others such as Homer 1, Fkbp1a, PPP1cb and PPP3ca have only one motif per 3′ UTR. In addition, one of the known targets of HuD, the p27 mRNA, contains HuD-binding motifs both in the 5′ and 3′ UTRs, consistent with a role of HuD in translation of this mRNA (48). Altogether these results indicate that the majority of HuD targets can be explained by the presence of these new motifs.
Affinity of HuD for the three motifs
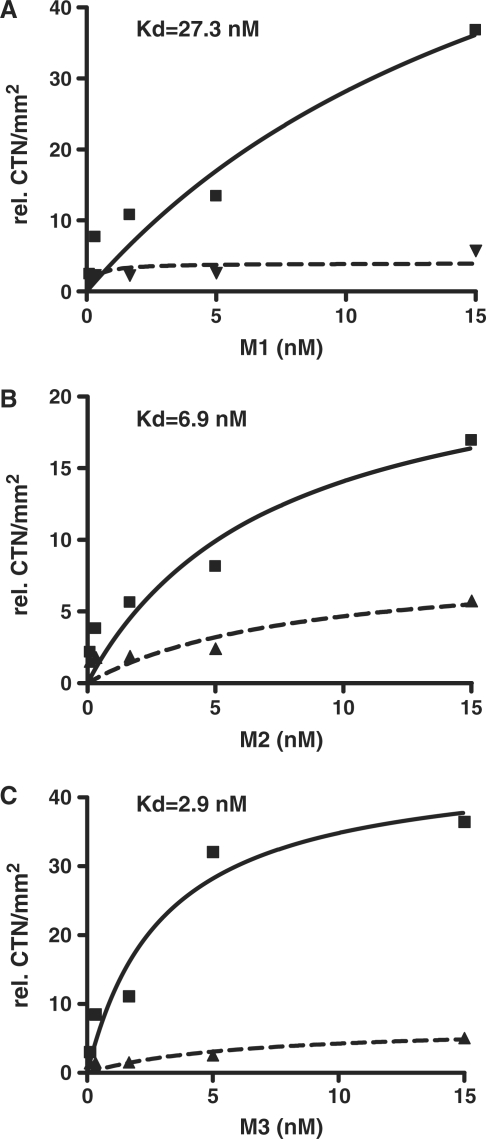
To directly test whether HuD binds these new motifs and to determine the binding affinities we performed in vitro binding assays using recombinant protein and synthetic radiolabeled ribo-oligonucleotides corresponding to the most likely sequence of each of the three motifs. The specific binding of HuD to each motif was calculated by subtracting non-specific binding, which was assessed by the addition of 100 times molar excess of unlabeled ribo-oligoribonucleotide. The affinities of recombinant HuD for each motif were then calculated using non-linear regression analyses. As shown in Figure 4, HuD interacts with different affinities with each of the three sequences, with motif 3 showing the highest affinity followed by motif 2 and the C-rich motif 1 showing the least affinity (Kd = 2.9 nM motif 3, 6.9 nM motif 2 and 27.3 nM motif 1). Taken together, these data provides experimental validation of the in silico identified HuD-binding motifs.
Figure 4.
Binding affinities of recombinant HuD for the three motifs. Panels show binding curves of motif 1 (A), motif 2 (B) and motif 3 (C). In vitro binding assays were performed with increasing amounts of radiolabeled oligoribonucleotides corresponding to the most likely sequence in each motif (CCCUCCCUCUCUC for motif 1, UUUUGUUUUGUUU for motif 2 and UUUUUUUUUUAAA for motif 3) and fixed amounts GST-HuD as indicated in ‘Materials and Methods’ section. Non-specific binding was measured using a 100 molar excess of cold oligoribonucleotides. Binding curves of total (solid line) and non-specific (dashed lines) used to calculate the Kd for each motifs.
Enrichment of HuD-binding motifs in genes upregulated in HuD-Tg mice
To further validate the biological significance of the three binding motifs we determined the frequency of each sequence in a dataset of genes upregulated in the brains of HuD overexpressor mice (HuD-Tg). As described previously, HuD-Tg mice have increased levels of HuD in neurons of the forebrain (14) and increased levels of GAP-43 and AChE mRNAs, two well characterized HuD targets (14,19). Microarray gene expression analysis of the HuD-Tg mice forebrain identified 646 probe sets representing 419 transcripts that had a ‘present’ call and whose expression was significantly upregulated in these mice (P < 0.05, t-test). Sequence analyses of this set of mRNAs revealed that the frequency of the three motifs (motif 1, 36.3%; motif 2, 36.3% and motif 3, 21.2%) was significantly increased in their 3′ UTRs (Figure 5). Also, we found that the 5′ UTRs of upregulated mRNAs had increased frequencies of motif 1 but decreased frequencies of motifs 2 and 3. Finally, our finding that many upregulated transcripts did not contain HuD-binding motifs suggest that some of the observed gene expression changes are indirect events regulated by HuD targets rather that direct HuD effects, which is expected in animals with constitutive HuD overexpression.
