Abstract
Activating Transcription Factor 3 (ATF3), a member of the ATF/CREB family, is induced rapidly by various stresses. Its induction mechanism and role in response to changes in cellular redox status, however, have not been elucidated. Here, we found that NF-E2-related factor 2 (Nrf2), a transcription factor known to bind to antioxidant response element (ARE) in promoters, transcriptionally upregulated ATF3 expression in astrocytes. Treatment with Nrf2 activators and oxidants provoked ATF3 induction in astrocytes, whereas its expression was reduced in Nrf2-depleted cells. We further demonstrated that the consensus ARE in the ATF3 promoter is critical for Nrf2-mediation by promoter analyses using an ATF3 promoter-driven luciferase construct and a chromatin immunoprecipitation assay. In addition, we found that Nrf2-dependent ATF3 induction contributed to the antioxidative and cytoprotective functions of Nrf2 in astrocytes. Taken together, our findings suggest that ATF3 is a new target for Nrf2 and has a cytoprotective function in astrocytes.
INTRODUCTION
Aerobic life is constantly challenged by oxidative stress. To cope with this stress, biological systems have devised defense mechanisms. The perturbation of this protective regulation causes the accumulation of reactive oxygen species (ROS), which contribute to the pathogenesis of various diseases including neurological and neurodegenerative disorders. Nrf2, a member of the Cap‘n’Collar subfamily of basic-region leucine zipper (bZIP) transcription factor family, plays a pivotal role in the defense system against oxidative stress. Nrf2 is normally degraded in the cytoplasm when it interacts with Keap1. However, oxidative stress leads to the dissociation of Nrf2 from Keap1, resulting in the accumulation of Nrf2 in the nucleus. Stabilized Nrf2 induces the expression of several cytoprotective and antioxidant enzymes by binding to antioxidant response element (ARE) in their promoter regions. In the nervous systems, Nrf2 is predominantly active in astrocytes (1). Preferential activation of Nrf2 in astrocytes confers neuronal protection against oxidative stress (2,3) and prevents the pathogenesis of familial amyotrophic lateral sclerosis and Parkinson’s disease (4,5).
ATF3, a bZIP-containing ATF/CREB family transcription factor, is a stress-responsive gene. The expression of ATF3 is rapidly induced upon exposure to numerous stresses including oxidative stress, while it remains at low level in normal conditions (6). It has been previously reported that JNK/SAPK, ERK/MAPK, and p38 signaling pathways are involved in the mechanism of ATF3 induction (7,8), suggesting that there are many upstream regulatory molecules for ATF3 induction. Since most of ATF3 inductions are linked to an increase in the expression of transcripts, the information on the regulation of the ATF3 promoter would provide insight for understanding the mechanisms of ATF3 expression. However, relatively little has been described in the context of the transcriptional control of the ATF3 promoter.
Here, we found that Nrf2 up-regulates ATF3 expression in astrocytes and Nrf2-binding site is located in the ATF3 promoter. Furthermore, we revealed that Nrf2-mediated ATF3 induction in astrocytes contributed to the cytoprotective function of Nrf2 against oxidative insults.
MATERIALS AND METHODS
Cell culture and chemicals
Human primary brain astrocytes were purchased from the Applied Cell Biology Research Institute (Kirkland, WA, USA) and were cultured in DMEM supplemented with 10% FBS (Invitrogen, Carlsbad, CA, USA). Mouse primary cerebral astrocytes were purified from neonatal ICR mice according to standard procedure (9). Mouse embryonic fibroblasts (MEFs) were isolated from Nrf2+/+ and Nrf2−/− mice (which were generously provided by Dr. Jeffrey A. Johnson, University of Wisconsin, WI, USA). Tert-butylhydroquinone (tBHQ) and butylated hydroxyanisole (BHA) were purchased from Fisher Scientific (Fairlawn, NY, USA) and sodium arsenite (AS) and phenazine methosulfate (PMS) were purchased from Sigma (St Louis, MO, USA).
Western blot analysis
Equal amounts of proteins were separated on 11% SDS–PAGE gels, transferred onto nitrocellulose membranes, and incubated with antibodies specific for Nrf2, ATF3, HO1 and β-actin (Santa Cruz Biotechnology, Santa Cruz, CA, USA). The expressions of ATF3 and β-actin were analyzed quantitatively using an image analysis software (NIH-Image J).
Immunocytochemistry
Cells were washed with phosphate buffered saline (PBS), fixed with 3.7% formaldehyde, and permeabilized with 0.5% Triton X-100 in PBS. After blocking with 1% blocking reagent (Roche) for 1 h, cells were incubated with the anti-ATF3 antibody overnight at 4°C, followed by incubation with an FITC-conjugated secondary antibody. Immunocytochemical analysis using the anti-phospho-histone H2A.X antibody (ser139) (Millipore, Billeria, MA, USA) was performed according to the manufacturer’s instructions. Human astrocytes were fixed with 5% acetic acid in ethanol for 5 min. After blocking with 3% BSA for 30 min, cells were incubated with anti-phospho-histone H2A.X antibody for 1 h, followed by incubation with an FITC-conjugated secondary antibody. After counterstaining with 2 µg/ml of 4′,6′-diamidine-2-phenylindole dihydrochloride (DAPI) (Roche), images were acquired using an Axiovert M200 microscope (Zeiss, Oberkochen, Germany).
RT–PCR and quantitative real-time PCR
Total RNA was isolated from astrocytes using the TRIzol reagent (Molecular Research Center, Cincinati, OH, USA). cDNAs were synthesized using an oligo-(dT) primer and MMLV reverse transcriptase (Promega, Madison, WI, USA) and were used in subsequent PCR reaction using specific primer sets for human ATF3 (5′-ATGATGCTTCAACACCCAGG-3′ and 5′-TTTCGGCACTTTGCAGCTG-3′), human Nrf2 (5′-AAACCAGTGGATCTGCCAAC-3′ and 5′-GACCGGGAATATCAGGAACA-3′) (10), human HO1 (5′-GCTACCTGGGTGACCTGTCT-3′ and 5′-GGGCAGAATCTTGCACTTTG-3′), human Sulfiredoxin1 (5′-CAAGGTGCAGAGCCTCGTGG-3′ and 5′-AGCTGGTGAGAGGGGTGCCC-3′), human NQO1 (5′-CATTCTGAAAGGCTGGTTTG-3′ and 5′-GGCTGCTTGGAGCAAAATAC-3′), and human β-actin (5′-GACTACCTCATGAAGATC-3′ and 5′-GATCCACATCTGCTGGAA-3′). PCR products were resolved on 1.5% agarose gels and visualized by ethidium bromide staining. Quantitative real-time PCR was carried out using commercially available SYBR green-based detection reagents (Roche, Branchburg, NJ, USA) and a Light Cycler apparatus (Roche).
siRNA transfection
siRNA duplexes targeting human Nrf2 (siNrf2, 5′-AAGAGUAUGAGCUGGAAAAAC-3′) (11), human ATF3 (siATF3#1, 5′-UGAAGAAGAUGAAAGGAAA-3′, siATF3#2, 5′-GAAACAAGAAGAAGGAGAA-3′), and GFP (siGFP, 5′-GGCUACGTCCAGGAGCGCA-3′) were synthesized by Dharmacon (Lafayette, CO, USA) or Samchully Pharm (Seoul, Korea). siRNAs were transfected using Oligofectamine (Invitrogen).
