Abstract
MicroRNAs (miRNAs) are 21–22 nucleotide regulatory small RNAs that repress message translation via base-pairing with complementary sequences in the 3′ untranslated region (3′UTR) of targeted transcripts. To date, it is still difficult to find a true miRNA target due to lack of a clear understanding of how miRNAs functionally interact with their targeted transcripts for efficient repression. Previous studies have shown that nucleotides 2 to 7 at the 5′-end of a mature miRNA, the ‘seed sequence’, can nucleate miRNA/target interactions. In the current study, we have validated that the RhoB mRNA is a bona fide miR-223 target. We have analyzed the functional activities of two miR223-binding sites within the RhoB 3′UTR. We find that the two miR-223 target sites in the RhoB 3′UTR contribute differentially to the total repression of RhoB translation. Moreover, we demonstrate that some AU-rich motifs located upstream of the distal miRNA-binding site enhance miRNA function, independent of the miRNA target sequences being tested. We also demonstrate that the AU-rich sequence elements are polar, and do not affect the activities of miRNAs whose sites lie upstream of these elements. These studies provide further support for the role of sequences outside of miRNA target region influencing miRNA function.
INTRODUCTION
Small regulatory RNAs are gaining attention for their important roles in spatially or temporally fine tuning target gene expression. Among the small regulatory RNAs, the miRNA family is the most extensively studied and their biogenesis and mechanisms of down-regulating gene expression represent some of the most exciting and fascinating areas in molecular biology (1,2).
Canonical miRNAs are generated from intronic or exonic capped, polyadenylated RNA polymerase II transcripts, termed primary miRNAs (pri-miRNA) (3–5). The primary transcripts are processed to ∼55–80 nt long precursors, partial hairpin-like duplexes termed pre-miRNAs, by the ribonuclease (RNase) III enzyme Drosha that partners with the RNA-binding protein DGCR8 (DiGeorge Critical Region 8) (4,6). Alternatively, there is a small percentage of pre-miRNAs that are generated by the action of the pre-mRNA splicing/de-branching machinery, termed the ‘miRtron pathway’ (7–9). In both pathways, pre-miRNAs are exported to the cytoplasm by the exportin-5/RAN-GTP complex (10). In the cytoplasm, the pre-miRNAs are processed once again into an miRNA/miRNA* duplex by the RNase III enzyme Dicer that partners with the RNA-binding protein TRBP (TAR RNA-binding protein) (11,12). Usually one of the two strands in the miRNA/miRNA* duplex is then incorporated into the RNA induced silencing complex (RISC), where the incorporated strand works as the guide for miRISC function. MicroRNAs in RISC bind to\ the 3′UTRs of transcripts harboring complementary ‘seed’ targets, ultimately resulting in translational repression (1,13) or in some cases degradation of the targeted mRNA in RNA processing bodies (P-bodies) (14,15).
In contrast to most plant miRNAs, which share near-perfect complementarity to their targeted sequences, most animal miRNAs usually form imperfect Watson–Crick base pairing with the target sequences. However, complete complementarity of six to seven nucleotides at the 5′-end of the miRNAs including nucleotides 2–7, the so-called ‘seed sequence’ has been shown to be crucial for miRNA function (16,17). For those binding sites with imperfect seed sequences, a strong 3′ base pairing could compensate for weak seed pairing to create more efficient miRNA-mediated target gene inhibition (16). On the basis of miRNA seed match hypothesis, it is estimated that, on average, an individual miRNA can target upwards of 200 transcripts (16,17). A recent prediction based on the targets of conserved vertebrate mammalian miRNAs predicts that the average number of targets per miRNA will exceed 300 (18). There are about 885 cloned or computer-predicted mature human miRNA sequences in the current human miRBase 13.0 (19), and it is estimated there may be as many as 1000 human miRNAs. Thus, 30% or more of the human transcriptome is potentially regulated by miRNAs (16,17). Conserved sequence motifs in mammalian 3′UTRs correlate well with miRNA target sites and it appears that 3′UTRs are under selective pressure to maintain miRNA interactions (18,20).
Since mammalian miRNAs interact with their targets by partial base pairing complementarity, the identification of miRNA targets has been a difficult undertaking. Given the high prevalence of six to seven nucleotide complementary sequences in the genome, identification of true targets for any given miRNA is a difficult task. Among the many factors that could affect translational repression mediated by miRISC are the sequence contexts of target sites, which can influence miRNA/mRNA-binding energies and Waston–Crick base pairing, the influence of flanking sequences on the accessibility of a target site, the occurrence of multiple target sites that provide additive or synergistic repression, and the relative position of the target sites within the 3′UTR (17,21). One intriguing problem is that not all seed matches in a given 3′ UTR of a validated target are effectively targeted [e.g. let-7 target sites in RAS (22), miR-150 target sites in Myb (23)]. Many additional unknown factors might exist that could affect miRNA targeting beside those aforementioned. It has been proposed that RNA-binding proteins (RBP) may play a role in translational repression. For example, puf-9 is required for let-7 repression of hbl-1 in C. elegans, Dnd1 counteracts the function of several miRNAs by binding to uridine-rich regions present in targeted mRNAs and prevents the loading of miRISC (24), HuR releases miR-122-mediated translational repression of the CAT-1 gene transcript (25), the poly(A)-binding protein (PABP) interacts directly with the P-body component GW182 for miRNA-mediated deadenylation of target messages (26), and the fragile X mental retardation-related protein 1 (FXR1) provocatively forms a miR-369-3p mediated translation activation complex with Ago2 when cells are in a translationally quiescent state (27). In silico studies have shown that the local structure around the target site plays a role in the efficiency of miRNA-mediated repression (28,29). Studies of the lsy-6 target COG-1 in C. elegans revealed that two sequence context features 3′ of the lsy-6-binding sites in the COG-1 3′ UTR are required for lsy-6 function and they need to be present in a specific configuration relative to the lys-6 targeting sites (30,31). Such contextual features could either represent RNA-binding protein (RBP)-binding sites or provide an appropriate structural configuration to favor or prevent RISC access. RBPs usually bind to specific sequence motifs. In silico studies have revealed that there are many types of regulatory RNA motifs in 3′UTRs (32). These sequences often overall with or include binding sites for HuR, TIA-1, and consist of AU rich elements (ARE) and GU rich elements (GRE). A significant portion of these have the core ARE sequences AUUUA and UAUUUAU (33). It is therefore of interest to determine whether or not specific sequence motifs in a given 3′UTR influence miRNA functional repression of that message. In the present study, we validate RhoB as a miR-223 target. We find that there are two miR-223 seed matches in this 3′UTR and they contribute differently to the total repression of RhoB translation by miR-223. We have analyzed the contributions of flanking sequences that may be responsible for these differences and our results reveal that some putative RBP motifs in the 3′UTR outside of the target site can influence the functionality of miRNAs. Our data support a model in which AU rich elements in 3′UTRs may be required for efficient miRNA mediated post-transcriptional regulation as previously predicted (34).
