Abstract
The nuclear cap-binding protein complex (CBC) participates in 5′ splice site selection of introns that are proximal to the mRNA cap. However, it is not known whether CBC has a role in alternative splicing. Using an RT–PCR alternative splicing panel, we analysed 435 alternative splicing events in Arabidopsis thaliana genes, encoding mainly transcription factors, splicing factors and stress-related proteins. Splicing profiles were determined in wild type plants, the cbp20 and cbp80(abh1) single mutants and the cbp20/80 double mutant. The alternative splicing events included alternative 5′ and 3′ splice site selection, exon skipping and intron retention. Significant changes in the ratios of alternative splicing isoforms were found in 101 genes. Of these, 41% were common to all three CBC mutants and 15% were observed only in the double mutant. The cbp80(abh1) and cbp20/80 mutants had many more changes in alternative splicing in common than did cbp20 and cbp20/80 suggesting that CBP80 plays a more significant role in alternative splicing than CBP20, probably being a platform for interactions with other splicing factors. Cap-binding proteins and the CBC are therefore directly involved in alternative splicing of some Arabidopsis genes and in most cases influenced alternative splicing of the first intron, particularly at the 5′ splice site.
INTRODUCTION
Alternative splicing is a widespread process that generates more than one spliced mRNA isoform from the same gene and expands both transcriptome and proteome diversity. Alternative events include alternative 5′ and 3′ splice site selection, intron retention, exon skipping and mutually exclusive exon splicing (1–3). Current estimates from both experimental and bioinformatic analyses are that 30–35% of Arabidopsis thaliana and rice genes undergo alternative splicing (4,5), while in human up to 95% of multi-exon genes undergo alternative splicing (6). The number of alternatively spliced plant genes is still likely to be an underestimate because of the relatively low EST coverage and depth of sequencing for many plant transcripts (5). In addition, many alternative splicing events occur only in specific cells and tissues, at specific stages of development and/or under certain physiological conditions and are therefore under-represented in EST databases. In plants, intron retention is the most frequent alternative event (45–56% of A. thaliana alternative splicing events) (4,7–10). Alternative 3′ and 5′ splice site selection accounts for ∼22 and 10% of events, respectively, and ∼4% have both 5′ and 3′ alternatively spliced sites. Only 8% of alternative splicing events in plants involve exon skipping (inclusion/exclusion of an exon) in contrast to animals where exon skipping is the most common form of alternative splicing (58% of genes) (8,11,12). The two major consequences of alternative splicing are to increase protein diversity by the inclusion or exclusion of peptide sequences or protein domains or to modulate gene expression through the production of mRNA isoforms which are degraded by nonsense-mediated decay (NMD). More than 75% of alternative splicing events are within the coding sequence of the gene and can generate proteins with new structures and biological functions (2,8,13). However, in both plants and animals, a significant number of AS events in the coding regions have a premature termination codon and are the potential targets of NMD (8,14,15). In plants, ∼21% of all alternative splicing events take place within the 5′ UTR (15%) or 3′ UTR (6%) which can affect transport and stability of mRNAs, create new initiation codons or polyadenylation sites, generate upstream open reading frames or shift the reading frame (7,11). Regulation of alternative splicing in plants has been reported for many genes and evolutionary conserved regulation of alternative splicing of plant SR proteins which themselves are splicing regulators points to important roles of alternative splicing in plant development (16). For example, the circadian clock RNA-binding protein, AtGRP7, autoregulates its transcript levels by binding its pre-mRNA to cause alternative splicing and generate an isoform that is turned over by NMD (17). The rice Waxy gene encodes a granule-bound starch synthase for which alternative splicing is temperature sensitive, potentially contributing to poor grain quality when seed maturation occurs at low temperature (18). Alternative splicing of the first intron of the isoaspartyl methyltransferase 2 gene creates different protein variants found in different subcellular compartments where they are involved in protein repair processes (19). Some transcripts can be alternatively spliced in plants in response to wounding or virus infection (20–22). Alternative splicing can also be regulated in a temperature-dependent manner, such as the A. thaliana SR1 splicing factor (23), or in a tissue-dependent manner, for example, tobacco RGP (24) and spinach and tobacco chloroplast ascorbate peroxidase transcripts (25).
The eukaryotic nuclear cap-binding complex (CBC) consists of two subunits (CBP20 and CBP80) that, as a complex, bind to the cap structure of RNA polymerase II transcripts (26). The cap and the CBC have multiple functions in mRNA biogenesis including splicing (26–31), 3′-end formation by stabilizing the interaction of the 3′-end processing machinery (32,33), nuclear export (34–37) and protection of the transcripts from nuclease degradation (38–39). Constitutive and alternative splicing is mediated by the spliceosome which is assembled on an intron in a stepwise process. An early event in spliceosome assembly is the interaction of U1snRNP with the 5′ splice site of the intron. In mammals and yeast, the CBC at the 5′-end of the pre-mRNA transcript promotes the initial interaction between U1snRNP and the 5′ splice site of the first intron in the transcript, and enhances the formation of spliced mRNAs (26–31,40–44). CBP proteins remain bound to the mRNA during the pioneer round of translation playing an essential role in mRNA quality control (45). Moreover, in mammalian cells, CBP80 recruits the NMD factor Upf1 and promotes the interaction of Upf1 with the NMD factor Upf2 (46).
The plant CBC also consists of two subunits: AtCBP20 and AtCBP80. AtCBP20 has a calculated molecular mass of 29.9 kDa and exhibits 68% identity and 82% similarity to its human orthologue and 53% identity and 77% similarity to the yeast protein. The larger CBP80 subunit is 96.5 kDa and exhibits lower identity (28 and 22%) and similarity (50 and 42%) to its human and yeast orthologues, respectively. AtCBP20 contains a canonical RNA binding domain (RBD) and AtCBP80 contains a protein–protein and protein–nucleic acid interaction domain, MIF4G (47). Additionally, in contrast to human and yeast, AtCBP20 has a long C-terminal extension with two nuclear localisation signals (NLSs) and is actively transported to the nucleus, while AtCBP80 can reach the nucleus only in a complex with AtCBP20. This feature distinguishes the mechanism of CBP transport in plants and animals where the nuclear import of human CBC requires a bipartite NLS on CBP80 (48). Moreover, neither CBP20 nor CBP80 is involved in NMD in A. thaliana (49).
Both plant CBC proteins are encoded by single-copy genes on Arabidopsis chromosomes V and II, respectively (47) and T-DNA insertion mutants which disrupt either the AtCBP20 or AtCBP80(ABH1) genes show identical phenotypes: slow growth and serrated leaf margins. Both mutants are abscisic acid (ABA) hypersensitive and can tolerate water deficiency much better than wild type plants (50–53). The mutants also affect flowering time and the processing and splicing of mRNA of factors involved in the regulation of flowering time in Arabidopsis is affected (41,54,55). The increase in the occurrence of unspliced introns demonstrates a further effect of the CBC on pre-mRNA splicing (40,41,56). Finally, the plant CBC has recently been shown to mediate the biogenesis of microRNAs (miRNA) (40,53,57). Both cbp20 and cbp80(abh1) mutants have reduced miRNA levels and increased pri-miRNA levels. CBP20 and CBP80 are suggested to bind to capped pri-miRNA transcripts and play role in their processing (53). The CBC interaction with the miRNA processing machinery appears to involve SERRATE (40) and is likely to facilitate the loading of the miRNA processing machinery onto pri-miRNA, in analogy with its role in recruiting the splicing commitment complex onto pre-mRNA (40,57). The role of CBP80 in miRNA-mediated RNA silencing pathway may also be conserved in animals (58). Finally, both proteins also participate in ta-siRNA biogenesis through regulating the biogenesis of miR173 and miR390 (53).