Plasmid constructs
ATF3 promoter regions were amplified from human genomic DNA by PCR using primer sets corresponding to −3646 (5′-CGGGGTACCACCACTACTGAGGGCTGACTGG-3′), −3150 (5′-CGGGGTACCACCACTACTGAGGGCTGACTGG-3′), 1250 (5′-CGGGGTACCAGGTCTCAGCTCGAATCTCGG-3′) and +3 (5′-CCCAAGCTTACTGTGGCTTGGAGAGCGTTG-3′) positions. PCR products were cloned into the pGL3-Basic vector. For construction of pATF3-ARE1-SV40 and pATF3-ARE2-SV40, fragments of approximately 200 bps that encompassed the putative ARE of the ATF3 promoter were amplified using the following primers: for pATF3-ARE1-luc, 5′-CGGGGTACCGCCACATCCTCAGAGAAATGTCAC-3′ (forward) and 5′-GGAAGATCTATTGTCAGGAAACAGGGGCAG-3′ (reverse); for pATF3-ARE2-luc, 5′-CGGGGTACCTAGATTTGGTTGCCACGAGTGG-3′ (forward) and 5′-GGAAGATCTGCAGAGACTTGAAGATTGCTGAGG-3′ (reverse). These PCR products were cloned into the pGL3-Promoter vector upstream of the SV40 promoter. Site-directed mutagenesis was performed by over-lapping PCR using the following primers: 5′-AAATAACTTACTTAAGGGTACACATAATCTAAGTGCTGAATC-3′, and reverse-complement primers (the mutated sequences were underlined). Mouse wild-type Nrf2, dominant-negative Nrf2 (Nrf2-DN, amino acids 296–581), and mouse Keap1 were amplified and cloned into the pCS2+ expression vectors.
Luciferase reporter assay
HeLa cells and human astrocytes were transfected using Lipofectamine Plus (Invitrogen). All cells were cotransfected with the β-galactosidase expression vector (pSV- β-galactosidase). Luciferase activity was measured using the Luciferase assay kit (Promega) and β-galactosidase activity was measured using O-nitrophenyl β-galactopyranoside.
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) was carried out using the ChIP kit (Upstate Biotechnology, Lake Placid, NY, USA). ARE of the ATF3 promoter was amplified by PCR using the following primers: for 1st PCR of ARE1, 5′-TACCACTACTGAGGGCTGACTGG-3′, and 5′-CATTGTCAGGAAACAGGGGC-3′; for 2nd PCR of ARE1, 5′-GTGCCACATCCTCAGAGAAATG-3′, and 5′-CACAGTAGGCACCAAAGTGTGG-3′; for PCR of ARE2, 5′-TAGATTTGGTTGCCACGAGTGG-3′ and 5′-GCAGAGACTTGAAGATTGCTGAGG-3′. Primer pairs corresponding to the ARE of the NQO1 promoter were used as a positive control (12).
ROS measurement and glutathione assay
To assess the generation of ROS, human primary astrocytes were incubated with 5 µM 2′,7′-dichlorodihydrofluorescein diacetate (DCFH-DA, Sigma) for 30 min at 37°C. Astrocytes were analyzed on a FACScan flow cytometer (Becton Dickinson, Bedford, MA, USA) and dichlorofluorescein (DCF) fluorescence was derived using the CellQuest® software. The Glutathione assay kit (Cayman Chemical, Ann Arbor, MI, USA) was used to measure total glutathione levels. Total glutathione [reduced glutathione (GSH) + oxidized glutathione (GSSG)] levels were measured at 405 nm, according to the manufacturer’s instructions.
Cell viability assay
Human astrocytes were treated with oxidants. After 4 h, we examined the cellular reduction of 4-[3-(4-iodophenyl)-2-(4-nitrophenyl)-2H-5-tetrazolio]-1,3-benzene disulfonate (WST-1) to formazan using the EZ-Cytox Enhanced cell viability assay kit (ITSBIO, Seoul, Korea). Cells were incubated in a culture medium containing 10% WST-1 for 1 h and the absorbance was measured at 450 nm.
TdT-mediated dUTP nick and labeling assay
To detect apoptotic cells, TdT-mediated dUTP nick and labeling (TUNEL) assay was performed using an in situ cell detection kit according to the manufacturer’s instructions (Roche). After fixation and permeabilization, human astrocytes were incubated with TUNEL reaction mixtures for 1 h. After counterstaining with DAPI, apoptotic cells were observed using an Axiovert M200 microscope (Zeiss). The percentage of apoptotic cells was calculated as the number of TUNEL-positive apoptotic nuclei divided by the total number of nuclei stained with DAPI.
Single-cell gel electrophoresis assay
Dissociated cells were mixed with low-melting agarose, dropped onto a glass slide, and dried. The slide was placed in lysis buffer for 2 h and electrophoresed at 12 V for 20 min. The slides were stained with 2 µg/ml ethidium bromide. The DNA was then visualized using a fluorescence microscope (Leica CTR5000, Cambridge, UK). The DNA damage in about 35 cells were analyzed using the Comet Assay χ program (Perceptive Instruments, Suffolk, UK).
Statistical analyses
All data are presented as mean ± standard error of the mean (SEM) of three independent experiments. Significance of differences was evaluated by unpaired Student’s t-test. NSP > 0.05, *P < 0.05, **P < 0.01.