Figure 5.
Increased frequency of HuD-binding motifs in mRNAs upregulated in the brains of HuD overexpressor mice. (A) Frequency of HuD-binding motif 1 in the 5′ UTR, CR and 3′ UTR of mouse genome (white bars), Affymetrix 430 2.0 chip genes (light grey bars), forebrain transcripts (dark grey bars) and mRNAs upregulated in HuD-Tg mice (black bars). (B) Same analysis for HuD-binding motif 2. (C) Frequency of motif 3 in the same gene datasets. ***P < 0.0001 HuD-Tg mice versus each of the three reference sets (genome, chip and forebrain), (a) P < 0.02 versus genome and chip, (b) P < 0.05 versus genome and chip and (c) P < 0.05 versus all three reference sets.
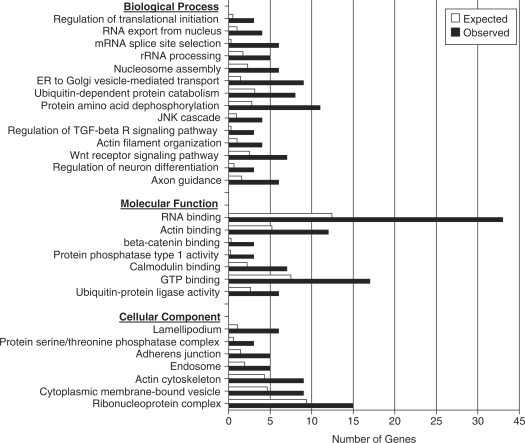
Gene ontology analysis of HuD targets
It has been proposed that mRNAs coordinately regulated by the same RBP comprise a post-transcriptional operon or ‘RNA regulon’ (49). To gain a further understanding of the biological processes regulated by HuD, subsequent studies examined the Gene Ontology distribution of HuD targets (Figure 6). Several gene ontology categories were significantly enriched in HuD targets relative to the genome. Amongst these categories were axon guidance and regulation of neuron differentiation, two processes that are known to be regulated by HuD in several models. Other biological processes that were targeted by HuD were unexpected such as regulation of translation initiation, RNA processing, and protein amino acid dephosphorylation. Likewise, we found that molecular categories such as RNA-binding, protein phosphatase activity type 1 activity, GTP-binding and actin-binding were significantly enriched in HuD targets. Analysis of cellular components revealed that lamellipodium, actin cytoskeleton and ribonucleoprotein complex, all components related to growth cone activity and axonal growth as well as neural plasticity, were significantly overpopulated with HuD targets. Finally, to put the set of HuD targets in a biological context we searched for canonical cellular pathways containing multiple targets. Among these, we found that the developmental wnt signaling pathway and the long-term potentiation pathway contain a number of HuD targets (Supplementary Figure S2). Likewise, analysis of cellular networks using Metacore software revealed that several networks such as those centered on β-actin and the translation factor eIF4E (Supplementary Figure 3) were overpopulated by HuD targets. These examples highlight the importance of HuD in the post-transcriptional control of gene regulation during neural development and synaptic plasticity.
Figure 6.
Gene Ontology analysis of HuD targets. The graph shows the number of HuD target genes observed in each gene ontology category (black bars) and the expected number in the genome (white bars). All the categories presented in the graph have P-values < 0.01.
DISCUSSION
The identification of the host of cellular targets regulated by specific RBPs is critical for understanding the biological function of these regulatory proteins. In this study, we used a two-step mRNA–RBP complex purification method and genome-wide bioinformatics analyses to identify the targets of the ELAV-like protein HuD, define its recognition motifs and characterize the cellular pathways regulated by this protein. HuD is expressed in neurons where it promotes axonal growth during nervous system development and participates in synaptic plasticity mechanisms in the mature brain (15,17,20,21,50–53). Not only is HuD present in neuronal cell bodies (14,18), but it is also localized to growth cones (54) and dendrites (51), two regions whereby neurons communicate with each other and the environment and sites of local post-transcriptional regulation (55,56). Consistent with the known functions of HuD, we found that its target mRNAs encoded proteins involved in processes critical for neuronal differentiation such as axon guidance, actin reorganization and wnt signaling. In addition, HuD targets were found to participate in novel pathways such as protein phosphatase regulation, ubiquitin ligation and mRNA transport, processing and translation.