The CBC is important in pre-mRNA splicing and other aspects of RNA processing in plants. To date, there is no evidence that the nuclear cap-binding complex is involved in alternative splicing and whether the plant CBC exerts an effect on splicing by a similar mechanism to animals—promoting spliceosome assembly on the first intron—is unknown. In this article, we have addressed the influence of the nuclear cap-binding complex on alternative splicing of 252 A. thaliana gene transcripts showing alternative splicing, using the T-DNA insertion knock-out mutants: the single cbp20 and cbp80(abh1) mutants and the double cbp20/80 mutant. In the cases that showed significant changes in AS, the mutants preferentially affect alternative splicing of the first intron and particularly at the 5′ splice site. Similar changes in the alternative splicing profiles of many gene transcripts were observed in all three cbp mutants suggesting that the CBC is directly involved in alternative splicing of these pre-mRNAs. Interestingly, our results revealed that AtCBP80 plays a more significant role in alternative splicing than AtCBP20.
MATERIALS AND METHODS
Plant material and growth conditions
Arabidopsis thaliana (wild-type Columbia and cbp mutant) seeds were placed in a bell-jar and sterilised using hydrochloric acid fumes generated from a solution containing 100 ml ACE bleach (5.25% v/v, Procter and Gamble) and 3 ml concentrated HCl. Sterilised seeds were sown in soil treated with Amistar fungicide (Syngenta), and were kept at 4°C for 72 h in the dark. After vernalisation, plants were grown in a growth chamber (SANYO MLR-350H) under controlled environmental parameters: 70% humidity, temperature 22°C, 16 h light/8 h dark photoperiod regime at 150–200 μE/m2. Five-week-old plants were harvested and leaf tissue was flash frozen in liquid nitrogen and stored at −80°C. The A. thaliana wild-type Columbia was originally obtained from Lehle Seeds; the cbp20 and cbp80(abh1) mutants were as described in Papp et al. (52) and Hugouvieux et al. (50), respectively. The cbp20/80 double mutant was obtained by crossing cbp20 and cbp80(abh1), and the homozygous line was isolated based on PCR assays.
RNA extraction and RT–PCR
Total RNA was extracted from 100 mg, five-week-old leaves using the RNeasy Plant Mini Kit (Qiagen) and quantified spectrophotometrically at 260 nm. RT–PCR experiments were performed on total RNA isolated after DNase RQ1 treatment, according to the manufacturer’s instructions (Promega). Reverse transcription by M-MLV RT (RNase H-) (Promega) was performed using oligo d(T)18 oligonucleotide as a primer. First, 5 µg of extracted RNA and 1 µl oligo d(T)18 (10 mM) were mixed in a total volume of 26 µl, incubated for 5 min at 65°C to melt secondary structures within the template, and cooled immediately for 10 min on ice. Subsequently, 8 μl M-MLV 5× Reaction Buffer, 1 μl M-MLV (RNase H-) (200 U/μl), 4 μl nucleotide mix (10 mM each dNTP) and 1 μl RNasin (40 U/μl) (Promega) were added to form a 40 μl volume reaction mix. The reaction mix was incubated for 1.5 h at 42°C and further incubated at 70°C for 10 min to inactivate the reverse transcriptase. The reverse transcription reaction was diluted to a final volume of 100 µl, and 1 µl cDNA was then aliquoted into a reaction tube along with 2.5 μl 10× PCR buffer with MgCl2 (Roche), 4 μl nucleotide mix (1.25 μM of each dNTP, Promega), 0.75 µl of combined alternative splicing event-specific primers (100 μM stock) and Taq DNA Polymerase (5 U/µl, Roche). A 25-μl volume PCR reaction mix was then subjected to the standard PCR reaction: 94°C for 2 min, followed by 24 cycles of 94°C for 15 s, 50°C for 30 s, 70°C for 1 min and completed with 10 min at 72°C. We have shown previously that 24 PCR cycles is in the exponential amplification range for mRNA transcripts of numerous genes with a range of expression levels (59). AS-specific primers were selected to amplify the expected alternative spliced mRNA isoforms, and gave RT–PCR products of sizes between 60 and 700 bp. In order to visualize the RT–PCR products, the forward primer was labelled with one of the following dyes: 6-FAM, VIC, NED or PET (Applied Biosystems). Primer sequences are given in Supplementary Table S1.
Splicing analysis
Labelled RT–PCR product (1 µl) from the RT–PCR reactions was mixed with 8.95 µl Hi Di Formamide (Applied Biosystems) and with 0.05 µl of GeneScan 500 LIZ or GeneScan 600 LIZ internal size standard (Applied Biosystems). Using an ABI3730 DNA Analyzer (Applied Biosystems), capillary electrophoresis of RT–PCR fragments was performed. Peak size and area data was analysed with GeneMapper or PeakScanner software (Applied Biosystems). RT–PCR products were accurately identified with ±1 nt resolution. The relative fluorescent peak areas for RT–PCR products with expected sizes for the alternatively spliced products were extracted, and a ratio for the AS events was calculated by dividing the value for the spliced products by the sum of the values for the alternatively spliced products. For an accurate measurement of AS ratios, three biological repeats were performed for all experiments. Mean alternative splicing efficiencies with standard errors were calculated for three separate biological repetitions. Means were compared by analysis of variance between wild type plants and the different CBP mutant lines and P-values generated. AS events with significant variation (P < 0.10) were selected.
RESULTS
Alternative splicing RT–PCR panel
The influence of nuclear cap-binding proteins was examined by analyzing 435 Arabidopsis alternative splicing events on a custom high resolution RT–PCR panel. The AS RT–PCR panel was based on that described previously which contained 89 AS events and seven controls (59). This panel was expanded to contain 428 AS events and seven controls. The AS events were in transcripts from genes encoding transcription factors (179), splicing factors (88), stress-related proteins (ABA) (51), stomatal ABA signaling (10), flowering time regulating proteins (16) and other miscellaneous proteins (91). The events were selected from either published AS events or from five different Arabidopsis/plant bioinformatics databases: ASIP (http://www.plantgdb.org/ASIP/EnterDB.php), TIGR (http://www.tigr.org/tdb/e2k1/ath1/), RIKEN (http://rarge.gsc.riken.jp/a_splicing/index.pl), ASTRA (http://alterna.cbrc.jp/) and TAIR 7.0 (http://www.arabidopsis.org/index.jsp). Seven genes were used as controls and amplified intronless regions (At5g03240, At5g60670, At3g61860), constitutively spliced introns (At3g12110, At5g13480) or a U12-dependent intron (At4g02560). Additionally, RuBisCo activase (At2g39730) is an alternative splicing control that consistently shows equal selection of two alternative 3′ splice sites in transcripts in all the tissues tested so far and was used as an alternative splicing control.