RESULTS
Nrf2 activators and oxidants induced ATF3 expression
To evaluate the role of Nrf2 in the regulation of ATF3 expression, we treated human and mouse primary astrocytes with the Nrf2 activators, tBHQ and BHA. Within the indicated concentration range, no apparent cytotoxicity was observed (data not shown). Immunoblotting results showed that tBHQ and BHA provoked a dose-dependent induction of ATF3 as well as heme oxygenase 1 (HO1) which is an Nrf2-target protein (Figure 1A). This result was confirmed by immunocytochemical analysis, which showed that the induced ATF3 was mainly localized in the nuclei of tBHQ-treated human astrocytes (Figure 1B). In contrast, unstimulated cells exhibited background staining. Moreover, RT–PCR and the quantitative real-time PCR analyses revealed that both tBHQ and BHA increased ATF3 mRNA levels significantly in human astrocytes (Figure 1C and D), indicating that the induction of ATF3 was caused by the upregulation of transcription.
Figure 1.
Nrf2 activators induce ATF3 expression in astrocytes. (A) Human primary astrocytes were treated with the indicated concentrations of tBHQ or BHA for 24 h, and mouse primary astrocytes were treated with the indicated concentrations of tBHQ for 24 h. Proteins were subjected to Western blot analysis. The bottom bar graph shows the quantitative analysis of ATF3 expression which was normalized to β-actin expression levels (ATF3/β-actin). The value obtained for the DMSO-treated sample was set as 100%. Data are means ± SEM of three independent experiments (*P < 0.05, **P < 0.01 compared with the DMSO-treated sample). (B) Human astrocytes were treated with 50 µM tBHQ for 18 h and immunocytochemistry was then performed using the anti-ATF3 antibody. ATF3 staining was concentrated in nuclei after treatment with tBHQ (arrows). Magnification: X400 (C) Human primary astrocytes were treated with the indicated concentrations of tBHQ or BHA for 24 h. The expression levels of ATF3 were assessed by RT–PCR. (D) Human primary astrocytes were treated with the indicated concentrations of tBHQ (left panel) or BHA (right panel) for 24 h. The expression levels of ATF3 and β-actin were assessed by quantitative real time PCR. The expression levels of ATF3 transcripts were normalized to β-actin mRNA levels. The value obtained for the DMSO-treated sample was set as 100%. Data are means ± SEM of three independent experiments (*P < 0.05, **P < 0.01 compared with the DMSO-treated sample).
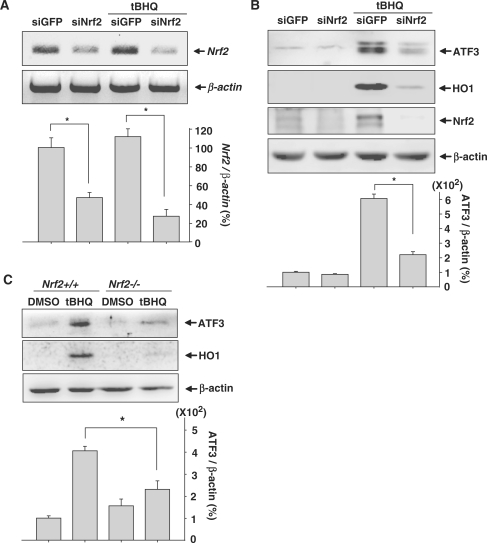
To further confirm that Nrf2 participated in tBHQ-induced ATF3 expression, we used siRNA targeting Nrf2 (siNrf2). RT–PCR analysis showed that transfection of human astrocytes with siNrf2 reduced the levels of Nrf2 mRNA expression (Figure 2A). As shown in Figure 2B, the depletion of Nrf2 in human astrocytes resulted in the reduction of ATF3 and HO1 induction. Additionally, when MEFs isolated from Nrf2+/+ and Nrf2−/− mice were cultured in the presence tBHQ, ATF3 and HO1 induction were considerably reduced in Nrf2-deficient MEFs (Figure 2C).These results suggest that Nrf2 plays an essential role in ATF3 induction after treatment with tBHQ.
Figure 2.
tBHQ-induced ATF3 expression is dependent on Nrf2. (A) Human astrocytes transfected with 75 nM siGFP or siNrf2 were treated with 100 µM tBHQ for 24 h. The expression levels of Nrf2 were assessed by RT–PCR. The bottom bar graph shows the quantitative analysis of Nrf2 expression which was normalized to β-actin expression levels (Nrf2/β-actin). The value obtained for the siGFP-transfected and DMSO-treated sample was set as 100%. Data are mean ± SEM of three independent experiments (*P < 0.05). (B) Human astrocytes transfected with 75 nM siGFP or siNrf2 were treated with 100 µM tBHQ. After 24 h incubation, Western blot was performed. The bottom bar graph shows the quantitative analysis of ATF3 levels which were normalized to β-actin expression levels (ATF3/β-actin). The value obtained for the siGFP-transfected and DMSO-treated sample was set as 100%. Data are mean ± SEM of three independent experiments (*P < 0.05). (C) MEFs isolated from Nrf2+/+ and Nrf2−/− mice were treated with 20 µM tBHQ for 24 h. Proteins were subjected to western blot analysis. The bottom bar graph shows the quantitative analysis of ATF3 levels which were normalized to β-actin expression levels (ATF3/β-actin). The value obtained for the DMSO-treated MEFs isolated from Nrf2+/+ was set as 100%. Data are mean ± SEM of three independent experiments (*P < 0.05).
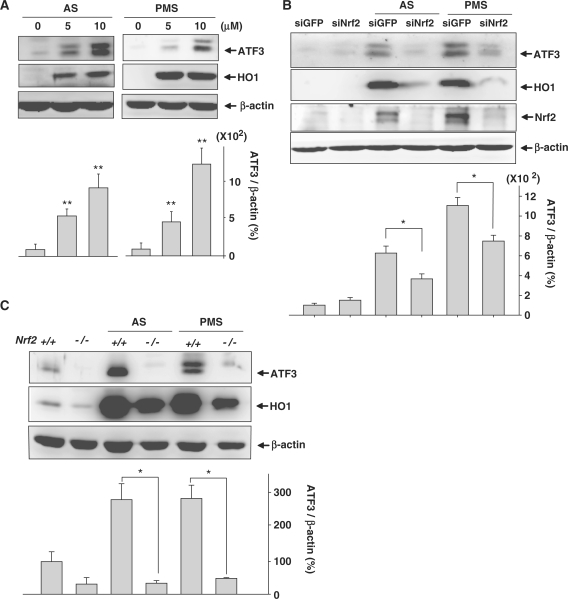
It is well known that oxidants induce the expression of Nrf2 target genes through the stabilization of Nrf2. To determine whether ATF3 was regulated by oxidants via Nrf2, human astrocytes were treated with the oxidants, sodium arsenite (AS) and phenazine methosulfate (PMS) which generate ROS inside cells. Figure 3A shows that both agents upregulated ATF3 and HO1 inductions in human astrocytes. Next, we determined whether Nrf2 was critical for oxidant-induced ATF3 expression. The depletion of Nrf2 in astrocytes reduced ATF3 and HO1 expression induced by treatment with AS or PMS (Figure 3B). Similarly, ATF3 expression was not induced by oxidants in Nrf2−/− MEFs (Figure 3C). The reduction of ATF3 induction in siNrf2-transfected astrocytes (Figure 3B) was less drastic than that observed in Nrf2−/− MEFs (Figure 3C). We suggest that, because transfection of human astrocytes with siNrf2 did not fully abolish the expression of Nrf2 mRNA, the reduction of ATF3 induction may be insufficient. Moreover, it is possible that ATF3 induction by oxidant treatment will be regulated by several pathways including the Nrf2/Keap1 pathway. Nevertheless, our findings showed that Nrf2 was one of the major pathways for the transcription of the ATF3 gene.