The methods used to isolate mRNAs bound to RNA-BPs typically utilize either immunoprecipitation of RNA–protein complexes, with or without prior cross-linking, or in vitro pull down assays of purified RNA with recombinant RNA-BPs. The problem with the latter method is that it does not take into consideration that mRNAs that interact in vitro with specific RNA-BPs may never do so in vivo as they could be localized to different subcellular fractions or different cell types in the tissue. To avoid this problem, Keene and colleagues devised a method to isolate mRNP complexes under conditions that preserve in vivo RNA-protein interactions (25,26). Given that HuD is known to interact with other RBPs including other members of the Hu protein family (30–32), it is possible that some of the mRNAs in the initial IP could be bound to other protein in the mRNP. Therefore, to isolate directly bound mRNAs, transcripts in the HuD IP were further purified using in vitro pull downs with recombinant protein and HuD targets were defined as the mRNAs with highest enrichment values. On average, HuD targets were found to contain longer 3′ UTRs comprising an increased proportion of ARE sequences and HuD-binding motifs. These motifs were also significantly enriched in a dataset of mRNAs upregulated in the forebrain of HuD overexpressor mice, validating the biological significance of these sequences.
All four ELAV-like Hu proteins have been shown to interact with ARE sequences via their first and second RRMs, while the third RRM binds to long poly A tails (57,58). Previous predictions of Hu protein binding motifs have been based upon the consensus sequences of target mRNAs (27,59). In addition to this criterion, in this study, we took advantage of the known crystal structure of the complex of HuD’s RRM I and II and Type I AREs and restricted our search to motifs that matched its seven nucleotide (YUNNYUY) minimal consensus sequence (34). Based on this sequence restriction and the 3′ UTR sequences of the most enriched targets, three new HuD recognition motifs were identified. Consistent with the ARE-binding motifs of other ELAV-like proteins (27,59), two of the HuD-binding motifs (motifs 2 and 3) are U-rich; with motif 2 having interspersed Gs similar to the sex-lethal binding site (47) and the recently identified GRE (9) and motif 3 matching class I and II ARE sequences. HuD was found to bind with high affinity to ribo-oligonucleotides containing motifs 2 and 3 sequences. In contrast, binding to motif 1, which is C-rich with interspersed Us, was about one order of magnitude lower than that of motif 3. Although none of the ELAV-binding proteins were shown to recognize pure C sequences, HuR was found to interact with a CU-rich element in the androgen receptor mRNA (46).
Analysis of the distribution of all three HuD-binding motifs in target mRNAs showed that these sequences were primarily localized to the 3′ UTR. Interestingly, these motifs were also present at higher frequencies in the 5′ UTRs of target mRNAs. Although the vast majority of instability conferring sequences described so far mapped to the 3′ UTR, a few of them were found in the 5′ UTR and coding region (10,11). Furthermore, for at least one mRNA, that encoding the cdk inhibitor p27, HuD binds to an IRES like sequence in its 5′ UTR repressing translation (48). As shown in Table 3, we found several putative HuD-binding motifs in both the 5′ UTR and 3′ UTRs of p27 mRNA. In addition to HuD, HuB was shown to increase translation of neurofilament M (60) and HuR to block translation of IGF-I receptor (61). Finally, not only is HuD co-localized with ribosomes in neuronal cell bodies (18), growth cones (54) and dendrites (51) but it also, as shown by a recent study, co-localizes with two important translational regulators, the poly(A)-binding protein (PABP) and the cap-binding protein (eIF4E) (62). Altogether, these results suggest that in addition to controlling mRNA stability, in some instances HuD and other Hu proteins could also regulate the rate of translation of specific mRNAs by interacting with sequences in the 5′ UTR.
Gene ontology and biological pathway analyses of putative HuD targets revealed several interesting features of this dataset. For instance, ∼7% of HuD targets were other RBPs, including HuD itself, HuB, HuR, polypyrimidine tract binding protein 2 (Ptbp2), CUG triplet repeat-binding protein 2 (Cugbp2), cytoplasmic polyadenylation element binding protein 3 (Cpeb3), poly(rC) binding protein 2 (Pcbp2), Pumillio 2, Staufen 2, Musashi 2 as well as several hnRNP proteins and splicing factors. Our finding that HuD binds its own mRNA and those of other ELAV-like proteins is similar to that observed for Drosophila ELAV, which has been shown to bind and downregulate the expression of its own mRNA (63). These findings also correlate with the decreases in HuB and HuR mRNAs observed in the brains of our HuD overexpressor mice (14). Likewise, HuR was shown to associate with its cognate mRNA and with mRNAs encoding other RBPs (40,64). Additional clues on the importance of HuD in gene regulation comes from our findings that HuD targets also encode proteins controlling translational initiation, RNA export from the nucleus, mRNA splice site selection and nucleosome assembly (Figure 6). These observations support a model in which HuD and other RNA-BPs form a complex and highly integrated network of post-transcriptional regulators.