The splicing profiles were determined by RT–PCR with fluorescently labelled primers for each AS event using total RNA from wild type plants and from the single mutants: cbp20 and cbp80(abh1) and the double mutant: cbp20/80. The double mutant has a similar phenotype to the single mutants: slow growth and serrated leaf margins, ABA hypersensitivity and greater resistance to water deficiency than wild type plants. PCR products were separated on an ABI3730 automated DNA sequencer and analysed by GeneMapper or PeakScanner software (Applied Biosystems). The ratio of the peak areas of the alternatively spliced isoforms for each gene in the wild type plant was compared to the ratio of the peak areas of products in the cbp mutants. Means and standard errors were calculated for three separate experiments.
Alternative splicing is affected in cbp mutants
Of the 435 genes tested, 99 genes did not show RT–PCR products. This is most likely to be due to very low level of expression of these particular genes in the plant tissue analysed (35-days-old leaves). For 84 cases only one splicing isoform was observed. This suggested that the alternative splicing events did not occur in the tissue and developmental stage analysed or represented extremely rare events even though supported by EST/cDNA sequences. As neither of the above groups provide alternative splicing information in the tissue analysed, these events were not included in further analysis.
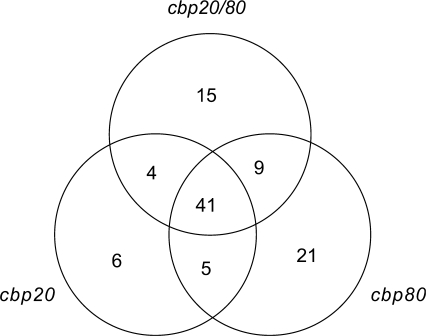
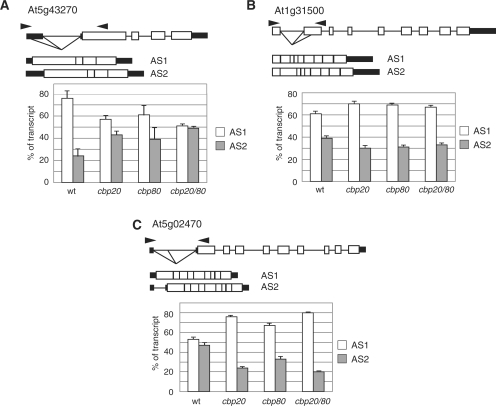
Of the 252 remaining different AS events 101 showed significant (P ≤ 0.10) changes in the ratio of alternatively spliced isoforms of over 3% between wild type plants and the cbp20, cbp80(abh1) or cbp20/80 mutants. Of these events, 41 were common to all three mutants, nine were found in both the cbp80(abh1) and cbp20/80 mutants, four were common to cbp20 and cbp20/80, five were common to cbp20 and cbp80(abh1), 15 were found only in the double mutant, and 6 and 21 only in the single cbp20 and cbp80(abh1) mutants, respectively (Figure 1). Different types of alternative splicing events were affected in the cbp mutants (Table 1). The majority of the alternative splicing events involved two alternative products (Table 2) and four events involved three alternative products (Table 3). Thus, the majority of AS events showing significant changes in alternative splicing profiles (41 events—41%) were found in all three cbp mutants. For example, in At5g43270, both single mutants showed a similar significant change in AS but the largest change was seen in the double mutant where the ratio of the two alternatively spliced isoforms changed from 76%:24% to 51%:49% (Figure 2A). In At1g31500, the degree of change was similar in all three mutants changing from 61%:39% to ∼70%:30% (Figure 2B). In At5g02470, there were different degrees of significant changes in all three mutants but again the biggest change in the ratio of AS1:AS2 was in the double mutant (from 53:47 to 80:20%) (Figure 2C). Knocking out the CBC could potentially have a general affect on mRNA stability leading to the observed changes in alternative splicing. We therefore analysed by microarray analysis the expression levels of genes in the cbp20 and cbp80 mutants compared to wild-type. Across all genes, >97% showed no differences in expression and in the genes containing the 101 significantly different alternative splicing events, only one had a reduced expression level and two had increased expression (data not shown). Thus, the CBC directly influences the selection of alternative splice sites and the extent and pattern of changes varied among the AS events and the different mutants.
Figure 1.
Distribution of alternative splicing events with significant changes in alternative splicing profiles in the cbp mutants.
Table 1.
Distribution of different alternative splicing events with significantly changed alternative splicing profile among the cbp mutants
| Alternative splicing event |
||||||
|---|---|---|---|---|---|---|
| Gene number | Alt 3′ splice site | Alt 5′ splice site | Exon skipping | Alternative position | Intron retention | |
| All cbp mutants | 41 | 20 | 18 | 3 | 0 | 0 |
| cbp80 plus cbp20/80 | 9 | 4 | 1 | 2 | 1 | 1 |
| cbp20/80 | 15 | 9 | 3 | 1 | 0 | 2 |
| cbp80 | 21 | 9 | 5 | 2 | 2 | 3 |
| cbp20 plus cbp80 | 5 | 5 | 0 | 0 | 0 | 0 |
| cbp20 | 6 | 3 | 3 | 0 | 0 | 0 |
| cbp20 plus cbp20/80 | 4 | 3 | 0 | 1 | 0 | 0 |
| Total | 101 | 53 | 30 | 9 | 3 | 6 |
Table 2.
Significant changes in alternative splicing isoform (two products) abundance in nuclear cap-binding complex mutants
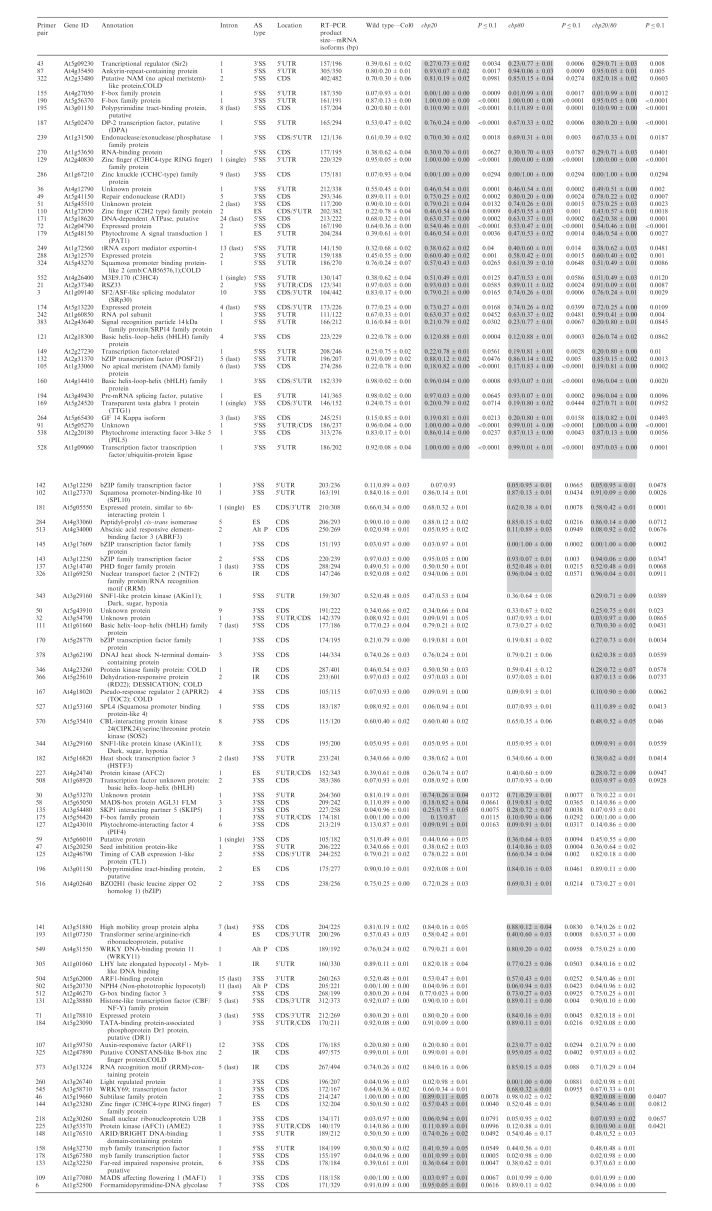 |
The relative abundance of alternatively spliced isoforms is presented as a ratio of the two products for wild type, for the single mutants cbp20 and cbp80, and the double mutant cbp20cbp80 (cbp20/80). The standard errors derived from two or three repeat experiments are given with the ratios. Significant changes between the wild type and mutants is measured at P ≤ 0.10. Only comparisons which show significant changes from wild type are presented with a P-value and are shaded grey. ES—exon skipping; 5′SS—alternative 5′ splice site; 3′SS—alternative 3′ splice site; AltP—events involving both alternative 5′ and 3′ splice sites; IR—intron retention; CDS—coding sequence; 5′UTR—5′ untranslated region; 3′UTR—3′ untranslated region.