MATERIALS AND METHODS
Cell culture and transfection
HEK293, Hela, and CEM cells were purchased from ATCC. 293 and Hela cells were maintained in high glucose (4.5 g/l) DMEM supplemented with 2 mM glutamine, 10% FBS and 2 mM penicillin/streptomycin. CEM cells were grown in RPMI1640 supplemented with 2 mM glutamine, 10% FBS and 2 mM penicillin/streptomycin.
Transfections of 293 and Hela cells were performed with Lipofectamine 2000 (Invitrogen) in 24 well plate formats when cells were at about 80% confluency. Each well was transfected with a plasmid-Lipofectamine 2000 complex mixture of 10 ng psiCheck2.2-based reporter plasmids and 100 ng fU1-miR based pri-miRNA expression vectors. Forty eight hours post transfection, the cells were lysed with 100 μl Passive Lysis Buffer (Promega) and Luciferase levels were analyzed from 20 μl lysate using the Dual Luciferase reporter assay (50 μl of each substrate reagents, Promega) on a Veritas Microplate Luminometer (Turner Biosystems). Transfection results were obtained by averaging the results from at least three individual transfections with two replicates of each measurement. Transfections of CEM cells were performed with a nucleofection system (Amaxa) by following the protocol provided for CEM cells.
Primary miRNA expression
The primary miRNA expression system and miRNA target reporter assays have been previously described (35,36). The pri-miRNA expression vector, fU1-miR, was constructed by cloning the U1 promoter and the U1 termination sequence in the MCS of Bluescript SK. Pri-miRNA was cloned by PCR from HEK293 cell genomic DNA, then inserted between the U1 promoter and U1 terminator in fU1-miR. An amplified pri-miRNA fragment covers at least 100 bases of flanking sequence on both ends of the stem-loop miRNA reported in miRBase. Expression of functional mature miRNAs was tested using two methods. One is the knockdown of Renilla luciferase (Rluc) in a psiCheck2.2 reporter bearing a fully complementary miRNA target sequence in the Rluc 3′UTR. The other is a northern blot to detect the processed mature miRNAs by transfection of the fU1-miR expressed pri-miRNAs in cultured cells.
Dual luciferase reporter assays
Details of the dual luciferase reporter system have been previously reported (36). Reporters harboring the RhoB 3′UTR or NF-1A 3′UTR were constructed by inserting annealed synthetic oligo target sequences or the PCR amplified 3′UTR fragments into the XhoI/NotI sites of the 3′UTR of Rluc in the psiCheck2.2 dual reporter vector (Promega). For the GCR1 and GCR2 based reporters, annealed oligos were first cloned into the Xho I/Spe I sites of psiCheck2.2 to create GCR1- and GCR2- based reporters without TS1, TS2 or NF1A. The correct constructs were then inserted into Sal I/Not I sites along with one of the mir-223 target sites in RhoB (TS1, TS2), or the miR-223 target site in NF-1A (NF1A), to create reporters with GC-rich or AU-rich Rluc 3′UTRs harboring miR-223 target sites
Firefly luciferase (Fluc) expressed from the same vector serves as an internal normalization control. Binding of miRISC to the target site in Rluc’s mRNA 3′UTR results in a concomitant reduction of translated Rluc protein, which can be detected by a luminescence based assay system. The ratio of Rluc/Fluc was used to measure the repression efficiency. The empty pri-miRNA expression vector fU1-miR was used as negative control.
Mutagenesis of miRNA target sites
Point mutations were introduced with the QuikChange site-directed mutagenesis kit II (Stratagene) by following the protocol provided in the kit. Mutations were confirmed by sequencing.
Immunobloting
For western blot analyses of RhoB, cells in six-well plates were washed with 2 mL PBS and lysed in 0.2 mL M-PER® Mammalian protein extraction reagent (Pierce, #78501). After microcentrifugation at top speed for 10 min, the supernatants were collected and a cocktail of protease inhibitors (Roche) was added. The protein concentration was quantified by the Bradford method (Bio-Rad, Protein assay dye). Twenty micrograms of total protein extract were electrophoresed in a 15% SDS–PAGE gel at 60 V for 3 h, and electro-blotted to a Hybond™-P PVDF transfer membrane (GE healthcare) for 90 min at 80 V or semi-dry electro-blotted for 30 min at 15 V. The membrane was blocked in 5% dry milk in TBS-T (0.05% Tween) for over 1 h at 4°C, and probed with a primary rabbit anti-RhoB antibody (Santa Cruz Biotechnology, Inc.) overnight at 4°C, followed by one hour incubation with AP-conjugated secondary anti-rabbit Abs (Santa Cruz Biotechnology). Expression was visualized using standard AP detection chemistry (ECL western blotting substrate, Pierce).
RNA isolation and northern blotting
RNA isolation and northern blotting were carried out as previously reported (37). Briefly, RNA was isolated with RNA STAT-60 (Tel-Test Inc.) and 20 µg of total RNA were loaded in a denaturing 12.5% SDS–PAGE gel. A DNA oligonucleotide probe complementary to the sequence of the miRNA was labeled with γ-32P-ATP. The hybridization was performed overnight in PerfectHyb™ Plus Hybridization Buffer from Sigma at 37°C.
RESULTS
RhoB was predicated as an miR-223 target by miRanda with two miR-223-binding sites in the RhoB 3′UTR (Figures 1 and 2a) (38). In order to validate RhoB as a true miR-223 target, we used reporter assays to validate the miR-223/RhoB 3′UTR interactions at the two target sites. These two target sites exhibited distinct differences in their susceptibility to miR-223 mediated repression.
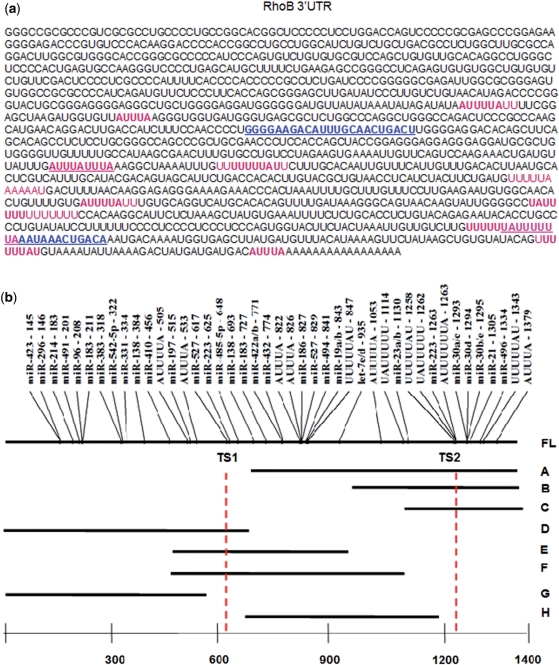
Figure 1.
RhoB 3′UTR sequence features and reporter constructs. (a) Human RhoB 3′UTR. Sequence of the two miR-223 target sites are in blue and underlined. AUUUA, UUUUUAU and UAUUUUU motifs are highlighted in pink. (b) Diagram shows miRanda and TargetScan predicted human miRNA target sites in the human RhoB 3′UTR. AU-rich motifs are also indicated. RhoB 3′UTR fragments that were cloned into the 3′UTR of Rluc in the psiCheck2.2 reporter for reporter assays are mapped with respect to their relative positions in the FL RhoB 3′UTR.
