Abstract
Life scientists are often interested to compare two gene sets to gain insight into differences between two distinct, but related, phenotypes or conditions. Several tools have been developed for comparing gene sets, most of which find Gene Ontology (GO) terms that are significantly over-represented in one gene set. However, such tools often return GO terms that are too generic or too few to be informative. Here, we present Martini, an easy-to-use tool for comparing gene sets. Martini is based, not on GO, but on keywords extracted from Medline abstracts; Martini also supports a much wider range of species than comparable tools. To evaluate Martini we created a benchmark based on the human cell cycle, and we tested several comparable tools (CoPub, FatiGO, Marmite and ProfCom). Martini had the best benchmark performance, delivering a more detailed and accurate description of function. Martini also gave best or equal performance with three other datasets (related to Arabidopsis, melanoma and ovarian cancer), suggesting that Martini represents an advance in the automated comparison of gene sets. In agreement with previous studies, our results further suggest that literature-derived keywords are a richer source of gene-function information than GO annotations. Martini is freely available at http://martini.embl.de.
INTRODUCTION
High-throughput experiments such as microarrays, mass spectrometry, or automated digital microscopy often produce a single list of genes associated with a specific phenotype or condition, and many computational tools have been developed to help biologists use such a list to gain insight into the underlying biological processes (1). Some of these tools even allow end-users to interactively explore gene sets, and to identify functional sub-clusters (2,3).
While a single gene set is probably the most common outcome of a single experiment, scientists are often interested to compare two sets defined by two distinct, but related, phenotypes or conditions. For example, a scientist may want to compare the set of genes associated with a primary cancer versus those associated with the metastatic form of the same cancer (4). Alternatively, a scientist might want to compare genes associated with a disease to the genes associated with the presence of a drug.
For this article, we briefly reviewed the available tools for analyzing gene lists; we found that most tools allow only a single input gene list, which is usually compared with the background of all remaining genes in the same organism. Only a subset of tools allow the user to explicitly ask the more specific question: ‘how do two gene sets differ?’ Clearly, the ability to answer this question is important and relevant to many life scientists today.
Of the tools that do address this question [e.g. FatiGO (5) and ProfCom (6)], most are based on GO (7), a controlled vocabulary of ∼30 000 terms for describing gene function. GO-based tools find GO terms that are significantly over-represented in one set of genes versus a second reference set. However, dependency on GO gives rise to some limitations (8). For many genes, GO annotations give a very incomplete description of function, e.g. human genes in Entrez (http://tinyurl.com/entrez-gene) have a median of only seven GO terms. As a result, GO-based tools sometimes produce disappointing results, finding only a few, or only very general, GO terms (e.g. see ‘Results’ section).
An alternative approach is to examine the literature cited in each gene entry, and extract keywords that can be used to describe gene function. In most cases, the literature associated with a gene gives a much richer description of function than the currently available GO terms. For example, human genes in Entrez have a median of nine Medline (http://pubmed.org) abstracts; clearly, nine abstracts will contain more information than just seven GO terms, although the exact number of keywords extracted per gene will depend on the size and scope of the keyword dictionary used. Indeed, in at least one previous study it has been reported that literature-based approaches give more sensitive and more specific results than using only GO terms (9).
Several such keyword-based methods have been described in the literature (9–14), however, we could only find two systems that are provided as automated, freely available services, namely CoPub (14) and Marmite (13). CoPub is based on a dictionary of 250 000 keywords, including gene and pathway names, GO terms, diseases, drugs and tissues. CoPub can only analyze a single gene set, and it is further restricted to only human, mouse or rat genes. Marmite is based on three types of keywords, namely diseases, chemicals and ‘word roots’, or generic ‘bio-terms’. Marmite can compare two gene sets, but is restricted to human genes only.
In this work we present Martini, a new, easy-to-use tool that allows end-users to compare two gene sets using a sensitive, keyword-based method. Martini can be used with genes from a large number of species, and it uses a rich keyword dictionary of over 3 million terms, including gene names, drugs, chemicals, diseases, symptoms, organisms and processes/bio-actions.
MATERIALS AND METHODS
Input data and initial processing
Martini requires the end-user to specify two input sets. By default Martini assumes the input sets are lists of Entrez gene IDs (http://tinyurl.com/entrez-gene), in which case Martini retrieves, for each gene, all PubMed IDs (http://pubmed.org) that are referred to in the Entrez entry including the GeneRIFs and interaction records. The mapping from genes to abstracts is retrieved offline using SRS (15) and stored in random access memory (RAM) to enable fast access while processing jobs. Any gene IDs that occur in both input sets are ignored for the purpose of subsequent analysis.