In agreement with the role of HuD in neural development and axonal outgrowth (24,53,65–68), gene ontology analyses also revealed that multiple HuD targets are associated with axon guidance and related processes such as regulation of actin dynamics and vesicle trafficking. Furthermore, other important developmental pathways such as those involving wnt, TGF-β and β-catenin signaling are also overpopulated with HuD targets. Finally, concurring with the known biological properties of HuD and other Hu proteins in synaptic plasticity (20,21) many HuD targets encode proteins implicated in these processes. For instance, HuD targets include protein phosphatases such as PPP1 and PPP3 (aka PP2B or calcineurin) and a regulator of PPP3 activity (FKBP1 a.k.a., FKB12), which are known to participate in long-term potentiation and long-term depression, two different forms of synaptic plasticity (69). Additionally, another HuD target, the metabotropic glutamate receptor 5 (mGluR5), is known to control localized protein synthesis in dendrites during activity-dependent synaptic remodeling (70). Along these lines, two recent studies demonstrated the activity-dependent transport of HuD to neuronal processes and corresponding increased binding of HuD to the mRNAs for the plasticity associated proteins homer 1a, GAP-43 and CaMKinIIα (62,71).
In conclusion, our results demonstrate that the ELAV-like protein HuD binds to three novel motifs in the 3′ UTRs of its target mRNAs and that multiple HuD targets encode proteins involved in post-transcriptional gene regulation, neuronal differentiation and synaptic remodeling. These findings support a post-transcriptional operon model in which HuD interacts with multiple mRNAs to regulate these complex biological processes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (grant number NS30255 to N.P.B.). Funding for open access charge: National Institutes of Health (grant NS30255 to N.P.B.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr Rebecca Hartley for critical reading of the manuscript and Dr Jack Keene and Neel Mukherjee for their help with the IP data analyses.
REFERENCES
- 1.Wilusz CJ, Wormington M, Peltz SW. The cap-to-tail guide to mRNA turnover. Nat. Rev. Mol. Cell Biol. 2001;2:237–246. doi: 10.1038/35067025. [DOI] [PubMed] [Google Scholar]
- 2.Bolognani F, Perrone-Bizzozero NI. RNA-protein interactions and control of mRNA stability in neurons. J. Neurosci. Res. 2008;86:481–489. doi: 10.1002/jnr.21473. [DOI] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 4.Shaw G, Kamen R. A conserved AU sequence from the 3′ untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell. 1986;46:659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- 5.Chen CY, Shyu AB. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem. Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- 6.Bakheet T, Williams BR, Khabar KS. ARED 3.0: the large and diverse AU-rich transcriptome. Nucleic Acids Res. 2006;34:D111–D114. doi: 10.1093/nar/gkj052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marquis J, Paillard L, Audic Y, Cosson B, Danos O, Le Bec C, Osborne HB. CUG-BP1/CELF1 requires UGU-rich sequences for high-affinity binding. Biochem. J. 2006;400:291–301. doi: 10.1042/BJ20060490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moraes KC, Wilusz CJ, Wilusz J. CUG-BP binds to RNA substrates and recruits PARN deadenylase. RNA. 2006;12:1084–1091. doi: 10.1261/rna.59606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vlasova IA, Tahoe NM, Fan D, Larsson O, Rattenbacher B, Sternjohn JR, Vasdewani J, Karypis G, Reilly CS, Bitterman PB, et al. Conserved GU-rich elements mediate mRNA decay by binding to CUG-binding protein 1. Mol. Cell. 2008;29:263–270. doi: 10.1016/j.molcel.2007.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen CY, Del Gatto-Konczak F, Wu Z, Karin M. Stabilization of interleukin-2 mRNA by the c-Jun NH2-terminal kinase pathway. Science. 1998;280:1945–1949. doi: 10.1126/science.280.5371.1945. [DOI] [PubMed] [Google Scholar]