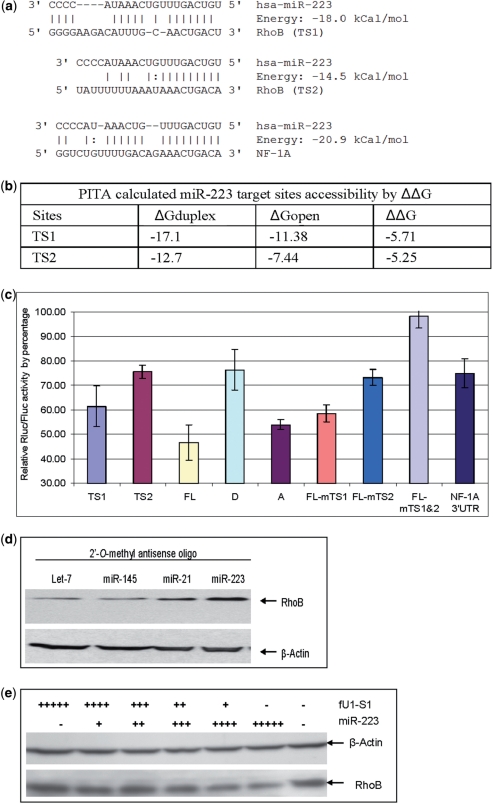
Figure 2.
RhoB is a miR-223 target and the two target sites contribute differentially to the total repression of RhoB translation. (a) miRnada predicted MiR-223 target sites in the 3′UTR of RhoB and NF-1A. (b) PITA calculated miR-223 target accessibility as measured by ΔΔG. TS1 has a slightly better accessibility than TS2. (c) Reporter assay of miR-223 inhibition of reporters carrying sequence fragments with miR-223 MREs. Rluc was fused with different fragments of the RhoB or NF-1A 3′UTR. The Y-axis represents relative expression of Rluc to Fluc when co-transfected with fU1-miR-223 and normalized to co-transfection with fU1-miR. Lane 1: MiR-223 target site one (TS1) short target sequence only; Lane 2: MiR-223 target site two (TS2) short target sequence only; Lane 3: FL RhoB 3′UTR (FL); Lane 4: First half of RhoB 3′UTR (D with TS1 only); Lane 5: Second half of RhoB 3′UTR (A) with TS2 only; Lane 6: FL with mutated TS1; Lane 7: FL with mutated TS2; Lane 8: FL with both TS1 and TS2 sites mutated; Lane 9: NF-1A 3′UTR. Each bar represents the average of at least three independent transfections with duplicate determinations for each construct. Error bars represent the standard deviation (SD). (d) Western blot result of RhoB expression in CEM cells when miR-223 function was blocked. CEM cells were transfected with a 2′-O-methyl anti-let-7 (lane 1), anti-miR-145 (lane 2), anti-miR-21 (lane 3), and anti-miR-223 (lane 4) oligo. Total cell extract were prepared 48 hours post transfection. The data show both anti-miR-223 and anti-miR-21 resulted in elevated RhoB protein levels while neither anti-let-7 nor anti-miR-145 antagomirs affected the RhoB protein level. Both miR-223 and miR-21 were predicted to target RhoB by miRanda and TargetScan (Figure 1b, miR-223 TS2 is located near miR-21 TS). (e) Western blot result of RhoB expression in Hela cells in the presence of miR-223. Hela cells were transfected with the miR-223 expression cassette. A U1 promoter-driven shRNA S1 (targeting the HIV Tat/Rev exon) was used as control. ‘−’ represents the absence of miR-223/S1 in the transfection. ‘+’ represents the present of miR-223/S1. The number of ‘+’s represents the amount of miR-223/S1 in the transfection. Total cell extracts were prepared 48 hours post transfection.
RhoB is a bona fide miR-223 target
The RhoB full-length (FL) 3′UTR was cloned into psiCheck2.2. The reporter with a FL RhoB 3′UTR (Figure 1b) along with the miR-223 artificial expression vector, fU1-miR-223, were co-transfected into HEK293 cells (miR-223 expression is not detectable in HEK293 by northern blotting, data not shown). A 60% repression of the RhoB 3′UTR reporter was observed (Figure 2c, lane ‘FL’). Under the same experimental conditions, the reporter with the FL 3′UTR of NF-1A, a previously demonstrated miR-223 target (39), was repressed only 30% (Figure 2c, lane ‘NF-1A 3′UTR’). Mutated seed sequences in both target sites in the RhoB 3′UTR abolished the repression (Figure 2c, lane ‘FL-mTS1&TS2’). These results demonstrate that miR-223 mediated miRISC interacts with the RhoB 3′UTR and effectively represses translation of the RhoB 3′UTR associated transcripts. To further validate RhoB as a bona fide miR-223 target, loss and gain of miR-223 function experiments were performed. For the loss of function experiment, a 2′-O-methyl-anti-miR-223 antagomir was electroporated into CEM cells, which express relatively high levels of miR-223 and low levels of RhoB (Data not presented); Western blotting showed that endogenous RhoB expression was increased when miR-223 was inhibited (Figure 2d). In the alternative approach of gain of function, ectopic expression of miR-223 was accomplished by transfecting fU1-miR-223 into Hela cells which express very low levels of endogenous miR-223 and high levels of RhoB protein (miR-223 expression in Hela cells is not detectable by northern blot analyses). The ectopic expression resulted in a dosage dependent reduction of endogenous RhoB protein in HeLa cells (Figure 2e). These data provide further validation that RhoB is a miR-223 target.
The two target sites in RhoB 3′UTR do not contribute equally to the repression and are influenced by the sequence context beyond the local target sequence
MiRanda predicted two miR-223 target sites in the RhoB 3′UTR (38). TargetScan offers the same prediction and lists site one (TS1) as non-conserved site and site two (TS2) as a conserved site (40) (Figures 1 and 2a). Double mutations in the two target sites showed that the repression from miR-223 was virtually eliminated (Figure 2c, lane ‘mTS1&TS2’). One interesting observation is that the individually mutated TS2 site resulted in a greater loss of repression than did the mutated TS1 site (Figure 2c, lane ‘FL-mTS1’ and ‘FL-mTS2’), although TS1 was predicated to have potentially more bases pairing with miR-223 and a calculated higher binding energy (Figure 2a). Conversely, when only the predicted short target sequences were cloned into psiCheck2.2 (TS1 versus TS2), miR-223 repressed the reporter with the TS1 site more effectively than the reporter with the TS2 site (Figure 2c, lane ‘TS1’ versus lane ‘TS2’). These data show that the two target sites, although both effective, function at different strengths in the RhoB 3′UTR. Thus, sequences outside of the target sites probably influence the effectiveness of miRNA translational repression.