Figure 3.
Oxidants provoke ATF3 induction via the action of Nrf2. (A) Proteins extracted from human astrocytes treated with the indicated concentrations of AS or PMS for 24 h were subjected to western blot analysis. The bottom bar graph shows the quantitative analysis of ATF3 levels which were normalized to β-actin expression levels (ATF3/β-actin). The value obtained for the DMSO-treated sample was set as 100%. Data are mean ± SEM of three independent experiments (*P < 0.05, **P < 0.01 compared with the DMSO-treated sample). (B) Human astrocytes transfected with 75 nM siGFP or siNrf2 were treated with 7.5 µM AS or 7.5 µM PMS. Western blot was performed after 24 h incubation. The bottom bar graph shows the quantitative analysis of ATF3 levels which were normalized to β-actin expression levels (ATF3/ β-actin). The value obtained for the siGFP-transfected and DMSO-treated sample was set as 100%. Data are mean ± SEM of three independent experiments (*P < 0.05). (C) MEFs isolated from Nrf2+/+ and Nrf2−/− mice were treated with 7.5 µM AS or 7.5 µM PMS for 24 h. Proteins were subjected to Western blot analysis. The bottom bar graph shows the quantitative analysis of ATF3 levels which were normalized to β-actin expression levels (ATF3/β-actin). The value obtained for the DMSO-treated MEFs isolated from Nrf2+/+ mice was set as 100%. Data are mean ± SEM of three independent experiments (*P < 0.05).
Identification of ARE in the ATF3 promoter
Nrf2 binds to ARE in promoter regions for the transcriptional activation of target genes. To determine whether the ATF3 promoter contains an ARE or not, we analyzed about 5-kb upstream region of the human ATF3 promoter using MOTIF search software. Thereafter, we found two potential ARE sites, termed ARE1 and ARE2, which were located from −3541 to −3533 and from −2975 to −2967, respectively (Figure 4A). ARE1 exhibited perfect match with the consensus ARE sequence, while ARE2 contained two mismatched bases.
Figure 4.
Human ATF3 promoter responds to Nrf2/Keap1. (A) Schematic representation of the putative AREs in the human ATF3 promoter. Two potential AREs, ARE1 and ARE2 were predicted. (B) Human astrocytes transfected with ATF3 promoter-luciferase construct (pATF3-3.6k-luc) were treated with 100 µM tBHQ. Luciferase and β-galactosidase activities were measured after 24 h incubation. Luciferase activities were normalized to β-galactosidase activities. The value obtained for the DMSO-treated sample was set as 100%. Data are mean ± SEM of three independent experiments (*P < 0.05 compared with the DMSO-treated sample). (C) HeLa cells were transfected with ATF3 promoter-luciferase construct (pATF3-3.6k-luc) in the combination with various amounts of pCS2 + (Mock) or pCS2 + Nrf2 (Nrf2). Luciferase and β-galactosidase activities were measured after 48 h. Luciferase activities were normalized to β-galactosidase activities. The value obtained for the Mock-transfected sample was set as 100%. Data are mean ± SEM of three independent experiments (*P < 0.05, **P < 0.01 compared with the Mock-transfected sample). (D and E) HeLa cells were transfected with ATF3 promoter-luciferase construct (pATF3-3.6k-luc) and pCS2 + Nrf2 (Nrf2) with the indicated amounts of pCS2 + Nrf2-DN (Nrf2-DN, D) or pCS2 + Keap1 (Keap1, E) expression vector mixtures. Luciferase and β-galactosidase activities were measured after 48 h. Luciferase activities were normalized to β-galactosidase activities. The value obtained for the Mock-transfected sample was set as 100%. Data are mean ± SEM of three independent experiments (*P < 0.05, **P < 0.01).
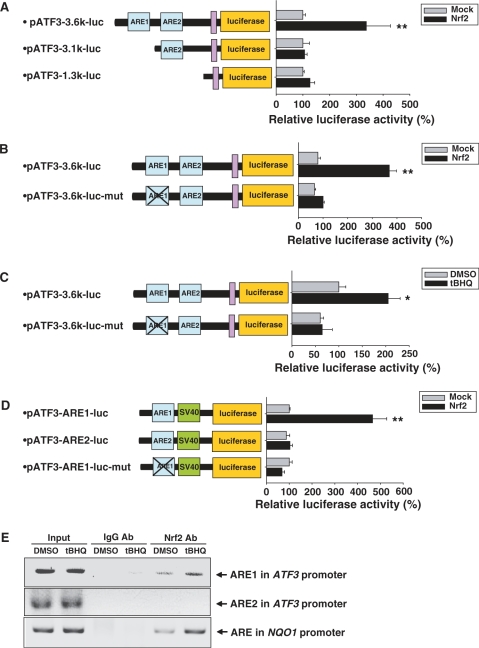
To verify whether these two putative ARE sites were functional for Nrf2, the ATF3 promoter encompassing both ARE sites was cloned into the luciferase reporter vector (pATF3-3.6k-luc). Luciferase activity in tBHQ-treated human astrocytes was almost twice as high as that measured in non-treated cells (Figure 4B). In addition, luciferase activity was also increased in proportion to increasing doses of exogenous Nrf2 (Figure 4C), whereas Nrf2-DN lacking a transactivating domain strongly suppressed the luciferase activity that was increased by the overexpression of Nrf2 (Figure 4D). Keap1 prevents Nrf2 activation by sequestering Nrf2 in the cytoplasm. Consequently, the overexpression of Keap1 almost completely repressed the transcriptional activity increased by the overexpression of Nrf2 (Figure 4E). These results indicate that the ATF3 promoter contains Nrf2-responsive element(s).