Alternatively, the end-user can specify a PubMed query, in which case Martini queries PubMed via Entrez Programming Utilities (http://tinyurl.com/eutils-help) and retrieves a list of PubMed IDs. As a third alternative, Martini allows the end-user to specify a list of PubMed IDs directly as input. Thus, for each of the three different types of input to Martini, the initial processing results in a list of PubMed IDs.
The next step is to convert each PubMed ID into a list of keywords. For this, we used the AKS2 literature analysis tool (http://tinyurl.com/bioalma-aks2), which is based on a keyword dictionary of over 3 million entries covering the following types: drugs, chemicals, genes, diseases, symptoms, bio-actions and other generic biologically-relevant keywords. In AKS2, this dictionary has been used to pre-tag keywords in Medline abstracts, resulting in an average of 32 keywords per abstract. In Martini, we extract this information into a hash table, linking each Medline abstract to a list of AKS2 keywords. By default, Martini uses all keyword types (genes, drugs, diseases, etc.), however the user can choose to exclude some types via the ‘Advanced’ option.
Martini relies on the literature that is linked to Entrez gene entries. In some cases, the Entrez gene entries have no associated abstracts. In other cases, some Medline entries contain only titles not abstracts. Another potential limitation is that AKS2 indexes only the latest 9 million Medline abstracts ignoring older entries. In addition, to reduce server load and processing time, the total number of entries in each input field of Martini is limited to either 25 000 genes or abstracts—if the user specifies more than this limit, the job will not run, and the user will be asked to reduce the size of the input sets.
Keyword enhancement
After the initial processing described above, Martini analyzes each keyword separately to test for statistically-significant over-representation in one input set compared with the other set, using the two-tailed version of Fisher’s exact test (16). If the user specified either a list of PubMed IDs or a query, Martini counts the numbers of abstracts in which the keyword occurs at least once. Alternatively, if the user specified genes as input, Martini counts the numbers of genes in which the keyword occurs at least once in any of the abstracts associated with each gene.
To account for the total number of keywords tested, we used the false discovery rate method of Benjamini and Hochberg (17) with α, the fraction of false positives considered acceptable, set to 5%. The Fisher p-value for each keyword was then adjusted using:
where n is the total number of entities in a set, and k is the rank of the largest p-value that satisfies the false discovery criteria, calculated separately for keywords associated with each of the two input sets. By default, Martini returns only those keywords for which the adjusted p-value is <5%. However, Martini also provides users access to all keywords found, including those with higher p-value. The Benjamini and Hochberg method assumes that all p-values are mutually independent, which is clearly not true since some words are very likely to appear together. However the method errs on the conservative side, hence we end up rejecting more words than we should. Ideally we would instead use a permutation-based approach, as some authors have in similar cases (18). However, currently this would not be feasible for Martini as it would require a significantly longer response time.
Comparisons with similar tools
We surveyed the literature for methods that can compare two gene lists; some of these methods have not been made available as free tools or services, and others were once available, but are no longer working. Several of the available tools have a rather complex user interface; these tools may have rich functional capabilities, but they do not provide end-users with a simple way to compare two gene sets. We found three tools that we considered to be comparable with Martini, namely FatiGO (5), Marmite (13) and ProfCom (6). For testing these tools we used default parameters, except for FatiGO, where we used all available database sources (GO terms, pathway names, etc.). We also tested CoPub (14), which uses a similarly rich keyword dictionary to Martini, but cannot compare two genes lists: instead CoPub effectively compares one list to the background of all genes from the same organism. However, we included results from CoPub for one dataset (cell cycle, see below) primarily to illustrate the benefit of using two datasets. For CoPub, we again used default parameters and the following search categories: ‘biological processes’, ‘Pathway’, ‘Drug’ and ‘Disease’.
In assessing the output of these tools, we attempted to manually assign each keyword or GO term into one of three categories: ‘true positive’, ‘false positive’ or ‘uninformative’. The criteria we used for these assignments are as follows: True positives were defined as terms that refer to processes or entities that are unambiguously correct, given the biological context of the dataset, determined by a manual literature search. False positives were terms that refer to processes or entities that are unambiguously incorrect, given the biological context. Finally, uninformative terms were simply those that are not clearly right (true positive) and not clearly wrong (false positive).
Datasets
To compare our work with other tools, we used several datasets described in this section—these datasets are also available at http://martini.embl.de.
Arabidopsis
To create a simple test dataset, we used the Arabidopsis Information Resource, TAIR (19), to find 269 Arabidopsis genes that matched the term ‘disease resistance’. We randomly selected a further 514 Arabidopsis genes that did not match this search term.