Since accessibility has been reported to affect the efficacy of miRNAs, the accessibilities of both sites were analyzed by Sfold (29) and PITA (28). From these analyses, TS1 had a slightly better accessibility than TS2 (PITA-calculated accessibilities measured by ΔΔG are shown in Figure 2b). Nevertheless, the narrow differences in accessibility cannot explain the large differences in the target repression. This conclusion is supported by data from flanking sequences (about 17 bases from both sides of TS1 or both sides of TS2) swapping experiments. First, TS1 or TS2 was cloned with the respective flanking sequences to make TS1-L and TS2-L reporters. Alternatively, TS1 was juxtaposed with TS2’s flanking sequence to make TS1-L212 reporter, and TS2 was juxtaposed with TS1’s flanking sequence to make TS2-L121 reporter (Figure 3a). Reporter assays revealed that TS1-L212 expression was inhibited more than TS1-L reporter expression (about 28% versus 18% reduction in reporter activity), and that TS2-L121 expression was inhibited less than TS2-L expression (about 12% reduction versus 22% reduction) (Figure 3b). None of the above constructs resulted in the 55% or 45 % Rluc reduction obtained with reporters harboring the FL RhoB 3′UTR or the 3′ half of the RhoB 3′UTR with the TS2 (Figure 2c, lane ‘FL’ and, ‘A’). Thus, the nearby flanking sequences of these two sites are only accountable for some of the differences in sensitivity to miR-223.
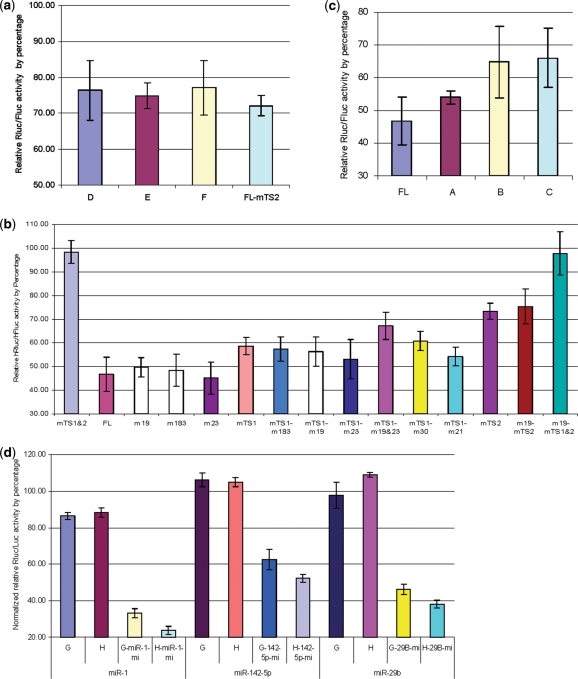
Figure 3.
Flanking sequence and 3′UTR context effects on miR-223 mediated inhibition. (a) Sequences used for reporter assay of flanking sequence effects. TS1 and TS2 represent miR-223 TS1 (in blue) and TS2 (in green) sequences cloned in psiCheck2.2, respectively. TS1-L and TS2-L represent miR-223 TS1 and TS2 with flanking sequences cloned inpsiCheck2.2, respectively. TS1-L212 represents the miR-223 TS1 sequence cloned in psiCheck2.2 with the TS2 flanking sequences (left in pink and right in cyan). TS2-L121 represents the miR-223 TS2 sequence cloned inpsiCheck2.2 with the TS1 flanking sequences (left in gray and right in red). (b) Reporter assay data showed the target and flanking sequences were not the major factors influencing the effectiveness of TS2. TS2 was actually worse than TS1 when used as short target sequences. Each bar represents the average of at least three independent transfections with duplicate determinations for each construct. Error bars represent the SD. (c) Reporter assays of 3′UTR sequence contexts outside of the miR-223 MREs and flanking sequences affecting miR-223 repression. The data show MREs fused with fragment H are more effective than fragment G. miR-223 target sequences in RhoB (TS1 and TS2) and NF-1A (NF1A) were cloned downstream of RhoB 3′UTR fragments G and H (Figure 1b). The data also show that target sites located downstream of fragment H were more effective. Each bar represents the average of at least three independent transfections with duplicate determinations for each construct. Error bars represent the SD.
We therefore examined sequences beyond the local region and observed that TS1 was located in the middle of the 3′UTR while TS2 was located towards the 3′-end of the 3′UTR. So the differences in susceptibility to RNAi of these two sites could be due to the short distance between TS2 and the AAUAAA (HEX), making it closer to the poly A tail. The distance between the target site and HEX has been suggested as being an important factor that can influence miRNA target effectiveness (21,41). The following experiments proved that this not to be the case for the miR223 sites in RhoB. To avoid a length bias, two pieces of the RhoB 3′UTR of almost the same length were separately cloned into the psiCheck2.2 vector. Fragment D was cloned as the first half of the 3′UTR that contains TS1 to make a reporter with TS1 close to the HEX, and fragment A was cloned as the second half of the 3′UTR that contains TS2 (Figure 1b). Reporter assays in HEK-293 cells showed that the reporter with fragment A was repressed more effectively by miR-223 (close to the levels observed with the FL RhoB 3′UTR) than the reporter carrying the fragment D (Figure 2c, lane A versus D).
Based on these data, we concluded that sequences beyond the immediate flanking region of the target site influence the effectiveness of miRNA function. We hypothesized that the sequence between TS1 and TS2 was responsible for differences in response to miR-223 interaction. To test this, the sequence upstream of TS1 and the sequence between TS1 and TS2 were separately cloned to make fragments G and H, respectively (Figure 1b). Target sequences of TS1 and TS2 were cloned downstream of fragments G and H to make G-TS1, G-TS2 and H-TS1, H-TS2. We also cloned the miR-223 target site in the NF-1A 3′UTR downstream of G and H to make G-NF1A and H-NF1A, respectively. Reporter assays showed that when any of the target sites was located downstream of fragment G, the repression was reduced from ∼60 to 40% in comparison to when the target sites were localized downstream of fragment H (Figure 3c). These data demonstrated that the sequence of fragment H enhances the repression and implies that the sequence in fragment H may increase the effectiveness of miR-223 mediated repression of RhoB expression through TS2. The mutation analyses of TS2 (Figure 2c, lane FL-mTS1) also implies that the sequence context does not affect the repression efficiency of sites which are located upstream, as there was little effect on TS1 function. To further investigate this, the fragments of E and F that covered TS1 and its downstream sequence in fragment H were cloned into psiCheck2.2. The reporter assays showed that the sequence in the H fragment did not affect the target sites located before it, demonstrating a polarity of the enhancing effect (Figures 1b and 4a).
Figure 4.