- 11.Grosset C, Chen CY, Xu N, Sonenberg N, Jacquemin-Sablon H, Shyu AB. A mechanism for translationally coupled mRNA turnover: interaction between the poly(A) tail and a c-fos RNA coding determinant via a protein complex. Cell. 2000;103:29–40. doi: 10.1016/s0092-8674(00)00102-1. [DOI] [PubMed] [Google Scholar]
- 12.Szabo A, Dalmau J, Manley G, Rosenfeld M, Wong E, Henson J, Posner JB, Furneaux HM. HuD, a paraneoplastic encephalomyelitis antigen, contains RNA-binding domains and is homologous to Elav and Sex-lethal. Cell. 1991;67:325–333. doi: 10.1016/0092-8674(91)90184-z. [DOI] [PubMed] [Google Scholar]
- 13.Akamatsu W, Fujihara H, Mitsuhashi T, Yano M, Shibata S, Hayakawa Y, Okano HJ, Sakakibara S, Takano H, Takano T, et al. The RNA-binding protein HuD regulates neuronal cell identity and maturation. Proc. Natl Acad. Sci. USA. 2005;102:4625–4630. doi: 10.1073/pnas.0407523102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bolognani F, Tanner DC, Merhege M, Deschenes-Furry J, Jasmin B, Perrone-Bizzozero NI. In vivo post-transcriptional regulation of GAP-43 mRNA by overexpression of the RNA-binding protein HuD. J. Neurochem. 2006;96:790–801. doi: 10.1111/j.1471-4159.2005.03607.x. [DOI] [PubMed] [Google Scholar]
- 15.Bolognani F, Tanner DC, Nixon S, Okano HJ, Okano H, Perrone-Bizzozero NI. Coordinated expression of HuD and GAP-43 in hippocampal dentate granule cells during developmental and adult plasticity. Neurochem. Res. 2007;32:2142–2151. doi: 10.1007/s11064-007-9388-8. [DOI] [PubMed] [Google Scholar]
- 16.Tanner DC, Qiu S, Bolognani F, Partridge LD, Weeber EJ, Perrone-Bizzozero NI. Alterations in mossy fiber physiology and GAP-43 expression and function in transgenic mice overexpressing HuD. Hippocampus. 2008;18:814–823. doi: 10.1002/hipo.20442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bolognani F, Qiu S, Tanner DC, Paik J, Perrone-Bizzozero NI, Weeber EJ. Associative and spatial learning and memory deficits in transgenic mice overexpressing the RNA-binding protein HuD. Neurobiol. Learn. Mem. 2007;87:635–643. doi: 10.1016/j.nlm.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 18.Anderson KD, Merhege MA, Morin M, Bolognani F, Perrone-Bizzozero NI. Increased expression and localization of the RNA-binding protein HuD and GAP-43 mRNA to cytoplasmic granules in DRG neurons during nerve regeneration. Exp. Neurol. 2003;183:100–108. doi: 10.1016/s0014-4886(03)00103-1. [DOI] [PubMed] [Google Scholar]
- 19.Deschenes-Furry J, Mousavi K, Bolognani F, Neve RL, Parks RJ, Perrone-Bizzozero NI, Jasmin BJ. The RNA-binding protein HuD binds acetylcholinesterase mRNA in neurons and regulates its expression after axotomy. J. Neurosci. 2007;27:665–675. doi: 10.1523/JNEUROSCI.4626-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perrone-Bizzozero N, Bolognani F. Role of HuD and other RNA-binding proteins in neural development and plasticity. J. Neurosci. Res. 2002;68:121–126. doi: 10.1002/jnr.10175. [DOI] [PubMed] [Google Scholar]
- 21.Pascale A, Amadio M, Quattrone A. Defining a neuron: neuronal ELAV proteins. Cell Mol. Life Sci. 2008;65:128–140. doi: 10.1007/s00018-007-7017-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chung S, Eckrich M, Perrone-Bizzozero N, Kohn DT, Furneaux H. The Elav-like proteins bind to a conserved regulatory element in the 3′-untranslated region of GAP-43 mRNA. J. Biol. Chem. 1997;272:6593–6598. doi: 10.1074/jbc.272.10.6593. [DOI] [PubMed] [Google Scholar]
- 23.Beckel-Mitchener AC, Miera A, Keller R, Perrone-Bizzozero NI. Poly(A) tail length-dependent stabilization of GAP-43 mRNA by the RNA-binding protein HuD. J. Biol. Chem. 2002;277:27996–28002. doi: 10.1074/jbc.M201982200. [DOI] [PubMed] [Google Scholar]
- 24.Anderson KD, Morin MA, Beckel-Mitchener A, Mobarak CD, Neve RL, Furneaux HM, Burry R, Perrone-Bizzozero NI. Overexpression of HuD, but not of its truncated form HuD I+II, promotes GAP-43 gene expression and neurite outgrowth in PC12 cells in the absence of nerve growth factor. J. Neurochem. 2000;75:1103–1114. doi: 10.1046/j.1471-4159.2000.0751103.x. [DOI] [PubMed] [Google Scholar]