Table 3.
Significant changes in alternative splicing isoform abundance in nuclear cap-binding complex mutants for genes with three alternative splicing isoforms
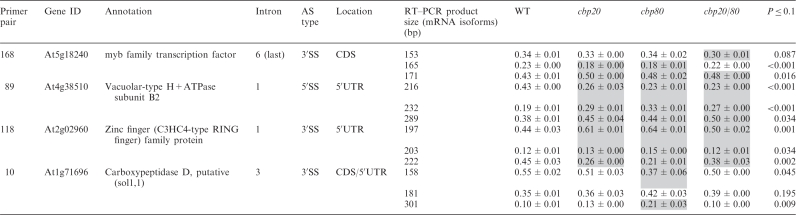 |
The relative abundance of the three alternatively spliced isoforms is presented for each of the three products for wild type, for the single mutants cbp20 and cbp80, and for the double mutant cbp20cbp80 (cbp20/80). Standard errors are the result of two or three biological repeats. Significant changes in the abundance of each product is determined by comparing across the wild type and all three mutants. Significance is measured at P ≤ 0.10. Products showing significant changes are shaded grey. 5′SS—alternative 5′ splice site; 3′SS—alternative 3′ splice site; CDS—coding sequence; 5′UTR—5′ untranslated region; 3′UTR—untranslated region.
Figure 2.
Gene and transcript stuctures of examples of alternative splicing events which change significantly in three cbp mutants. The exon/intron structure of the gene is shown with those of the alternatively spliced transcripts (AS1 and AS2). The proportion of each transcript in wild type (wt) plant and the cbp20, cbp80(abh1) and cbp20/80 mutants is shown as a percentage in the histograms. (A) At5g43270, (B) At1g31500 and (C) At5g02470. Boxes—exons; black boxes—untranslated regions (UTRs); horizontal lines—introns; diagonal lines below gene structure—alternative splicing event; standard errors of the means are indicated.
Alternative splicing is mainly affected in the cbp80 and cbp20/80 double mutants
Forty-one AS events were significantly changed in all three cbp mutants. Of the remaining 60 events which showed changes in either two out of three mutants or only in one of them (Figure 1), only six events involved the cbp20 mutant while 21 and 15 events involved either the cbp80(abh1) or the cbp20/80 mutants, respectively (Figure 1). In addition, nine events with significant changes in alternative splicing were common to the cbp80 and cbp20/80 mutants such that the majority of events affected in the cbp mutants were found in the cbp80 and cbp20/80 double mutants.
cbp mutants preferentially affect alternative splicing in the first intron
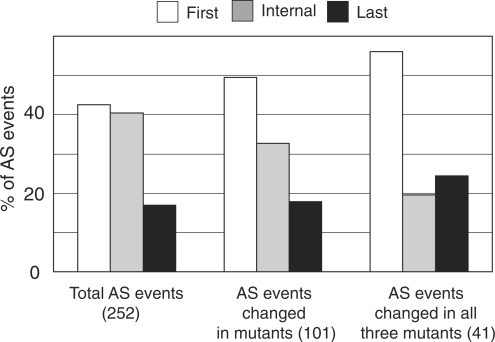
The CBC in animal and yeast systems promotes an efficient interaction between U1snRNP and the first intron, and enhances production of spliced mRNAs (29,30). To examine whether Arabidopsis nuclear cap-binding proteins also affected the first intron of transcripts, we compared the position of the introns affected by alternative splicing in the total number of events (252), the events which showed a significant change at least one of the mutants (101) (Figure 1) and those events which showed a significant change in alternative splicing in all three mutants (41). Among the 252 events showing alternative splicing on the RT–PCR panel, 107 events (42%) involved the first intron in the gene transcript, 102 (40%) involved an internal intron and 43 (17%) involved the last intron in the transcript (Figure 3). In the group of 101 AS events with a significantly different alternative splicing profile in at least one mutant 50% are alternatively spliced in the first intron, whereas 33 and 18% are alternatively spliced in the internal intron and the last intron of transcripts, respectively. Interestingly, of the 41 events which showed significant changes in alternative splicing in all three mutants, the first intron was alternatively spliced in 23 of the events (56%) (Figure 3). Only 19.5% involved internal introns and 24% involved the last intron. Thus, the percentage of alternatively spliced first and internal introns differs significantly between the total number of AS events analysed and those with significant changes in the three cbp mutants. The percentage of AS events involving the last intron increased in the AS events which were significantly different in all three mutants (Figure 3). These results indicate that for those genes where mutation of the CBC affected alternative splicing, the CBC is preferentially involved in alternative splicing of the first intron. In most cases (93%), the alternative first introns are located within the 5′UTRs or span the 5′UTR and coding sequence of the genes studied.
Figure 3.
Percentage distribution of the position of alternative splicing events. The positions of the alternatively spliced introns (first intron, internal intron, last intron) are presented for the total events showing alternative splicing (252 AS events), AS events which changed in at least one mutant plant (101 AS events) and those which showed significant changes in all three cbp mutants (41 AS events).
cbp mutants preferentially affect alternative 5′ splice site selection
Since the CBC promotes an efficient interaction between U1snRNP and the 5′ site of the first intron during constitutive splicing, we asked whether the CBC can also influence the selection between distal or proximal alternative 5′ splice sites during alternative splicing. First, we compared the number of events involving alternative 5′ splice site selection among all of the events showing alternative splicing (252), in those where AS profiles changed in at least one mutant (101) or in all three mutants (41). Of the 252 AS events, 75 (30%) involved selection of alternative 5′ splice sites (Figure 4A). Similarly, of the 101 AS events changed in at least one mutant plant only 30 (30%) of AS events involved 5′ splice sites. Interestingly, a greater proportion of the events where alternative splicing changed in all three mutants (18 of the 41 events—44%), involved alternative 5′ splice sites with fewer events involving alternative 3′ splice site selection (Figure 4A).
Figure 4.
Percentage distribution of alternative splicing phenotypes. The alternative splicing phenotypes are presented for (A) the total events showing alternative splicing (252 AS events), AS events which changed in at least one mutant plant (101 AS events) and those which showed significant changes in all three cbp mutants (41 AS events) and (B) the total events involving the first intron (107 AS events), AS events changed in the first intron in at least one mutant plant (50 AS events) and those with significant changes in the three cbp mutants which involved the first intron (23 AS events). ES—exon skipping; Alt 5′—alternative 5′ splice site; Alt 3′—alternative 3′ splice site; AltP—events involving both alternative 5′ and 3′ splice sites; IR—intron retention.