Reporter assays for sequence context features that affect miRNA function. (a) Reporter assays show polarity for the sequence context effects. Specific sequences affect downstream MRES, not upstream MREs. Compare the repression difference between TS1 with the upstream sequence (fragment D) and TS1 with the downstream sequence (fragments E and F). mTS2 represents the repression of target exptession from TS1 in reporters carrying the FL with a mutated TS2 site. Each bar represents the average of at least three independent transfections with duplicate determinations for each construct. Error bars represent the SD. (b) Reporter assays to study the effects of mutations in other conserved miRNA target sites in the RhoB 3′UTR. The influence of mutant miR-19, 183 or 23 targets is marginal when compared with the wild-type FL RhoB 3′UTR. The same conclusion can be drawn when comparing all mutants (m19, m183, m23, m30, m21, m19 and m23) in FL with mutated TS1. Each bar represents the average of at least three independent transfections with duplicate determinations for each construct. Error bars represent the SD. (c) Reporter assays determining miRNA target enhancer sequences. The data show that serial deletions in the RhoB 3′UTR reduce the inhibition of target expression as the UTR is progressively shortened. The FL UTR is required for efficient repression by miRISC. Each bar represents the average of at least three independent transfections with duplicate determinations for each construct. Error bars represent the SD. (d) Reporter assays for determining the effects of RhoB fragments on the function of non-miRNA 223targets. The data show that the H fragment has a similar effect on miR-1, miR-142-5p and miR-29b target sequences. Therefore, H’s effect is independent of the miRNA target sequence. Each bar represents the average of at least three independent transfections with duplicate determinations for each construct. Error bars represent the SD.
TargetScan predicted several conserved miRNA target sites located in fragment H and near TS2 (40) (Figure S1). To rule out the possibility of a synergistic, coordinate effect from other miRNAs, mutations were made in the conserved miRNA-binding sites (Figure S4). Reporter assays showed that mutated miR-19a/b, miR-23a/b, miR-183, miR-30 and miR-21-binding sites had some affects on the strong repression mediated by TS2, but none of them affected inhibition by greater than 10%. We thus conclude that other miRNA interactions are not a major factor influencing the potency of TS2 (Figure 4b).
In order to locate the specific sequence or region that may contribute to the potency, progressive deletions were performed on fragment H. The data from these experiments revealed that the longer the sequence before TS2, the better the TS2 function (Figure 4c). Since the specific sequence could be a non-canonical miR-223 target sequence without a perfect seed sequence match, we tested fragment H’s effect on other miRNAs to rule out that this effect is miR-223 specific. Artificially designed miRNA response elements (MRE) for miR-1, miR-142-5p and miR-29b were cloned downstream of fragments H and G. The MREs were designed so as not to base-pair at positions 11, 12, 13, and the last two nucleotides of the miRNA. The results obtained with these analyses show that fragment H has a similar effect on these three miRNAs (Figure 4d). We conclude that the repression effect of the sequence context is not specific to miR-223.
The TTP-binding motif (ARE) and the CPEB-binding motif (CPE) are enhancer sequences for miRNA mediated target knockdown
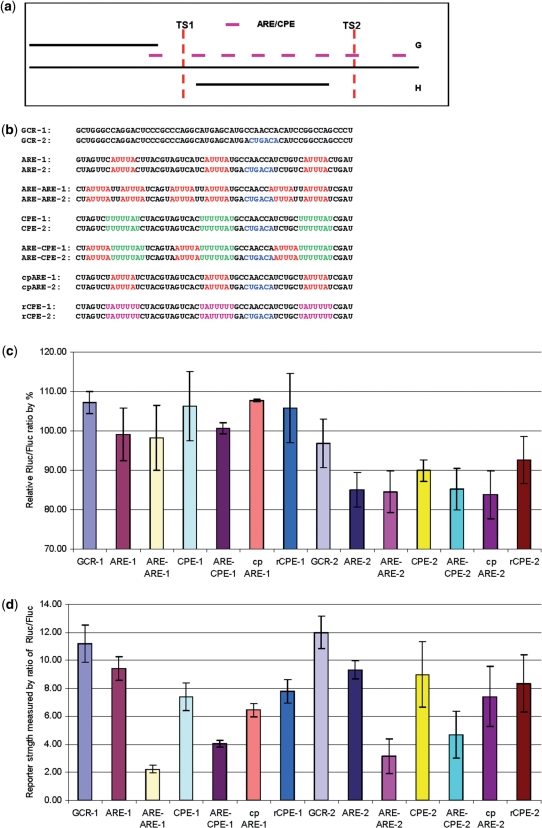
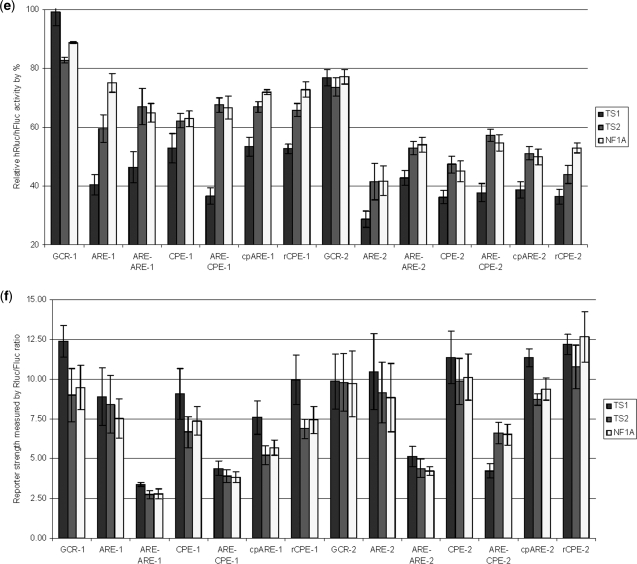
Translational repression is a common mechanism to regulate protein expression. We hypothesized that translational repressors can function as enhancers for miRNA-mediated repression. Several translational repressors are listed in a recent review (42). Among those, CPEB (CPE-binding protein), TTP (Tristetraprolin) and eIF4E are also found in humans. Sequence comparisons of fragments H and G show that H is more AU-rich than G (Figure S3), with 63% AU in H versus 38% in G (Figure S2). Most of the TTP and CPEB-binding motifs (43,44) are located in H (Figure 5a). We next investigated whether the AU-rich sequence itself can enhance miRNA function. To investigate this question, we designed a variety of AU-rich sequences with different potential TTP and CPEB-binding motifs to test their effects on miRNA-mediated translational repression (Figure 5b). We first designed two GCR (GC-rich) sequences GCR-1 and GCR-2. To test the effects of multiple MREs’, we embedded a miR-223 seed sequence ‘ACUGAC’ in GCR-2. Next, we modified sequences in GCR-1 and GCR-2 to make them AU-rich and to contain three identical copies of the following motifs: ARE (AUUUA) for ARE-1 and ARE-2, ARE-ARE (AUUUAUUAUUUA) for ARE-ARE-1 and ARE-ARE-2, CPE (UUUUUAU) for CPE-1 and CPE-2, ARE-CPE (AUUUAUUUUUAU) for ARE-CPE-1 and ARE-CPE-2, cpARE (UAUUUAU) for cpARE-1 and cpARE-2, rCPE (UAUUUUU) for rCPE-1 and rCPE-2 (Figure 5b). The second and third motifs were separated by the miR-223 seed sequence in all the ‘-2’ reporters. Reporter assays were performed to monitor the repression mediated by miR-223 on these constructs. The results showed that negligible reporter knockdown from reporters without the miR-223 seed and about 10–20% repression of reporters with the embedded seed. Hence, we conclude that one seed sequence can lead to about 10–20% repression and ARE/CPE-containing sequences result in stronger repression (Figure 5c). Since the inserted ARE/CPE motif could affect the stability of the reporters, the Rluc mRNA stability was calculated as the ratio of Rluc/Fluc in the control transfection with fU1-miR. Indeed, the ratio decreased by 70–80% from ARE to ARE-ARE (Figure 5d). Interestingly, the repression in ARE-ARE reporters was similar to the ARE reporters, implying it is the motif, not the content of A/U, which contributes to the effectiveness of target sites harboring this motif. Similar results were obtained for both sets of constructs with 3′ fusions of one of the TS1, TS2 or NF1A sites (Figure 5e and f). These results also show that efficient repression is ARE motif specific, since both sets of ARE reporters performed better than cpARE, which has one ‘U’ added to both sides of an ARE. We therefore conclude that miRNAs more efficiently target 3′UTRs bearing ARE and CPE motifs. When we compared the three target sites, TS1 always performed better in ARE/CPE constructs than did TS2. This verified that the target sequence does matter and it also implies that the location of target sites within the 3′UTR is important for function. It can also be implied from these data that the first position pairing of the miRNA/target sequence A:U match (‘U’ is the first base at the 5′-end of miRNA and ‘A’ is its paired base in the target mRNA) in miRNAs/mRNA may not be an important factor in the miR-223 targets, since the first position for TS1 is U:U, but both TS2 and NF1A are A:U (Figure 2a). Among the three target sites, NF1A has the highest binding energy in the miRNA/mRNA duplex, but NF1A performed poorly when compared with TS1 and barely matched TS2, which has the lowest binding energy. These results suggest that the binding energy of miRNA/mRNA duplexes is not always a reliable factor in target identification.