- 25.Tenenbaum SA, Carson CC, Lager PJ, Keene JD. Identifying mRNA subsets in messenger ribonucleoprotein complexes by using cDNA arrays. Proc. Natl Acad. Sci. USA. 2000;97:14085–14090. doi: 10.1073/pnas.97.26.14085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tenenbaum SA, Lager PJ, Carson CC, Keene JD. Ribonomics: identifying mRNA subsets in mRNP complexes using antibodies to RNA-binding proteins and genomic arrays. Methods. 2002;26:191–198. doi: 10.1016/S1046-2023(02)00022-1. [DOI] [PubMed] [Google Scholar]
- 27.Lopez de Silanes I, Zhan M, Lal A, Yang X, Gorospe M. Identification of a target RNA motif for RNA-binding protein HuR. Proc. Natl Acad. Sci. USA. 2004;101:2987–2992. doi: 10.1073/pnas.0306453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Halbeisen RE, Galgano A, Scherrer T, Gerber AP. Post-transcriptional gene regulation: from genome-wide studies to principles. Cell Mol. Life Sci. 2008;65:798–813. doi: 10.1007/s00018-007-7447-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keene JD, Tenenbaum SA. Eukaryotic mRNPs may represent posttranscriptional operons. Mol. Cell. 2002;9:1161–1167. doi: 10.1016/s1097-2765(02)00559-2. [DOI] [PubMed] [Google Scholar]
- 30.Kasashima K, Sakashita E, Saito K, Sakamoto H. Complex formation of the neuron-specific ELAV-like Hu RNA-binding proteins. Nucleic Acids Res. 2002;30:4519–4526. doi: 10.1093/nar/gkf567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saito K, Fujiwara T, Katahira J, Inoue K, Sakamoto H. TAP/NXF1, the primary mRNA export receptor, specifically interacts with a neuronal RNA-binding protein HuD. Biochem. Biophys. Res. Commun. 2004;321:291–297. doi: 10.1016/j.bbrc.2004.06.140. [DOI] [PubMed] [Google Scholar]
- 32.Atlas R, Behar L, Elliott E, Ginzburg I. The insulin-like growth factor mRNA binding-protein IMP-1 and the Ras-regulatory protein G3BP associate with tau mRNA and HuD protein in differentiated P19 neuronal cells. J. Neurochem. 2004;89:613–626. doi: 10.1111/j.1471-4159.2004.02371.x. [DOI] [PubMed] [Google Scholar]
- 33.Chung S, Jiang L, Cheng S, Furneaux H. Purification and properties of HuD, a neuronal RNA-binding protein. J. Biol. Chem. 1996;271:11518–11524. doi: 10.1074/jbc.271.19.11518. [DOI] [PubMed] [Google Scholar]
- 34.Wang X, Tanaka Hall TM. Structural basis for recognition of AU-rich element RNA by the HuD protein. Nat. Struct. Biol. 2001;8:141–145. doi: 10.1038/84131. [DOI] [PubMed] [Google Scholar]
- 35.Bailey TL, Gribskov M. Combining evidence using p-values: application to sequence homology searches. Bioinformatics. 1998;14:48–54. doi: 10.1093/bioinformatics/14.1.48. [DOI] [PubMed] [Google Scholar]
- 36.Bailey TL, Williams N, Misleh C, Li WW. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34:W369–W373. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang B, Schmoyer D, Kirov S, Snoddy J. GOTree Machine (GOTM): a web-based platform for interpreting sets of interesting genes using Gene Ontology hierarchies. BMC Bioinformatics. 2004;5:16. doi: 10.1186/1471-2105-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang B, Kirov S, Snoddy J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005;33:W741–W748. doi: 10.1093/nar/gki475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mukherjee N, Lager PJ, Freidersdorf M, Thompson M, Keene JD. Coordinated posttranscriptional mRNA population dynamics during T-Cell activation. Mol. Syst. Biol. 2009;5:288. doi: 10.1038/msb.2009.44. Epub 28 July 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Okazaki N, Imai K, Kikuno RF, Misawa K, Kawai M, Inamoto S, Ohara R, Nagase T, Ohara O, Koga H. Influence of the 3′-UTR-length of mKIAA cDNAs and their sequence features to the mRNA expression level in the brain. DNA Res. 2005;12:181–189. doi: 10.1093/dnares/dsi001. [DOI] [PubMed] [Google Scholar]