Second, we looked at the number of alternative 5′ splice site selection events among those involving the first intron. These groups consisted of 107 of the 252 total AS events, 50 of the 101 AS events changed in at least one cbp mutant and 23 of the 41 AS events with significant changes in all three mutants. Of the 107 first intron events, 36 (34%) involved alternative 5′ splice site selection. Similarly, of the 50 AS events changed in at least one cbp mutant, 16 (32%) involved 5′ splice site selection. Finally, of the 23 first intron events with significant AS changes in all three cbp mutants, 11 (48%) involved alternative 5′ splice site selection (Figure 4B). Thus, both the total number of alternative splicing events which are changed in all three mutants and those which affect the first intron are enriched in alternative 5′ splice site selection. This change in distribution suggests that CBC proteins are preferentially, but not exclusively, involved in the regulation of alternative 5′ splice site selection in the first intron.
As the CBC preferentially affects alternative 5′ splice selection in the first intron of transcripts, we examined whether the CBC also influenced the selection of the alternative 5′ splice site. If the CBC promoted usage of the 5′ splice site nearest to it, these events would be expected to predominantly use the 5′ splice site proximal to the cap. A total of 16 AS events involved alternative 5′ splice sites in the first intron and showed significant changes in all three mutants (11 events) or in one mutant (five events) (Table 4). Of these, seven preferentially used the 5′ splice site closest to the cap in wild type plants, seven used the cap-distal site and two used both sites equally. This suggests that there is no preference for selection of either cap-proximal or cap-distal sites in first intron. The splicing behaviour of these events in the absence of the CBC followed two basic and subtly different patterns. In the first case, in the wild type, the CBC preferentially promoted selection of either the cap-proximal or the cap-distal site to generate more of the major alternative splicing isoform. In the knock-out mutants, the use of these sites were reduced increasing the amount of the minor isoform. Thus, although both sites were used, the CBC preferentially used one or other of the 5′ splice sites. Six of the 16 events followed this pattern (Table 4—labelled as ‘Major’). In the second case, one splice site was used preferentially in the wild-type as above, but in the mutants, use of this splice site increased showing that the CBC actively promoted usage of the alternative splice site which generates the minor splice isoform. This occurred in eight of the remaining cases (Table 4—labelled ‘Minor’). In two events studied, it was impossible to point out the major splicing isoform since both alternative variants occur in equal amounts in wild type plants. However, the ratio between both splicing isoforms changed significantly in the cbp mutants; in one case use of the proximal 5′ss increased, and in another, use of the proximal 5′ss decreased. Our results show that there is no preference for the CBC to promote usage of the 5′ splice site closest to it, and the CBC, therefore can promote selection of one or other or both (or more) alternative 5′ splice sites to increase the levels of alternative isoforms.
Table 4.
Use of alternative 5′ splice sites in first introns with significant changes in nuclear cap-binding complex mutants
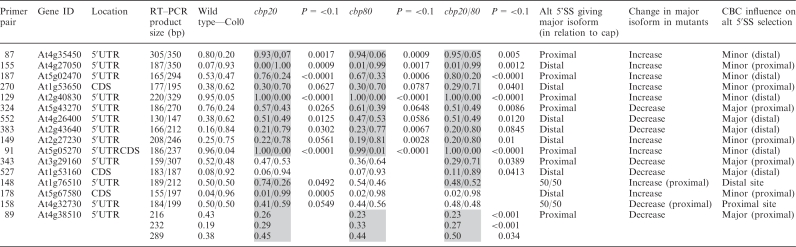 |
The relative abundance of alternatively spliced isoforms is presented as a ratio of the two products for wild type, for the single mutants cbp20 and cbp80, and for the double mutant cbp20cbp80 (cbp20/80). Significant changes between the wild type and mutants is measured at P ≤ 0.10. Only comparisons which show significant changes from wild type are presented with a P-value and are shaded grey. CDS—coding sequence; 5′UTR—5′ untranslated region; 3′UTR—3′ untranslated region.
Since in animals, the CBC promotes efficient splicing of the first intron during constitutive splicing we asked whether it also affects the efficiency of intron excision in plants. To address this, we looked at the six intron retention events where there was a significant change in splicing in at least one cbp mutant. Of these, two events involved retention of the first intron (At1g01060 and At4g23260) and two of the second intron (At5g25610 and At2g47890). Although significant in only the cbp80 or the double mutant, all four showed a decrease in splicing of the intron. The other two intron retention events affected the fifth and sixth introns (At3g13224 and At1g69250) and splicing efficiency increased in the mutants affected. Although only a small number of intron retention events showed significant changes, those involving introns near the 5′ end of the mRNA were less efficiently spliced in at least one mutant suggesting that the CBC may influence the efficiency of removal of such introns.
DISCUSSION
The nuclear CBC in animal systems promotes an efficient interaction between the U1snRNP and the first intron of a pre-mRNA transcript, thereby enhancing spliceosome assembly and the formation of spliced mRNAs. Using knock-out mutants of the Arabidopsis cap-binding proteins and an alternative splicing RT–PCR panel, we have examined the influence of the CBC on alternative splicing of multiple different pre-mRNAs. The CBC significantly affected (P ≤ 0.10) the alternative splicing of 101 events from the 252 analysed and the events involved introns at different positions in the various transcripts. However, the cbp mutants preferentially affected splicing within the first intron in over half of the AS events which had significant changes in the mutants. In addition, this effect was preferentially exerted at the 5′ splice site consistent with the model for animal systems that the CBC promotes 5′ splice site selection of the first intron. Clearly many internal or last introns also showed an influence of the CBC on splicing/alternative splicing. Indeed, when considering events affected in all three mutants, the proportion of last introns affected increased as well as the proportion of first introns. The effect of the CBC on introns other than those towards the 5′-end of the mRNA may be the result of indirect regulation of other splicing factors affecting the splicing of these (and also some first introns) or may reflect three-dimensional interactions in the pre-mRNA. For example, interactions between the cap and polyadenylation proteins to circularize mRNAs increase translational efficiency (60) and have been suggested to be involved in NMD (61). In addition, plant genes and particularly introns are much smaller than those in animal genes (10) potentially permitting a range of interactions between the CBC and other regions of the transcript.
An effect of the CBC on alternative splicing has not been shown previously. Consideration of the model of the CBC promoting the interaction between the U1snRNP and 5′ splice site of the first intron predicts that, when alternative 5′ splice sites are available, the CBC would promote splicing by recruiting of U1snRNP to one or other of these sites and potentially to the 5′ splice site nearest to the cap. Our analysis of alternative 5′ splice site selection in first introns with significant changes in AS in the cbp mutants showed firstly that there was no preference for use of the cap-proximal alternative 5′ splice sites, and secondly, that the CBC either showed a preference for one or other site or actively promoted use of a minor site. Thus, the CBC affected usage of both major and minor alternative 5′ splice sites. This suggests that while the CBC preferentially affects alternative 5′ splice site selection in the first intron, it can promote usage of both alternative splice sites and thereby influence the levels of alternative isoforms. The choice of site and degree of use are therefore most likely to reflect the strength of the splice sites, possibly the distance of the sites from the cap, and the presence of splicing enhancer or silencer sequences and interactions of other splicing factors. This is the first demonstration that the CBC can affect alternative splicing and is again consistent with the model of the CBC mediating interactions between the U1 snRNP and 5′ splice site of the first intron.