Next, we investigated whether the two ARE sites of the ATF3 promoter were important for Nrf2-mediated activation. Figure 5A shows that the overexpression of Nrf2 failed to induce the increase in transcriptional activity of pATF3-1.3k-luc construct lacking both ARE sites and pATF3-3.1k-luc construct containing only ARE2, whereas Nrf2 increased the transcriptional activity of pATF3-3.6k-luc construct containing both ARE sites, suggesting that ARE1 may be important for Nrf2 mediation. The mutated ARE1 markedly reduced the Nrf2-mediated activation of the ATF3 promoter (Figure 5B). In addition, we showed that tBHQ treatment did not increase luciferase activity in human astrocytes transfected with mutant type ATF3 promoter (pATF3-3.6k-luc-mut) (Figure 5C). These results suggest that ARE1 was essential for the Nrf2-dependent transcriptional activation of the ATF3 gene. This finding was further confirmed in experiments using a luciferase-construct containing either ARE1 or ARE2 (pATF3-ARE1-luc or pATF3-ARE2-luc, respectively). The pATF3-ARE1-luc construct containing only ARE1 exhibited the Nrf2-mediated increase in luciferase activity, whereas the pATF3-ARE2-luc construct containing only ARE2 did not (Figure 5D). In addition, the site-specific mutation of the ARE1 sequence in the pATF3-ARE1-luc construct (pATF3-ARE1-luc-mut) completely abolished the Nrf2-mediated increase in luciferase activity (Figure 5D). Finally, we evaluated whether Nrf2 was able to bind ARE1 in vivo. ChIP analysis revealed that tBHQ treatment increased Nrf2 binding to ARE1 of the ATF3 promoters as well as ARE of the NAD(P)H: quinine oxidoreductase (NQO1) promoter which was used as a positive control (Figure 5E). These results suggest that Nrf2 binds to ARE1 located in the ATF3 promoter, thereby inducing ATF3 expression.
Figure 5.
Identification of ARE in human ATF3 promoter. (A) HeLa cells were transfected with ATF3 promoter-luciferase constructs (pATF3-3.6k-luc or pATF3-3.1k-luc or pATF3-1.3k-luc) in combination with pCS2 + (Mock) or pCS2 + Nrf2 (Nrf2). Luciferase and β-galactosidase activities were measured after 48 h. Luciferase activities were normalized to β-galactosidase activities. Relative luciferase activity is shown at the right side. The value obtained for the Mock-transfected sample was set as 100%. Data are mean ± SEM of three independent experiments (*P < 0.05, **P < 0.01 compared with the Mock-transfected sample). (B) Mutant ATF3 promoter-luciferase constructs (pATF3-3.6k-luc-mut) were generated by site-directly mutagenesis of ARE1 sequences in pATF3-3.6k-luc (5′-TGACACAGC-3′ → 5′-GTACACATA-3′, mutated sequences were underlined). HeLa cells were transfected with wild-type or mutant ATF3 promoter-luciferase constructs in combination with pCS2+ (Mock) or pCS2 + Nrf2 (Nrf2). Luciferase and β-galactosidase activities were measured after 48 h. Luciferase activities were normalized to β-galactosidase activities. Relative luciferase activity is shown at the right side. The value obtained for the pATF3-3.6k-luc- and Mock-transfected sample was set as 100%. Data are mean ± SEM of three independent experiments (*P < 0.05, **P < 0.01 compared with the pATF3-3.6k-luc- and Mock-transfected sample). (C) Human astrocytes transfected with wild-type (pATF3-3.6k-luc) or mutant ATF3 promoter-luciferase construct (pATF3-3.6k-luc-mut) were treated with 100 µM tBHQ. Luciferase and β-galactosidase activities were measured after 24 h incubation. Luciferase activities were normalized to β-galactosidase activity. Relative luciferase activity is shown at the right side. The value obtained for the pATF3-3.6k-luc-transfected and DMSO-treated sample was set as 100%. Data are mean ± SEM of three independent experiments (*P < 0.05 compared with the pATF3-3.6k-luc-transfected and DMSO-treated sample). (D) HeLa cells were transfected with ATF3 promoter-luciferase constructs (pATF3-ARE1-luc or pATF3-ARE2-luc or pATF3-ARE1-luc-mut) in combination with pCS2 + (Mock) or pCS2 + Nrf2 (Nrf2). Luciferase and β-galactosidase activities were measured after 48 h. Luciferase activities were normalized to β-galactosidase activities. Relative luciferase activity is shown at the right side. The value obtained for the pATF3-ARE1-luc- and Mock-transfected sample was set as 100%. Data are mean ± SEM of three independent experiments (*P < 0.05, **P < 0.01 compared with the pATF3-ARE1-luc- and Mock-transfected sample). (E) Human astrocytes were treated with 100 µM tBHQ for 1 h. Chromatin immunoprecipitation (ChIP) assay was performed then. The ARE present in the NQO1 promoter was used as a positive control.
ATF3 contributed to antioxidative and cytoprotective functions of Nrf2 in astrocytes
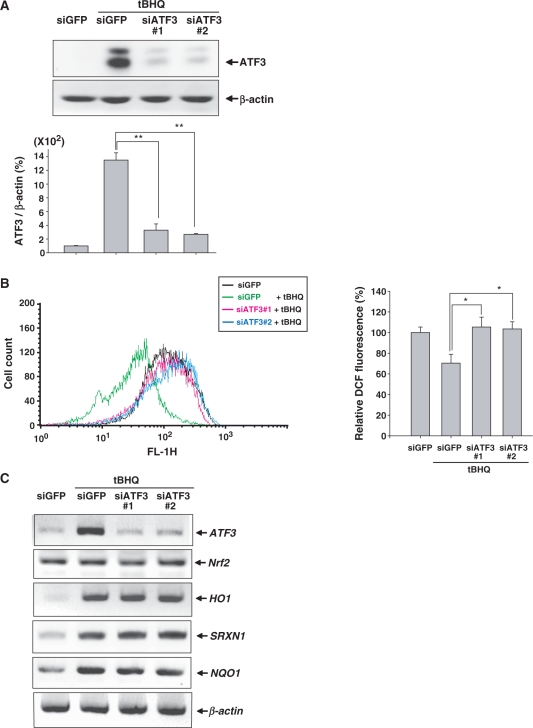
To assess the importance of ATF3 induction in astrocytes, we used two siRNAs targeting ATF3 (siATF3#1 and siATF3#2) for the knockdown of tBHQ-induced ATF3 expression (Figure 6A). It was previously reported that Nrf2 has an antioxidative function via the modulation of cellular ROS levels (13). Therefore, we assessed whether the Nrf2-dependent induction of ATF3 affected cellular ROS levels employing 2′,7′-dichlorodihydrofluorescein diacetate (DCFH-DA) which is sensitive to oxidation by cellular ROS. Figure 6B shows that tBHQ decreased ROS levels in astrocytes. However, the tBHQ-mediated reduction of cellular ROS levels was decreased in ATF3-depleted astrocytes, which implies that ATF3 may be required for the antioxidative functions of Nrf2. Furthermore, we examined the effect of ATF3 induction on the expression of Nrf2 target genes because ATF3 could exert its antioxidative effect via the transcriptional modulation of the expression of Nrf2 target genes. However, the absence of ATF3 did not affect the mRNA expression of Nrf2 and Nrf2 target genes including antioxidant enzymes HO-1 and sulfiredoxin 1 (SRXN1), and the detoxifying enzyme NQO1 (Figure 6C). Additionally, the immunocytochemical analysis using anti-Nrf2 antibody showed that the depletion of ATF3 did not affect the immunoreactivity for Nrf2 which was stabilized by treatment with tBHQ (Supplementary Figure S1). These findings suggest that ATF3 contributed to the antioxidative function of Nrf2 in astrocytes although ATF3 did not modulate the transcription of Nrf2 and Nrf2-target genes.