Figure 5.
AU-rich motif affects on miRNA targeting. (a) The distribution of AREs and CPEs in the RhoB 3′UTR. More AREs (AUUUA) or CPEs (UUUUUAU) are located in RhoB2 (Figure 1a). There are six AREs/CPEs in fragment H while there is only one in fragment G. (b) GC-rich or AU-rich sequences designed for reporter assay. The ‘−2’ reporters have one embedded miR-223 seed ‘ACUGAC’. GCR-1 and GCR-2 are two GC rich sequences; ARE-1 and ARE-2 are AU-rich sequences with three identical ‘AUUUA’ motifs; ARE-ARE-1 and ARE-ARE-2 are AU-rich sequences with three identical ‘AUUUAUUAUUUA’ motifs; CPE-1 and CPE-2 are AU-rich sequences with three identical ‘UUUUUAU’ motifs; ARE-CPE-1 and ARE-CPE-2 are AU-rich sequences with three identical ‘AUUUAUUUUUAU’ motifs; cpARE-1 and cpARE-2 are AU-rich sequences with three identical ‘UAUUUAU’ motifs; rCPE-1 and rCPE-2 are AU-rich sequences with three identical ‘UAUUUUU’ motifs. (c) Target knockdown by miR-223 in GCR and AU-rich reporters without TS1, TS2 or NF1A. Each reporter was transfected with fU1-miR-223, the same reporter that was transfected with fU1-miR was used as control. There is little knockdown from miR-223 for all the GCR-1-derived reporters. There is ∼10% knockdown bymiR-223 for most of the GCR-2 derived reporters that carry ARE/CPE elements. Each bar represents the average of at least three independent transfections with duplicate determinations for each construct. Error bars represent the SD. (d) Rluc mRNA stability in GCR and AU-rich reporters without TS1, TS2 or NF1A. The stability was calculated as the ratio of Rluc/Fluc in reporters that was transfected with fU1-miR. Each bar represents the average of at least three independent transfections with duplicate determinations for each construct. Error bars represent the SD. (e) Target knockdown by miR-223 in GCR and AU-rich reporters with TS1, TS2 or NF1A. The GCR and AU-rich reporters were further fused with one of the miR-223 target sequence TS1, TS2 or NF1A at the 3′-end. Each reporter was transfected with fU1-miR-223, the same reporter that was transfected with fU1-miR was used as control. Each bar represents the average of at least three independent transfections with duplicate determinations for each construct. Error bars represent the SD. (f) Rluc mRNA stability in GCR1, GCR2 and AU-rich constructs with TS1, TS2 or NF1A. The stability was calculated as the ratio of Rluc/Fluc in reporters that were transfected with fU1-miR. Each bar represents the average of at least three independent transfections with duplicate determinations for each construct. Error bars represent the SD.
DISCUSSION
Ever since the first miRNA was discovered in C. elegan and was found to play an essential role in the timing of worm development (45,46), it is now a widely accepted concept that miRNAs are important regulatory elements in development, apoptosis, and disease generation and progression. MicroRNAs, although harboring limited base pairing with their targets, can perform important functions equivalent to those of transcription factors. In general, miRNAs silence transcripts by guiding miRISC binding to the 3′UTR of a target gene, thus disrupting translation initiation and/or progression (47–49). MicroRNA associated targets can also be reversibly sequestered in P-bodies (25). Alternatively, miRNAs can promote sequestering of transcripts to P-bodies for decapping, deadenylation, and degradation (15,50,51). MicroRNAs can also fine tune gene expression that is regulated by transcription factors. Transcription factors themselves are usually major targets of miRNAs. By targeting transcription factors, miRNAs could form gene expression regulatory circuits with transcription factors to regulate target gene expression. Therefore, the key in the study of miRNA function is to identify true targets, a task that has not been easy.
Theoretically there are hundreds of potential targets for any miRNA but only a small number of targets for all the miRNAs have thus far been identified and validated (validated targets are collected in TarBase) (52). Additional evidence supports the model that the miRNA/mRNA interaction is far more complicated than just base pairing. The local structure as well as the entire 3′UTR sequence may contribute to miRNA function (28,29). RBP-binding motifs in the target transcript 3′UTR may influence miRISC function (24,25,27). As an example from the GCR sequence tested in this study, one seed sequence interaction site only leads to about 10% repression of the expression from the target transcript. Therefore, to achieve 50% or more repression of non-AU-rich 3′UTRs, it may require several target sites to achieve such a level of repression. In contrast, only one or two target sites in an AU-rich 3′UTR may be enough to allow significant miRNA mediated inhibition of translation.
It is now a standard practice in miRNA functional studies to use reporters with the target 3′UTR and reporters with mutated seed sequences to validate the direct interaction between miRNAs and target mRNAs. Our results imply that it is important to use the FL 3′UTR instead of a short MRE to perform these tests (36). As we have demonstrated, isolated, short MREs may give misleading results. A recent paper by Lal et al. reported miR-24 targeting of E2F2, MYC, and other cell-cycle genes via binding to ‘seedless’ 3′UTR MREs, further supporting the importance of using FL 3′UTRs for reporter assays (53). In our opinion, the number of true or functionally effective direct targets for a single miRNA maybe much lower, perhaps a small fraction of the estimated 200 average targets for any miRNA (16). A combination of five to six miRNAs targeting the same UTR may be required for optimal miRNA function.