Comparison of the effects of the two single mutants and the double mutant showed that on the basis of the number of alternatively spliced events analysed here, more AS events were affected by mutation of AtCBP80(ABH1) than AtCBP20. This suggests that the larger subunit of the plant nuclear CBC plays a more significant role in this complex during splicing/alternative splicing regulation. Given that in plants, CBP20 contains the NLSs and that nuclear import of CBP80 is thought to depend on CBP20, we expected that mutation of CBP20 would be more severe in its effects on splicing. However, although in the single mutants, no AtCBP20 or AtCBP80 is detected, the protein levels of the remaining AtCBP80 and AtCBP20 respectively vary. Interestingly, in the cbp80(abh1) mutant, not only is the CBP80 protein absent but also the level of AtCBP20 protein is much reduced suggesting that in the absence of AtCBP80, the AtCBP20 subunit is unstable (48). Thus, the cbp80 mutant is very similar to the double mutant in terms of the levels of CBP20 and CBP80 proteins and the splicing phenotypes of these mutants. In contrast, in the cbp20 mutant which affected fewer of the alternative splicing events, AtCBP20 was absent but the AtCBP80 protein was present only at slightly reduced level to wild-type. This suggests that in the absence of AtCBP20, the larger subunit may still be able to interact in the splicing process albeit less efficiently. In yeast, CBP20-deficient (cbp20-Δ) mutants accumulated amounts of yCBP80 similar to those observed in the wild type strain, whereas the cbp80-Δ strain accumulated four-fold less yCBP20 than the wild type, suggesting that yCBP20 is unstable in the absence of yCBP80 (44,62). This observation suggests that protein interaction with CBP80 may modulate the stability of its partners. Assuming that AtCBP80 can interact with other splicing factors, it may still recruit factors to particular pre-mRNAs and thereby exert a stronger influence on splicing than AtCBP20. It was previously shown that the level of the CBC in the nucleus is precisely regulated (48) and the composition of the protein bound to the cap changed dynamically during a growth cycle in Arabidopsis, playing a role in the regulation of gene expression (63). Finally, that the single and double mutants are viable [similar to yeast (42,62)], and that many splicing events are unaffected in the mutants in this analysis, suggests the splicing of some introns is more dependent on the CBC than others. This may reflect the strength of intron splicing signals and of splicing enhancer sequences which can recruit splicing factors without the CBC.
Some splicing alterations were unique to the cbp20 and the cbp80(abh1) mutants which may reflect variation in the degree of dependence of intron splicing on the cap or proteins associated with the CBPs or varying ability of other proteins to compensate in the absence of one or other of the CBPs. Similarly, some AS changes were observed in the double mutant only. It is also possible that loss of one or other or both CBPs affects the splicing/alternative splicing of transcripts of genes encoding specific factors required for splicing of certain genes thereby indirectly affecting splicing of these genes. The fact that alternative splicing of transcription and splicing factors may be regulated by the CBC complex can explain, at least partially, the growth, developmental and physiological phenotypes of the mutants. In conclusion, in addition to constitutive splicing, the CBC is involved in alternative splicing in plants. In both cases, the CBC preferentially influences the first intron, particularly at its 5′ splice site during alternative splicing in plants.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Polish Ministry of Science and Higher Education [PBZ-MNiSW-2/3/2006/5.2 to Z.S.K, PBZ 01-003-00-07 to A.J. and 312/N-COST/2008/0 to A.J. and Z.S.K.]; the Scottish Government—Rural and Environment Research and Analysis Directorate (RERAD) [WP114]; and the EU FP6 Programme Network of Excellence on Alternative Splicing (EURASNET) [LSHG-CT-2005-518238 to A.J. and J.W.S.B.]. Funding for open access charge: Department of Gene Expression and Faculty of Biology, Adam Mickiewicz University, Poznan, Poland.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Graveley BR. Alternative splicing: increasing diversity in the proteomic world. Trends Genet. 2001;17:100–107. doi: 10.1016/s0168-9525(00)02176-4. [DOI] [PubMed] [Google Scholar]
- 2.Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 3.Blencowe BJ. Alternative splicing: new insights from global analyses. Cell. 2006;126:37–47. doi: 10.1016/j.cell.2006.06.023. [DOI] [PubMed] [Google Scholar]
- 4.Campbell MA, Haas BJ, Hamilton JH, Mount SM, Buell CR. Comprehensive analysis of alternative splicing in rice and comparative analysis with Arabidopsis. BMC Genomics. 2006;7:327–343. doi: 10.1186/1471-2164-7-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xiao YL, Smith SR, Ishmael N, Redman JC, Kumar N, Monaghan EL, Ayele M, Haas BJ, Wu HC, Town CD. Analysis of cDNAs of hypothetical genes on Arabidopsis chromosome 2 reveals numerous transcript variants. Plant Physiol. 2005;139:1323–1337. doi: 10.1104/pp.105.063479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 7.Ner-Gaon H, Halachmi R, Savaldi-Goldstein S, Rubin E, Ophir R, Fluhr R. Intron retention is a major phenomenon in alternative splicing in Arabidopsis. Plant J. 2004;39:877–885. doi: 10.1111/j.1365-313X.2004.02172.x. [DOI] [PubMed] [Google Scholar]
- 8.Wang BB, Brendel V. Genome-wide comparative analysis of alternative splicing in plants. Proc. Natl Acad. Sci. USA. 2006;103:7175–7180. doi: 10.1073/pnas.0602039103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baek JM, Han P, Iandolino A, Cook DR. Characterization and comparison of intron structure and alternative splicing between Medicago truncatula, Populus trichocarpa, Arabidopsis and rice. Plant Mol. Biol. 2008;67:499–510. doi: 10.1007/s11103-008-9334-4. [DOI] [PubMed] [Google Scholar]
- 10.Barbazuk WB, Fu Y, McGinnis KM. Genome-wide analyses of alternative splicing in plants: opportunities and challenges. Genome Res. 2008;18:1381–1392. doi: 10.1101/gr.053678.106. [DOI] [PubMed] [Google Scholar]
- 11.Reddy ASN. Alternative splicing of pre-messenger RNAs in plants in the genomic era. Annu. Rev. Plant Biol. 2007;58:267–294. doi: 10.1146/annurev.arplant.58.032806.103754. [DOI] [PubMed] [Google Scholar]