Figure 6.
ATF3 exerts an antioxidative function in astrocytes. (A) Human astrocytes were transfected with 50 nM siGFP or siATF3#1 or siATF3#2 and were then treated with 100 µM tBHQ for 24 h. Proteins were subjected to Western blot analysis. The bottom panel shows the relative ATF3 expression which was normalized to β-actin expression levels. The value obtained for the siGFP-transfected and DMSO-treated sample was set as 100%. Data are mean ± SEM of three independent experiments (*P < 0.05, **P < 0.01). (B) Human astrocytes were transfected with 50 nM siGFP, siATF3#1 or siATF3#2 and were then treated with 100 µM tBHQ for 24 h. Intracellular ROS levels were measured by adding 5 µM DCFH-DA, followed by analysis using a flow cytometer. The right bar graph shows the relative DCF fluorescence. The value obtained for the siGFP-transfected and DMSO-treated sample was set as 100%. Each value represented the mean ± SEM of three independent experiments. Data are mean ± SEM of three independent experiments (*P < 0.05, **P < 0.01). (C) Human astrocytes were transfected with 50 nM siGFP, siATF3#1 or siATF3#2 and were then treated with 100 µM tBHQ for 24 h. The expression levels of ATF3, Nrf2, HO1, SRXN1, NQO1 and β-actin were assessed by RT–PCR.
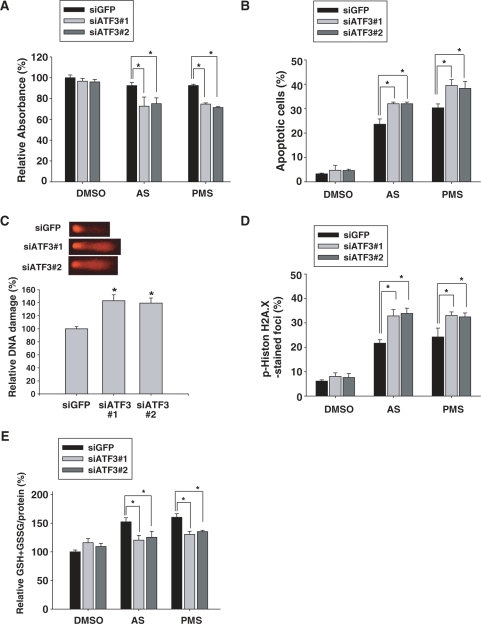
Next, we speculated that ATF3 might affect cellular viability against oxidative stress. To address this, we used siATF3 to silence the expression of ATF3 induced by treatment with AS and PMS. WST assay was performed to examine cytotoxicity after treatment with AS or PMS for 24 h. As shown in Figure 7A, the absorbance was decreased in ATF3-deficient astrocytes treated with oxidants, which implies that oxidant-induced cytotoxicity was increased by the knockdown of ATF3. To examine apoptosis, we performed a TUNEL assay which detects DNA fragmentation resulting from apoptosis. The number of apoptotic cells generated by treatment with oxidants was increased in ATF3-depleted cells (Figure 7B and Supplementary Figure S2). Next, we determined the effect of ATF3 on DNA damage by performing a single-cell gel electrophoresis assay to visualize single- and double- strand DNA breaks. As illustrated in Figure 7C, DNA damage was enhanced in ATF3-depleted cells. This result was confirmed by the immunocytochemical analysis using the anti-phospho histone H2A.X antibody. Phosphorylation of histone H2A.X is an indication of the early DNA damage response (14). We found that the number of foci generated by the treatment with oxidants was increased in ATF3-depleted cells (Figure 7D and Supplementary Figure S3). All together, these results demonstrated that the knockdown of ATF3 enhanced DNA damages under oxidative stress conditions in human astrocytes.
Figure 7.
ATF3 contributes to protective activity of Nrf2 in astrocytes. (A) Human astrocytes were transfected with 50 nM siGFP, siATF3#1 or siATF3#2 and were then treated with 7.5 µM AS or 7.5 µM PMS for 24 h. WST assay was performed to examine cytotoxicity. The absorbance observed for the siGFP-transfected and DMSO-treated sample was set as 100%. Data are means ± SEM of three independent experiments (*P < 0.05). (B) Human astrocytes were transfected with 50 nM siGFP, siATF3#1 or siATF3#2 and were then treated with 7.5 µM AS or 7.5 µM PMS for 24 h. TUNEL assay was performed to examine the apoptosis induced by oxidants. The proportion of apoptotic cells was calculated as the number of TUNEL-positive apoptotic nuclei divided by the total number of nuclei stained with DAPI. Data are mean ± SEM of three independent experiments (*P < 0.05). (C) Human astrocytes were transfected with 50 nM siGFP, siATF3#1 or siATF3#2 and were then treated with 7.5 µM AS for 24 h. A single-cell gel electrophoresis assay was performed to assess DNA damage in individual cells. The more damaged and broken pieces of DNA were shown in ATF3-depleted cells (upper panel). We analyzed and calculated the DNA damage in ∼35 cells (bottom panel) as described in Materials and Methods section. The value for the siGFP-transfected sample was set as 100%. Data are means ± SEM of three independent experiments (*P < 0.05 compared with the siGFP-transfected sample). (D) Human astrocytes were transfected with 50 nM siGFP, siATF3#1 or siATF3#2 and were then treated with 7.5 µM AS or 7.5 µM PMS for 24 h. To examine DNA damages, immunocytochemical analysis was performed using the anti-phospho-histone H2A.X antibody. The number of phospho-histone H2A.X-positive foci was divided by the total number of nuclei stained with DAPI. Data are mean ± SEM of three independent experiments (*P < 0.05). (E) Human astrocytes were transfected with 50 nM siGFP, siATF3#1 or siATF3#2 and were then treated with 7.5 µM AS or 7.5 µM PMS for 24 h. Total intracellular glutathione was quantified by measuring the levels of GSH and GSSG, which were normalized to the amounts of total proteins. The amount of GSH + GSSG/the amount of protein in the siGFP-transfected and DMSO-treated sample (20.52 nmol/mg) was set as 100%. Data are mean ± SEM of three independent experiments (*P < 0.05).