- 42.Wedgwood S, Lam WK, Pinchin KM, Markham AF, Cartwright EJ, Coletta PLMamm. Characterization of a brain-selective transcript of the Adenomatous polyposis coli tumor suppressor gene. Genome. 2000;11:1150–1153. doi: 10.1007/s003350010201. [DOI] [PubMed] [Google Scholar]
- 43.Suda S, Nibuya M, Suda H, Takamatsu K, Miyazaki T, Nomura S, Kawai N. Potassium channel mRNAs with AU-rich elements and brain-specific expression. Biochem. Biophys. Res. Commun. 2002;291:1265–1271. doi: 10.1006/bbrc.2002.6592. [DOI] [PubMed] [Google Scholar]
- 44.Pelka GJ, Watson CM, Christodoulou J, Tam PP. Distinct expression profiles of Mecp2 transcripts with different lengths of 3′UTR in the brain and visceral organs during mouse development. Genomics. 2005;85:441–452. doi: 10.1016/j.ygeno.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 45.Kong J, Ji X, Liebhaber SA. The KH-domain protein alpha CP has a direct role in mRNA stabilization independent of its cognate binding site. Mol. Cell Biol. 2003;23:1125–1134. doi: 10.1128/MCB.23.4.1125-1134.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yeap BB, Voon DC, Vivian JP, McCulloch RK, Thomson AM, Giles KM, Czyzyk-Krzeska MF, Furneaux H, Wilce MC, Wilce JA, et al. Novel binding of HuR and poly(C)-binding protein to a conserved UC-rich motif within the 3′-untranslated region of the androgen receptor messenger RNA. J. Biol. Chem. 2002;277:27183–27192. doi: 10.1074/jbc.M202883200. [DOI] [PubMed] [Google Scholar]
- 47.Handa N, Nureki O, Kurimoto K, Kim I, Sakamoto H, Shimura Y, Muto Y, Yokoyama S. Structural basis for recognition of the tra mRNA precursor by the sex-lethal protein. Nature. 1999;398:579–585. doi: 10.1038/19242. [DOI] [PubMed] [Google Scholar]
- 48.Kullmann M, Gopfert U, Siewe B, Hengst L. ELAV/Hu proteins inhibit p27 translation via an IRES element in the p27 5′UTR. Genes Dev. 2002;16:3087–3099. doi: 10.1101/gad.248902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Keene JD. RNA regulons: coordination of post-transcriptional events. Nat. Rev. Genet. 2007;8:533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- 50.Quattrone A, Pascale A, Nogues X, Zhao W, Gusev P, Pacini A, Alkon DL. Posttranscriptional regulation of gene expression in learning by the neuronal ELAV-like mRNA-stabilizing proteins. Proc. Natl Acad. Sci. USA. 2001;98:11668–11673. doi: 10.1073/pnas.191388398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bolognani F, Merhege MA, Twiss J, Nora IP-B. Dendritic localization of the RNA-binding protein HuD in hippocampal neurons: association with polysomes and upregulation during contextual learning. Neurosci. Lett. 2004;371:152–157. doi: 10.1016/j.neulet.2004.08.074. [DOI] [PubMed] [Google Scholar]
- 52.Pascale A, Gusev PA, Amadio M, Dottorini T, Govoni S, Alkon DL, Quattrone A. Increase of the RNA-binding protein HuD and posttranscriptional up-regulation of the GAP-43 gene during spatial memory. Proc. Natl Acad. Sci. USA. 2004;101:1217–1222. doi: 10.1073/pnas.0307674100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Anderson KD, Sengupta J, Morin M, Neve RL, Valenzuela CF, Perrone-Bizzozero NI. Overexpression of HuD accelerates neurite outgrowth and increases GAP-43 mRNA expression in cortical neurons and retinoic acid-induced embryonic stem cells in vitro. Exp. Neurol. 2001;168:250–258. doi: 10.1006/exnr.2000.7599. [DOI] [PubMed] [Google Scholar]
- 54.Smith CL, Afroz R, Bassell GJ, Furneaux HM, Perrone-Bizzozero NI, Burry RW. GAP-43 mRNA in growth cones is associated with HuD and ribosomes. J. Neurobiol. 2004;61:222–235. doi: 10.1002/neu.20038. [DOI] [PubMed] [Google Scholar]
- 55.Giuditta A, Chun JT, Eyman M, Cefaliello C, Bruno AP, Crispino M. Local gene expression in axons and nerve endings: the glia-neuron unit. Physiol. Rev. 2008;88:515–555. doi: 10.1152/physrev.00051.2006. [DOI] [PubMed] [Google Scholar]
- 56.Zeitelhofer M, Macchi P, Dahm R. Perplexing bodies: the putative roles of P-bodies in neurons. RNA Biol. 2008;5:244–248. doi: 10.4161/rna.6948. [DOI] [PubMed] [Google Scholar]