- 12.Simpson CG, Lewandowska D, Fuller J, Maronova M, Kalyna M, Davidson D, McNicol J, Raczynska D, Jarmolowski A, Barta A, et al. Alternative splicing in plants. Biochem. Soc. Trans. 2008;36:508–510. doi: 10.1042/BST0360508. [DOI] [PubMed] [Google Scholar]
- 13.Stamm S, Ben-Ari S, Rafalska I, Tang Y, Zhang Z, Toiber D, Thanaraj TA, Soreq H. Function of alternative splicing. Gene. 2005;344:1–20. doi: 10.1016/j.gene.2004.10.022. [DOI] [PubMed] [Google Scholar]
- 14.Pan Q, Saltzman AL, Kim YK, Misquitta C, Shai O, Maquat LE, Frey BJ, Blencowe BJ. Quantitative microarray profiling provides evidence against widespread coupling of alternative splicing with nonsense-mediated decay to control gene expression. Genes Dev. 2006;20:153–158. doi: 10.1101/gad.1382806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ni JZ, Grate L, Donohue JP, Preston C, Nobida N, O’Brien G, Shiue L, Clark TA, Blume JE, Ares M. Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes Dev. 2007;21:708–718. doi: 10.1101/gad.1525507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kalyna M, Lopato S, Voronin V, Barta A. Evolutionary conservation and regulation of particular alternative splicing events in plant SR proteins. Nucleic Acids Res. 2006;34:4395–4405. doi: 10.1093/nar/gkl570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schöning JC, Streitner C, Meyer IM, Gao Y, Staiger D. Reciprocal regulation of glycine-rich RNA-binding proteins via an interlocked feedback loop coupling alternative splicing to nonsense-mediated decay in Arabidopsis. Nucl. Acids Res. 2008;36:6977–6987. doi: 10.1093/nar/gkn847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Larkin PD, Park WD. Transcript accumulation and utilization of alternate and non-consensus splice sites in rice granule-bound starch synthase are temperature-sensitive and controlled by a single-nucleotide polymorphism. Plant. Mol. Biol. 1999;40:719–727. doi: 10.1023/a:1006298608408. [DOI] [PubMed] [Google Scholar]
- 19.Dinkins RD, Majee SM, Nayak NR, Martin D, Xu Q, Belcastro MP, Houtz RL, Beach CM, Downie AB. Changing transcriptional initiation sites and alternative 5′- and 3′-splice site selection of the first intron deploys Arabidopsis protein isoaspartyl methyltransferase2 variants to different subcellular compartments. Plant J. 2008;55:1–13. doi: 10.1111/j.1365-313X.2008.03471.x. [DOI] [PubMed] [Google Scholar]
- 20.Titarenko E, Rojo E, León J, Sánchez-Serrano JJ. Jasmonic acid-dependent and -independent signaling pathways control wound-induced gene activation in Arabidopsis thaliana. Plant Physiol. 1997;115:817–826. doi: 10.1104/pp.115.2.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bove J, Kim CY, Gibson CA, Assmann SM. Characterization of wound-responsive RNA-binding proteins and their splice variants in Arabidopsis. Plant Mol. Biol. 2008;67:71–88. doi: 10.1007/s11103-008-9302-z. [DOI] [PubMed] [Google Scholar]
- 22.Dinesh-Kumar SP, Baker BJ. Alternatively spliced N resistance gene transcripts: their possible role in tobacco mosaic virus resistance. Proc. Natl Acad. Sci. USA. 2000;97:1908–1913. doi: 10.1073/pnas.020367497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lazar G, Goodman HM. The Arabidopsis splicing factor SR1 is regulated by alternative splicing. Plant Mol. Biol. 2000;42:571–581. doi: 10.1023/a:1006394207479. [DOI] [PubMed] [Google Scholar]
- 24.Hirose T, Sugita M, Sugiura M. cDNA structure, expression and nucleic acid-binding properties of three RNA-binding proteins in tobacco: occurrence of tissue-specific alternative splicing. Nucleic Acids Res. 1993;21:3981–3987. doi: 10.1093/nar/21.17.3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoshimura K, Yabuta Y, Ishikawa T, Shigeoka S. Identification of a cis element for tissue-specific alternative splicing of chloroplast ascorbate peroxidase pre-mRNA in higher plants. J. Biol. Chem. 2002;277:40623–40632. doi: 10.1074/jbc.M201531200. [DOI] [PubMed] [Google Scholar]
- 26.Izaurralde E, Lewis J, McGuigan C, Jankowska M, Darzynkiewicz E, Mattaj IW. A nuclear cap binding protein complex involved in pre-mRNA splicing. Cell. 1994;78:657–668. doi: 10.1016/0092-8674(94)90530-4. [DOI] [PubMed] [Google Scholar]
- 27.Ohno M, Sakamoto H, Shimura Y. Preferential excision of the 5′ proximal intron from mRNA precursors with two introns as mediated by the cap structure. Proc. Natl Acad. Sci. USA. 1987;84:5187–5191. doi: 10.1073/pnas.84.15.5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patzelt E, Thalmann E, Hartmuth K, Blaas D, Kuechler E. Assembly of pre-mRNA splicing complex is cap dependent. Nucleic Acids Res. 1987;15:1387–1399. doi: 10.1093/nar/15.4.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lewis JD, Izaurralde E, Jarmolowski A, McGuigan C, Mattaj IW. A nuclear cap-binding complex facilitates association of U1 snRNP with the cap-proximal 5′ splice site. Genes Dev. 1996;10:1683–1698. doi: 10.1101/gad.10.13.1683. [DOI] [PubMed] [Google Scholar]
- 30.Lewis JD, Görlich D, Mattaj IW. A yeast cap binding protein complex (yCBC) acts at an early step in pre-mRNA splicing. Nucleic Acids Res. 1996;24:3332–3336. doi: 10.1093/nar/24.17.3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lewis JD, Izaurralde E. The role of the cap structure in RNA processing and nuclear export. Eur. J. Biochem. 1997;247:461–469. doi: 10.1111/j.1432-1033.1997.00461.x. [DOI] [PubMed] [Google Scholar]
- 32.Cooke C, Alwine JC. The cap and the 3′ splice site similarly affect polyadenylation efficiency. Mol. Cell Biol. 1996;16:2579–2584. doi: 10.1128/mcb.16.6.2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Flaherty SM, Fortes P, Izaurralde E, Mattaj IW, Gilmartin GM. Participation of the nuclear cap binding complex in pre-mRNA 3′ processing. Proc. Natl Acad. Sci. USA. 1997;94:11893–11898. doi: 10.1073/pnas.94.22.11893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hamm J, Mattaj IW. Monomethylated cap structures facilitate RNA export from the nucleus. Cell. 1990;63:109–118. doi: 10.1016/0092-8674(90)90292-m. [DOI] [PubMed] [Google Scholar]
- 35.Izaurralde E, Stepinski J, Darzynkiewicz E, Mattaj IW. A cap binding protein that may mediate nuclear export of RNA polymerase II-transcribed RNAs. J. Cell Biol. 1992;118:1287–1295. doi: 10.1083/jcb.118.6.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jarmolowski A, Boelens WC, Izaurralde E, Mattaj IW. Nuclear export of different classes of RNA is mediated by specific factors. J. Cell Biol. 1994;124:627–635. doi: 10.1083/jcb.124.5.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Izaurralde E, Lewis J, Gamberi C, Jarmolowski A, McGuigan C, Mattaj IW. A cap-binding protein complex mediating U snRNA export. Nature. 1995;376:709–712. doi: 10.1038/376709a0. [DOI] [PubMed] [Google Scholar]