GSH is the most abundant antioxidant. It protects cells by scavenging ROS in stress conditions. Next, we asked whether ATF3 was able to regulate the amount of GSH in response to oxidants. The levels of GSH increased after AS and PMS treatments (Figure 7E), suggesting that the upregulation of GSH has a counter-effect on oxidant-induced damage. However, the depletion of ATF3 reduced the upregulation of GSH, which implies that ATF3 is required for the generation of intracellular GSH. Therefore, these data suggest that ATF3 induction in astrocytes may contribute to the cytoprotective function of Nrf2 against oxidative insults by modulating GSH levels. The experiments depicted in Figure 7 were performed using non-transfected control cells, in which the values were not different from those observed in siGFP-transfected control cells (data not shown).
DISCUSSION
Cellular responses to oxidative stress are critical for the maintenance of homeostasis. Nrf2 is rapidly activated by oxidative reagents and ATF3 is induced by a variety of stresses including ROS. Although Nrf2 and ATF3 are similarly reactive against the oxidative stress, there is no direct evidence of Nrf2-dependent ATF3 induction. Here, we reported that ATF3 expression in astrocytes is upregulated by the binding of Nrf2 to ARE of the ATF3 promoter and may contribute to the antioxidative activity of Nrf2.
Two classical Nrf2 activators, tBHQ and BHA, upregulated the ATF3 expression in astrocytes (Figure 1). However, tBHQ did not affect ATF3 expression in other type of brain cells including human brain endothelial cells or mouse primary neuronal cells (data not shown), which suggests the presence of astrocyte-specific mechanism for the Nrf2-mediated induction of ATF3 expression. Several studies showed that Nrf2 activity is higher in astrocytes than in neurons (1–3). For instance, ARE-responsive genes NQO1 and GST are more abundant and effective in astrocytes than in neurons. Similar to what was observed for NQO1 and GST, ATF3 induction by tBHQ in astrocytes seemed to be due to the strong Nrf2 activity. Moreover, we showed that ATF3 induction in astrocytes contributes to the cytoprotective function of Nrf2 (Figure 7). Our findings are inconsistent with a previous study showing that ATF3 forms, a heterodimer with Nrf2 at the ARE, are present in the promoter regions of Nrf2 target genes and represses the function of Nrf2 to induce phase 2 enzymes in epithelial cells (15). In its system, however, ATF3 was expressed highly under normal conditions, whereas, in general, the expression of ATF3 is rarely detected in unstimulated conditions. Therefore, we suggest that the differences in the characteristics of cells may determine the function of ATF3. In support of this suggestion, both protective and proapoptotic effects of ATF3 in stressful conditions have been reported (16–20). In particular, a recent report showed that the molecular properties of specific cells determine the role of ATF3 as an oncogene or as a tumor suppressor (21). Although ATF3 has various functions, our findings demonstrated that, in astrocytes, Nrf2 mediated the upregulation of ATF3 and that the synergistic action of the Nrf2-ATF3 complex promoted the protective effect of astrocytes.
In the nervous system, ATF3 is a marker of cellular stress. Focal cerebral ischemia induces the biphasic expression of ATF3 (22) and sciatic nerve injury leads to the upregulation of ATF3 in glial cells (23). Furthermore, evidence suggests that ATF3 induction in the brain is a survival response to physiological dysfunctions. For instance, ATF3 expression rescues damaged cells in the nervous system (19) and promotes the regeneration of injured dorsal root ganglion peripheral neurons (20). The mechanism that underlies the protective capacity of ATF3 in the nervous systems, however, remains unclear. We propose that ATF3 provides a protective function to astrocytes under pathological conditions by modulating the redox status and glutathione levels. However, as we did not fully establish how ATF3 modulated redox status and glutathione levels in astrocytes, experiments aimed at elucidating the action of ATF3 target genes or the binding partners of ATF3 may be helpful for understanding the role of ATF3 in astrocytes.
In conclusion, we showed that ATF3 was a new member of the Nrf2/ARE regulated gene family and forms the part of the cytoprotective system in astrocytes. Therefore, we suggest that the synergistic activation of Nrf2 and ATF3 in astrocytes may provide a new intervention against nervous diseases caused by oxidative stress.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: The Korea Science and Engineering Foundation (KOSEF) grant funded by the Ministry of Education, Science and Technology (MEST) through the Creative Research Intiative program (grant R16-2004-001-01001-0-2008).