- 57.Abe R, Sakashita E, Yamamoto K, Sakamoto H. Two different RNA binding activities for the AU-rich element and the poly(A) sequence of the mouse neuronal protein mHuC. Nucleic Acids Res. 1996;24:4895–4901. doi: 10.1093/nar/24.24.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ma WJ, Chung S, Furneaux H. The Elav-like proteins bind to AU-rich elements and to the poly(A) tail of mRNA. Nucleic Acids Res. 1997;25:3564–3569. doi: 10.1093/nar/25.18.3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gao FB, Carson CC, Levine T, Keene JD. Selection of a subset of mRNAs from combinatorial 3′ untranslated region libraries using neuronal RNA-binding protein Hel-N1. Proc. Natl Acad. Sci. USA. 1994;91:11207–11211. doi: 10.1073/pnas.91.23.11207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Antic D, Lu N, Keene JD. ELAV tumor antigen, Hel-N1, increases translation of neurofilament M mRNA and induces formation of neurites in human teratocarcinoma cells. Genes Dev. 1999;13:449–461. doi: 10.1101/gad.13.4.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Meng Z, King PH, Nabors LB, Jackson NL, Chen CY, Emanuel PD, Blume SW. The ELAV RNA-stability factor HuR binds the 5′-untranslated region of the human IGF-IR transcript and differentially represses cap-dependent and IRES-mediated translation. Nucleic Acids Res. 2005;33:2962–2979. doi: 10.1093/nar/gki603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tiruchinapalli DM, Ehlers MD, Keene JD. Activity-dependent expression of RNA binding protein HuD and its association with mRNAs in neurons. RNA Biol. 2008;5:157–168. doi: 10.4161/rna.5.3.6782. [DOI] [PubMed] [Google Scholar]
- 63.Samson ML. Evidence for 3′ untranslated region-dependent autoregulation of the Drosophila gene encoding the neuronal nuclear RNA-binding protein ELAV. Genetics. 1998;150:723–733. doi: 10.1093/genetics/150.2.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pullmann R, Jr, Kim HH, Abdelmohsen K, Lal A, Martindale JL, Yang X, Gorospe M. Analysis of turnover and translation regulatory RNA-binding protein expression through binding to cognate mRNAs. Mol. Cell Biol. 2007;27:6265–6278. doi: 10.1128/MCB.00500-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Okano HJ, Darnell RB. A hierarchy of Hu RNA binding proteins in developing and adult neurons. J. Neurosci. 1997;17:3024–3037. doi: 10.1523/JNEUROSCI.17-09-03024.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dobashi Y, Shoji M, Wakata Y, Kameya T. Expression of HuD protein is essential for initial phase of neuronal differentiation in rat pheochromocytoma cells. Biochem. Biophys. Res. Commun. 1998;244:226–229. doi: 10.1006/bbrc.1998.8247. [DOI] [PubMed] [Google Scholar]
- 67.Kasashima K, Terashima K, Yamamoto K, Sakashita E, Sakamoto H. Cytoplasmic localization is required for the mammalian ELAV-like protein HuD to induce neuronal differentiation. Genes Cells. 1999;4:667–683. doi: 10.1046/j.1365-2443.1999.00292.x. [DOI] [PubMed] [Google Scholar]
- 68.Mobarak CD, Anderson KD, Morin M, Beckel-Mitchener A, Rogers SL, Furneaux H, King P, Perrone-Bizzozero NI. The RNA-binding protein HuD is required for GAP-43 mRNA stability, GAP-43 gene expression, and PKC-dependent neurite outgrowth in PC12 cells. Mol. Biol. Cell. 2000;11:3191–3203. doi: 10.1091/mbc.11.9.3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Winder DG, Sweatt JD. Roles of serine/threonine phosphatases in hippocampal synaptic plasticity. Nat. Rev. Neurosci. 2001;2:461–474. doi: 10.1038/35081514. [DOI] [PubMed] [Google Scholar]
- 70.Huber KM, Roder JC, Bear MF. Chemical induction of mGluR5- and protein synthesis–dependent long-term depression in hippocampal area CA1. J. Neurophysiol. 2001;86:321–325. doi: 10.1152/jn.2001.86.1.321. [DOI] [PubMed] [Google Scholar]
- 71.Tiruchinapalli DM, Caron MG, Keene JD. Activity-dependent expression of ELAV/Hu RBPs and neuronal mRNAs in seizure and cocaine brain. J. Neurochem. 2008a;107:1529–1543. doi: 10.1111/j.1471-4159.2008.05718.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.