In our tests, the AU-rich sequence is not limited to the TTP-binding motif. Therefore, we do not believe that miR-16 has the same role as reported before: TTP interacts with Ago/eiF2C family members to complex with miR16, and assists in the targeting of ARE-containing RNAs (54). In addition to the AU-rich sequence making the tertiary structure more accessible for miRISC, ARE-binding proteins (ARE-BP) may bind to AU-rich regions and affect the repression efficiency of downstream MREs. One possible explanation is that ARE-BPs bind to AREs [classification of AREs is reviewed in ref. (43)] and activate miRISC targeted mRNAs for deadenylation, decapping or 3′- to 5′-exonucleolytic decay (55,56). A possible explanation for why the AU-rich sequence must be placed before the MRE is that the uridines may play a role in the miRNA-mediated repression. It has been shown that miRNA-directed cleavage products have short uridine tails, which are added downstream of the cleavage sites, and the 3′ uridine addition is correlated with decapping and 5′ shortening of the cleaved products (57). It has also been reported in a cell-free system that a poly(U) tail enhances ‘decapping’ by binding to the LSM protein complex, which associates with decapping factors (58,59). The recently discovered poly(U) polymerases (PUPs) make the addition of poly(U) an interesting possible regulator of mRNA stability (60–62). The CPE sequence’s role in our test is not very clear since the CPE and rCPE performed approximately the same. Recently, a set of rules that can be used to predict the translational behavior of the CPE/CPEB was published. The number and relative position of CPEs and Pumilio-binding elements (PBE), with respect to the AAUAAA, defined a combined code for whether CPEB will repress or activate translation (63). It has been proposed that CPEB could play a role in miRNA-mediated repression (44). MicroRNAs can also upregulate translation when AGO2 and FXR1 form an activation complex at AREs in quiescent cells (27). CPEB may be a perfect candidate to form a complex with AGOs that can switch repression to activation, or vice versa. The number and relative positions of MRE, CPE, PBE, ARE, HEX, and their roles in translation need to be addressed in future experiments. Class III AREs have also been reported to up-regulate translation in reporters fused with the c-jun 3′UTR in cycling cells (64). It is not yet known whether this is miRNA-mediated.
Our results could possibly provide some insights as to why miRNAs primarily target the 3′UTR or why the 3′UTR performs the regulatory role. It has been reported by Cora et al. that ‘A’s and ‘U’s are more enriched in human and mouse 3′UTRs than in 5′UTRs or coding sequences, with 27% ‘A’, 22% ‘G’ or ‘C’, and 29% ‘U’ in Human, and 26.4% ‘A’, 22.4% ‘C’, 22.5% ‘G’, and 28.7% ‘U’ in mouse (65). In the same report, the authors also identified 45 regulatory elements. Among them, 13 were poly(A) signals, 3 were AREs and 6 were CPEs. So, besides poly(A) signals, 25% of the remaining regulatory elements are AREs or CPEs. A separate in silico study by Yoon et al. showed a large portion of over-represented sequences located in 3′UTRs contain the sequence motifs AUUUA and UAUUUAU, the two basic core sequences of AREs (66). A separate study conducted by Robins et al. showed ∼75% of miRNA targets are in the AU-rich 3′UTR population and genes with AU-rich 3′UTRs are preferentially associated with transcription and translation events (34).
In conclusion, our results demonstrate that RhoB is a bona fide target for miR223. In addition the data presented support a general mechanism in which miRNAs may selectively target distinct populations of transcripts harboring AU rich elements in the appropriate polarity to the miRNA-binding site, perhaps in concert with proteins which bind to AU–rich sequences in the 3′UTR.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [AI29329 to J.R., HL07470 to J.R.]. Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors wish to thank Kaming Pang and Brain Luk for critical reading of this manuscript.
REFERENCES
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Jackson RJ, Standart N. How do microRNAs regulate gene expression? Sci. STKE. 2007;2007:re1. doi: 10.1126/stke.3672007re1. [DOI] [PubMed] [Google Scholar]
- 3.Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 2002;21:4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 5.Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10:1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell. 2007;130:89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC. Mammalian mirtron genes. Mol. Cell. 2007;28:328–336. doi: 10.1016/j.molcel.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haase AD, Jaskiewicz L, Zhang H, Laine S, Sack R, Gatignol A, Filipowicz W. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep. 2005;6:961–967. doi: 10.1038/sj.embor.7400509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 14.Liu J, Valencia-Sanchez MA, Hannon GJ, Parker R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat. Cell Biol. 2005;7:719–723. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rehwinkel J, Behm-Ansmant I, Gatfield D, Izaurralde E. A crucial role for GW182 and the DCP1:DCP2 decapping complex in miRNA-mediated gene silencing. RNA. 2005;11:1640–1647. doi: 10.1261/rna.2191905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 17.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, Lander ES, Kellis M. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 23.Xiao C, Calado DP, Galler G, Thai TH, Patterson HC, Wang J, Rajewsky N, Bender TP, Rajewsky K. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell. 2007;131:146–159. doi: 10.1016/j.cell.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 24.Kedde M, Strasser MJ, Boldajipour B, Vrielink JA, Slanchev K, le Sage C, Nagel R, Voorhoeve PM, van Duijse J, Orom UA, et al. RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell. 2007;131:1273–1286. doi: 10.1016/j.cell.2007.11.034. [DOI] [PubMed] [Google Scholar]
- 25.Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 26.Fabian MR, Mathonnet G, Sundermeier T, Mathys H, Zipprich JT, Svitkin YV, Rivas F, Jinek M, Wohlschlegel J, Doudna JA, et al. Mammalian miRNA RISC Recruits CAF1 and PABP to Affect PABP-Dependent Deadenylation. Mol. Cell. 2009;35:868–880. doi: 10.1016/j.molcel.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 28.Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat. Genet. 2007;39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- 29.Long D, Lee R, Williams P, Chan CY, Ambros V, Ding Y. Potent effect of target structure on microRNA function. Nat. Struct. Mol. Biol. 2007;14:287–294. doi: 10.1038/nsmb1226. [DOI] [PubMed] [Google Scholar]
- 30.Didiano D, Hobert O. Molecular architecture of a miRNA-regulated 3′ UTR. RNA. 2008;14:1297–1317. doi: 10.1261/rna.1082708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Didiano D, Hobert O. Perfect seed pairing is not a generally reliable predictor for miRNA-target interactions. Nat. Struct. Mol. Biol. 2006;13:849–851. doi: 10.1038/nsmb1138. [DOI] [PubMed] [Google Scholar]