- 38.Furuichi Y, LaFiandra A, Shatkin AJ. 5′-Terminal structure and mRNA stability. Nature. 1997;266:235–239. doi: 10.1038/266235a0. [DOI] [PubMed] [Google Scholar]
- 39.Balatsos NA, Nilsson P, Mazza C, Cusack S, Virtanen A. Inhibition of mRNA deadenylation by the nuclear cap binding complex (CBC) J. Biol. Chem. 2006;281:4517–4522. doi: 10.1074/jbc.M508590200. [DOI] [PubMed] [Google Scholar]
- 40.Laubinger S, Sachsenberg T, Zeller G, Busch W, Lohmann JU, Rätsch G, Weigel D. Dual roles of the nuclear cap-binding complex and SERRATE in pre-mRNA splicing and microRNA processing in Arabidopsis thaliana. Proc. Natl Acad. Sci. USA. 2008;105:8795–8800. doi: 10.1073/pnas.0802493105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kuhn JM, Breton G, Schroeder JI. mRNA metabolism of flowering-time regulators in wild-type Arabidopsis revealed by a nuclear cap binding protein mutant, abh1. Plant J. 2007;50:1049–1062. doi: 10.1111/j.1365-313X.2007.03110.x. [DOI] [PubMed] [Google Scholar]
- 42.Uemura H, Jigami Y. GCR3 encodes an acidic protein that is required for expression of glycolytic genes in Saccharomyces cerevisiae. J. Bacteriol. 1992;174:5526–5532. doi: 10.1128/jb.174.17.5526-5532.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Colot HV, Stutz F, Rosbash M. The yeast splicing factor Mud13p is a commitment complex component and corresponds to CBP20, the small subunit of the nuclear cap-binding complex. Genes Dev. 1996;10:1699–1708. doi: 10.1101/gad.10.13.1699. [DOI] [PubMed] [Google Scholar]
- 44.Shen EC, Stage-Zimmermann T, Chui P, Silver PA. 7The yeast mRNA-binding protein Npl3p interacts with the cap-binding complex. J. Biol. Chem. 2000;275:23718–23724. doi: 10.1074/jbc.M002312200. [DOI] [PubMed] [Google Scholar]
- 45.Ishigaki Y, Li X, Serin G, Maquat LE. Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell. 2001;106:607–617. doi: 10.1016/s0092-8674(01)00475-5. [DOI] [PubMed] [Google Scholar]
- 46.Hosoda N, Kim YK, Lejeune F, Maquat LE. CBP80 promotes interaction of Upf1 with Upf2 during nonsense-mediated mRNA decay in mammalian cells. Nat. Struct. Mol. Biol. 2005;12:893–901. doi: 10.1038/nsmb995. [DOI] [PubMed] [Google Scholar]
- 47.Kmieciak M, Simpson CG, Lewandowska D, Brown JWS, Jarmolowski A. Cloning and characterization of two subunits of Arabidopsis thaliana nuclear cap-binding complex. Gene. 2002;283:171–183. doi: 10.1016/s0378-1119(01)00859-9. [DOI] [PubMed] [Google Scholar]
- 48.Kierzkowski D, Kmieciak M, Piontek P, Wojtaszek P, Szweykowska-Kulinska Z, Jarmolowski A. The Arabidopsis CBP20 targets the cap-binding complex to the nucleus, and is stabilized by CBP80. Plant J. 2009;59:813–823. doi: 10.1111/j.1365-313X.2009.03915.x. [DOI] [PubMed] [Google Scholar]
- 49.Dzikiewicz-Krawczyk A, Piontek P, Szweykowska-Kulinska Z, Jarmolowski A. The nuclear cap-binding protein complex is not essential for nonsense-mediated mRNA decay (NMD) in plants. Acta Biochim. Pol. 2008;55:825–828. [PubMed] [Google Scholar]
- 50.Hugouvieux V, Kwak JM, Schroeder JI. An mRNA cap binding protein, ABH1, modulates early abscisic acid signal transduction in Arabidopsis. Cell. 2001;106:477–487. doi: 10.1016/s0092-8674(01)00460-3. [DOI] [PubMed] [Google Scholar]
- 51.Hugouvieux V, Murata Y, Young JJ, Kwak JM, Mackesy DZ, Schroeder JI. Localization, ion channel regulation, and genetic interactions during abscisic acid signaling of the nuclear mRNA cap-binding protein, ABH1. Plant Physiol. 2002;130:1276–1287. doi: 10.1104/pp.009480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Papp I, Mur LA, Dalmadi A, Dulai S, Koncz C. A mutation in the cap binding protein 20 gene confers drought tolerance to Arabidopsis. Plant Mol. Biol. 2004;55:679–686. doi: 10.1007/s11103-004-1680-2. [DOI] [PubMed] [Google Scholar]
- 53.Kim S, Yang JY, Xu J, Jang IC, Prigge MJ, Chua NH. Two cap-binding proteins CBP20 and CBP80 are involved in processing primary MicroRNAs. Plant Cell. Physiol. 2008;49:1634–1644. doi: 10.1093/pcp/pcn146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bezerra IC, Michaels SD, Schomburg FM, Amasino RM. Lesions in the mRNA cap-binding gene ABA HYPERSENSITIVE 1 suppress FRIGIDA-mediated delayed flowering in Arabidopsis. Plant J. 2004;40:112–119. doi: 10.1111/j.1365-313X.2004.02194.x. [DOI] [PubMed] [Google Scholar]
- 55.Kuhn JM, Hugouvieux V, Schroeder JI. mRNA cap binding proteins: effects on abscisic acid signal transduction, mRNA processing, and microarray analyses. Curr. Top. Microbiol. Immunol. 2008;326:139–150. doi: 10.1007/978-3-540-76776-3_8. [DOI] [PubMed] [Google Scholar]
- 56.Szarzynska B, Sobkowiak L, Pant BD, Balazadeh S, Scheible WR, Mueller-Roeber B, Jarmolowski A, Szweykowska-Kulinska Z. Gene structures and processing of Arabidopsis thaliana HYL1-dependent pri-miRNAs. Nucleic Acids Res. 2009;37:3083–3093. doi: 10.1093/nar/gkp189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gregory BD, O'M;alley RC, Lister R, Urich MA, Tonti-Filippini J, Chen H, Millar AH, Ecker JR. A link between RNA metabolism and silencing affecting Arabidopsis development. Dev. Cell. 2008;14:854–866. doi: 10.1016/j.devcel.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 58.Parry DH, Xu J, Ruvkun G. A whole-genome RNAi Screen for C. elegans miRNA pathway genes. Curr. Biol. 2007;17:2013–2022. doi: 10.1016/j.cub.2007.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Simpson CG, Fuller J, Maronova M, Kalyna M, Davidson D, McNicol J, Barta A, Brown JWS. Monitoring changes in alternative precursor messenger RNA splicing in multiple gene transcripts. Plant J. 2008;53:1035–1048. doi: 10.1111/j.1365-313X.2007.03392.x. [DOI] [PubMed] [Google Scholar]
- 60.Tarun SZ, Jr, Sachs AB. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 1996;15:7168–7177. [PMC free article] [PubMed] [Google Scholar]
- 61.Brogna S, Wen J. Nonsense-mediated mRNA decay (NMD) mechanisms. Nat. Struct. Mol. Biol. 2009;16:107–113. doi: 10.1038/nsmb.1550. [DOI] [PubMed] [Google Scholar]
- 62.Fortes P, Kufel J, Fornerod M, Polycarpou-Schwarz M, Lafontaine D, Tollervey D, Mattaj IW. Genetic and physical interactions involving the yeast nuclear cap-binding complex. Mol. Cell Biol. 1999;19:6543–6553. doi: 10.1128/mcb.19.10.6543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bush MS, Hutchins AP, Jones AM, Naldrett MJ, Jarmolowski A, Lloyd CW, Doonan JH. Selective recruitment of proteins to 5′ cap complexes during the growth cycle in Arabidopsis. Plant J. 2009;59:400–412. doi: 10.1111/j.1365-313X.2009.03882.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.