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Ahlgren-Beckendorf JA, Reising AM, Schander MA, Herdler JW, Johnson JA. Coordinate regulation of NAD(P)H: quinone oxidoreductase and glutathione-S-transferases in primary cultures of rat neurons and glia: role of the antioxidant/electrophile responsive element. Glia. 1999;25:131–142. [PubMed] [Google Scholar]
- 2.Shih AY, Johnson DA, Wong G, Kraft AD, Jiang L, Erb H, Johnson JA, Murphy TH. Coordinate regulation of glutathione biosynthesis and release by Nrf2-expressing glia potently protects neurons from oxidative stress. J. Neurosci. 2003;23:3394–3406. doi: 10.1523/JNEUROSCI.23-08-03394.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kraft AD, Johnson DA, Johnson JA. Nuclear factor E2-related factor 2-dependent antioxidant response element activation by tert-butylhydroquinone and sulforaphane occurring preferentially in astrocytes conditions neurons against oxidative insult. J. Neurosci. 2004;24:1101–1112. doi: 10.1523/JNEUROSCI.3817-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vargas MR, Johnson DA, Sirkis DW, Messing A, Johnson JA. Nrf2 activation in astrocytes protects against neurodegeneration in mouse models of familial amyotrophic lateral sclerosis. J. Neurosci. 2008;28:13574–13581. doi: 10.1523/JNEUROSCI.4099-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen PC, Vargas MR, Pani AK, Smeyne RJ, Johnson DA, Kan YW, Johnson JA. Nrf2-mediated neuroprotection in the MPTP mouse model of Parkinson's; disease: critical role for the astrocyte. Proc. Natl Acad. Sci. USA. 2009;106:2933–2938. doi: 10.1073/pnas.0813361106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hai T, Wolfgang CD, Marsee DK, Allen AE, Sivaprasad U. ATF3 and stress responses. Gene Expr. 1999;7:321–335. [PMC free article] [PubMed] [Google Scholar]
- 7.Inoue K, Zama T, Kamimoto T, Aoki R, Ikeda Y, Kimura H, Hagiwara M. TNFalpha-induced ATF3 expression is bidirectionally regulated by the JNK and ERK pathways in vascular endothelial cells. Genes Cells. 2004;9:59–70. doi: 10.1111/j.1356-9597.2004.00707.x. [DOI] [PubMed] [Google Scholar]
- 8.Lu D, Chen J, Hai T. The regulation of ATF3 gene expression by mitogen-activated protein kinases. Biochem. J. 2007;401:559–567. doi: 10.1042/BJ20061081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shih AY, Li P, Murphy TH. A small-molecule-inducible Nrf2-mediated antioxidant response provides effective prophylaxis against cerebral ischemia in vivo. J. Neurosci. 2005;25:10321–10335. doi: 10.1523/JNEUROSCI.4014-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leonard MO, Kieran NE, Howell K, Burne MJ, Varadarajan R, Dhakshinamoorthy S, Porter AG, O'F;arrelly C, Rabb H, Taylor CT. Reoxygenation-specific activation of the antioxidant transcription factor Nrf2 mediates cytoprotective gene expression in ischemia-reperfusion injury. FASEB J. 2006;20:2624–2626. doi: 10.1096/fj.06-5097fje. [DOI] [PubMed] [Google Scholar]
- 11.Padmanabhan B, Tong KI, Ohta T, Nakamura Y, Scharlock M, Ohtsuji M, Kang MI, Kobayashi A, Yokoyama S, Yamamoto M. Structural basis for defects of Keap1 activity provoked by its point mutations in lung cancer. Mol. Cell. 2006;21:689–700. doi: 10.1016/j.molcel.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 12.Dhakshinamoorthy S, Jain AK, Bloom DA, Jaiswal AK. Bach1 competes with Nrf2 leading to negative regulation of the antioxidant response element (ARE)-mediated NAD(P)H: quinone oxidoreductase 1 gene expression and induction in response to antioxidants. J. Biol. Chem. 2005;280:16891–16900. doi: 10.1074/jbc.M500166200. [DOI] [PubMed] [Google Scholar]
- 13.Frohlich DA, McCabe MT, Arnold RS, Day ML. The role of Nrf2 in increased reactive oxygen species and DNA damage in prostate tumorigenesis. Oncogene. 2008;27:4353–4362. doi: 10.1038/onc.2008.79. [DOI] [PubMed] [Google Scholar]
- 14.Yih LH, Hsueh SW, Luu WS, Chiu TH, Lee TC. Arsenite induces prominent mitotic arrest via inhibition of G2 checkpoint activation in CGL-2 cells. Carcinogenesis. 2005;26:53–63. doi: 10.1093/carcin/bgh295. [DOI] [PubMed] [Google Scholar]
- 15.Brown SL, Sekhar KR, Rachakonda G, Sasi S, Freeman ML. Activating transcription factor 3 is a novel repressor of the nuclear factor erythroid-derived 2-related factor 2 (Nrf2)-regulated stress pathway. Cancer Res. 2008;68:364–368. doi: 10.1158/0008-5472.CAN-07-2170. [DOI] [PubMed] [Google Scholar]
- 16.Turchi L, Aberdam E, Mazure N, Pouyssegur J, Deckert M, Kitajima S, Aberdam D, Virolle T. Hif-2alpha mediates UV-induced apoptosis through a novel ATF3-dependent death pathway. Cell Death Differ. 2008;15:1472–1480. doi: 10.1038/cdd.2008.74. [DOI] [PubMed] [Google Scholar]
- 17.Yoshida T, Sugiura H, Mitobe M, Tsuchiya K, Shirota S, Nishimura S, Shiohira S, Ito H, Nobori K, Gullans SR, et al. ATF3 protects against renal ischemia-reperfusion injury. J. Am. Soc. Nephrol. 2008;19:217–224. doi: 10.1681/ASN.2005111155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu D, Wolfgang CD, Hai T. Activating transcription factor 3, a stress-inducible gene, suppresses Ras-stimulated tumorigenesis. J. Biol. Chem. 2006;281:10473–10481. doi: 10.1074/jbc.M509278200. [DOI] [PubMed] [Google Scholar]
- 19.Nakagomi S, Suzuki Y, Namikawa K, Kiryu-Seo S, Kiyama H. Expression of the activating transcription factor 3 prevents c-Jun N-terminal kinase-induced neuronal death by promoting heat shock protein 27 expression and Akt activation. J. Neurosci. 2003;23:5187–5196. doi: 10.1523/JNEUROSCI.23-12-05187.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seijffers R, Mills CD, Woolf CJ. ATF3 increases the intrinsic growth state of DRG neurons to enhance peripheral nerve regeneration. J. Neurosci. 2007;27:7911–7920. doi: 10.1523/JNEUROSCI.5313-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yin X, Dewille JW, Hai T. A potential dichotomous role of ATF3, an adaptive-response gene, in cancer development. Oncogene. 2008;27:2118–2127. doi: 10.1038/sj.onc.1210861. [DOI] [PubMed] [Google Scholar]
- 22.Ohba N, Maeda M, Nakagomi S, Muraoka M, Kiyama H. Biphasic expression of activating transcription factor-3 in neurons after cerebral infarction. Brain Res. Mol. Brain Res. 2003;115:147–156. doi: 10.1016/s0169-328x(03)00181-5. [DOI] [PubMed] [Google Scholar]
- 23.Hunt D, Hossain-Ibrahim K, Mason MR, Coffin RS, Lieberman AR, Winterbottom J, Anderson PN. ATF3 upregulation in glia during Wallerian degeneration: differential expression in peripheral nerves and CNS white matter. BMC Neurosci. 2004;5:9. doi: 10.1186/1471-2202-5-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.