- 32.Turi A, Loglisci C, Salvemini E, Grillo G, Malerba D, D'E;lia D. Computational annotation of UTR cis-regulatory modules through Frequent Pattern Mining. BMC Bioinformatics. 2009;10(Suppl. 6):S25. doi: 10.1186/1471-2105-10-S6-S25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoon K, Ko D, Doderer M, Livi CB, Penalva LO. Over-represented sequences located on 3′ UTRs are potentially involved in regulatory functions. RNA Biol. 2008;5:255–262. doi: 10.4161/rna.7116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robins H, Press WH. Human microRNAs target a functionally distinct population of genes with AT-rich 3′ UTRs. Proc. Natl Acad. Sci. USA. 2005;102:15557–15562. doi: 10.1073/pnas.0507443102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun G, Yan J, Noltner K, Feng J, Li H, Sarkis DA, Sommer SS, Rossi JJ. SNPs in human miRNA genes affect biogenesis and function. RNA. 2009;15:1640–1651. doi: 10.1261/rna.1560209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun G, Rossi JJ. Problems associated with reporter assays in RNAi studies. RNA Biol. 2009;6:1–6. doi: 10.4161/rna.6.4.9218. [DOI] [PubMed] [Google Scholar]
- 37.Sun G, Li H, Rossi JJ. Cloning and detecting signature microRNAs from mammalian cells. Methods Enzymol. 2007;427:123–138. doi: 10.1016/S0076-6879(07)27007-7. [DOI] [PubMed] [Google Scholar]
- 38.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fazi F, Rosa A, Fatica A, Gelmetti V, De Marchis ML, Nervi C, Bozzoni I. A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPalpha regulates human granulopoiesis. Cell. 2005;123:819–831. doi: 10.1016/j.cell.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 40.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 41.Hon LS, Zhang Z. The roles of binding site arrangement and combinatorial targeting in microRNA repression of gene expression. Genome Biol. 2007;8:R166. doi: 10.1186/gb-2007-8-8-r166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Mol. Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 43.Barreau C, Paillard L, Osborne HB. AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res. 2005;33:7138–7150. doi: 10.1093/nar/gki1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Richter JD. CPEB: a life in translation. Trends Biochem. Sci. 2007;32:279–285. doi: 10.1016/j.tibs.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 45.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 46.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 47.Doi N, Zenno S, Ueda R, Ohki-Hamazaki H, Ui-Tei K, Saigo K. Short-interfering-RNA-mediated gene silencing in mammalian cells requires Dicer and eIF2C translation initiation factors. Curr. Biol. 2003;13:41–46. doi: 10.1016/s0960-9822(02)01394-5. [DOI] [PubMed] [Google Scholar]
- 48.Humphreys DT, Westman BJ, Martin DI, Preiss T. MicroRNAs control translation initiation by inhibiting eukaryotic initiation factor 4E/cap and poly(A) tail function. Proc. Natl Acad. Sci. USA. 2005;102:16961–16966. doi: 10.1073/pnas.0506482102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pillai RS, Bhattacharyya SN, Artus CG, Zoller T, Cougot N, Basyuk E, Bertrand E, Filipowicz W. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- 50.Chu CY, Rana TM. Translation Repression in Human Cells by MicroRNA-Induced Gene Silencing Requires RCK/p54. PLoS Biol. 2006;4:e210. doi: 10.1371/journal.pbio.0040210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu J, Rivas FV, Wohlschlegel J, Yates JR, 3rd, Parker R, Hannon GJ. A role for the P-body component GW182 in microRNA function. Nat. Cell Biol. 2005;7:1261–1266. doi: 10.1038/ncb1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sethupathy P, Corda B, Hatzigeorgiou AG. TarBase: A comprehensive database of experimentally supported animal microRNA targets. RNA. 2006;12:192–197. doi: 10.1261/rna.2239606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lal A, Navarro F, Maher CA, Maliszewski LE, Yan N, O'D;ay E, Chowdhury D, Dykxhoorn DM, Tsai P, Hofmann O, et al. miR-24 Inhibits cell proliferation by targeting E2F2, MYC, and other cell-cycle genes via binding to “seedless” 3′UTR microRNA recognition elements. Mol. Cell. 2009;35:610–625. doi: 10.1016/j.molcel.2009.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jing Q, Huang S, Guth S, Zarubin T, Motoyama A, Chen J, Di Padova F, Lin SC, Gram H, Han J. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell. 2005;120:623–634. doi: 10.1016/j.cell.2004.12.038. [DOI] [PubMed] [Google Scholar]
- 55.Fenger-Gron M, Fillman C, Norrild B, Lykke-Andersen J. Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Mol. Cell. 2005;20:905–915. doi: 10.1016/j.molcel.2005.10.031. [DOI] [PubMed] [Google Scholar]
- 56.Lykke-Andersen J, Wagner E. Recruitment and activation of mRNA decay enzymes by two ARE-mediated decay activation domains in the proteins TTP and BRF-1. Genes Dev. 2005;19:351–361. doi: 10.1101/gad.1282305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shen B, Goodman HM. Uridine addition after microRNA-directed cleavage. Science. 2004;306:997. doi: 10.1126/science.1103521. [DOI] [PubMed] [Google Scholar]
- 58.Song MG, Kiledjian M. 3′ Terminal oligo U-tract-mediated stimulation of decapping. Rna. 2007;13:2356–2365. doi: 10.1261/rna.765807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tharun S, He W, Mayes AE, Lennertz P, Beggs JD, Parker R. Yeast Sm-like proteins function in mRNA decapping and decay. Nature. 2000;404:515–518. doi: 10.1038/35006676. [DOI] [PubMed] [Google Scholar]
- 60.Mellman DL, Gonzales ML, Song C, Barlow CA, Wang P, Kendziorski C, Anderson RA. A PtdIns4,5P2-regulated nuclear poly(A) polymerase controls expression of select mRNAs. Nature. 2008;451:1013–1017. doi: 10.1038/nature06666. [DOI] [PubMed] [Google Scholar]
- 61.Kwak JE, Wickens M. A family of poly(U) polymerases. RNA. 2007;13:860–867. doi: 10.1261/rna.514007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rissland OS, Mikulasova A, Norbury CJ. Efficient RNA polyuridylation by noncanonical poly(A) polymerases. Mol. Cell Biol. 2007;27:3612–3624. doi: 10.1128/MCB.02209-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pique M, Lopez JM, Foissac S, Guigo R, Mendez R. A combinatorial code for CPE-mediated translational control. Cell. 2008;132:434–448. doi: 10.1016/j.cell.2007.12.038. [DOI] [PubMed] [Google Scholar]
- 64.Barreau C, Watrin T, Beverley Osborne H, Paillard L. Protein expression is increased by a class III AU-rich element and tethered CUG-BP1. Biochem. Biophys. Res. Commun. 2006;347:723–730. doi: 10.1016/j.bbrc.2006.06.177. [DOI] [PubMed] [Google Scholar]
- 65.Cora D, Di Cunto F, Caselle M, Provero P. Identification of candidate regulatory sequences in mammalian 3′ UTRs by statistical analysis of oligonucleotide distributions. BMC Bioinformatics. 2007;8:174. doi: 10.1186/1471-2105-8-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yoon K, Ko D, Doderer M, Livi CB, Penalva LOF. Over-represented sequences located on 3′UTR are potentially involved in regulatory functions. RNA Biol. 2008;5:1–8. doi: 10.4161/rna.7116. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.