Cell cycle
This dataset consisted of 600 periodically expressed human genes identified by Jensen et al. (20) from the original dataset of Whitfield et al. (21). Based on when in the cell cycle each gene is most highly expressed, Jensen et al. (20) assigned each gene to a specific ‘peak time’, expressed as a percentage of cell-cycle progress, with 100% (equivalently 0%) corresponding to the moment of cell division. To divide this dataset into two input sets (A and B), we used a window of width 10% and slid this window in steps of 1% around the cell cycle. For example, genes occurring from 1 to 10% of the cycle were assigned to set A, and the remaining genes from 11 to 100% were assigned to set B. Next, genes from 2 to 11% were assigned to set A, and so on. In addition, we partitioned the 600 genes into four subsets corresponding to the classic cell-cycle phases and used these subsets for a four-state comparison.
Melanoma
This dataset consisted of 290 genes highly expressed in metastatic melanoma, and 899 genes highly expressed in primary melanoma based on microarray analysis (4).
Ovarian cancer
This data set consisted of 160 genes associated with clear-cell ovarian cancer, and 105 genes associated with non-clear-cell ovarian cancer, which includes serous and endometrioid ovarian cancers grouped together (22).
Cyclic keyword layout
Keywords and GO terms determined using the cell-cycle dataset were arranged in a circle using a layout algorithm developed for this work, and written in Mathematica (23). The algorithm first places each word along a circular arc that spans the exact region of the cell cycle in which the word is significantly over-represented. Next, the algorithm determines the radius at which to print each word. This is determined primarily based on the character density, i.e. number of characters in each keyword divided by the arc length. Thus, shorter words are placed closer to the center. Finally, the radial position for some words is modified slightly to avoid overlaps with neighboring words.
RESULTS
Arabidopsis dataset
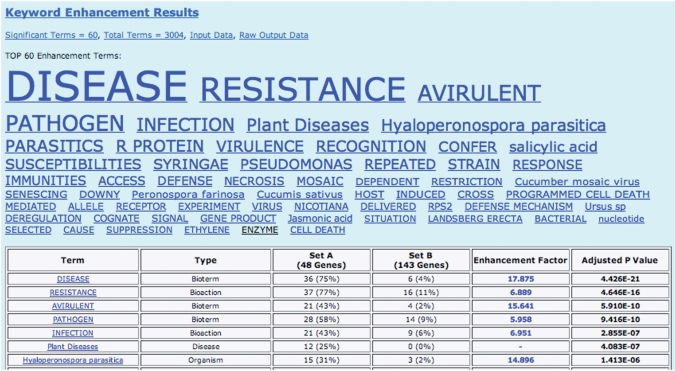
Figure 1 shows the output of a typical keyword analysis with Martini. In this case, Martini was given two input sets of genes—the first set contained 269 Arabidopsis genes known to be associated with disease resistance mechanisms; the second set consisted of 514 genes with no clear link to disease. Martini found 60 keywords that were significantly over-represented in either of the two input sets (Figure 1). Manually checking each keyword, we considered the majority (48 out of 60) to be true positives, i.e. to be clearly related to disease resistance mechanisms in plants. For example, Pseudomonas is a common plant pathogen, and salicylic acid is a phytohormone that is used by plants in triggering the defense-signaling pathway.
Figure 1.
Martini keyword output for the Arabidopsis dataset. All significantly enhanced keywords are shown first as a ‘keyword cloud’, where the size of each keyword is proportional to its statistical significance. The keywords assigned to input sets A or B are colored blue or black, respectively. Below the keyword cloud, the significant keywords are shown again in a table form, including: the number of times each keyword occurs in each set; the enhancement factor (i.e. the ratio of the previous numbers); finally, the table gives an adjusted p-value, which is an estimation of the likelihood that the given level of keyword enhancement occurred by chance. Note that the total number of genes or abstracts shown in this table may be slightly less than the number in the user-defined input. This may happen for two reasons: first, depending on the user’s choice of genes or abstracts as input, Martini will remove common items; secondly, some abstracts may not have been indexed in AKS2, and hence they are not counted.
The 12 keywords that were not true positives were: access, allele, cause, cognate, cross, enzyme, experiment, gene product, nucleotide, selected, situation and ursus sp. We considered that none of these satisfied the criteria for false positives (see ‘Methods’ section), hence we classified them as ‘uninformative’. Most of these 12 are too generic to be properly considered as ‘keywords’, and in future versions of Martini we plan to automatically blacklist such uninformative terms.
For comparison, the Arabidopsis datasets were also analyzed using FatiGO, Marmite and ProfCom, and in each case exactly zero terms were found.
Table 1 shows the time taken for Martini keyword enhancement. Generally, the time taken scales better than linearly with input size, however datasets involving many well-studied genes will be slower than this estimate.
Table 1.
Martini performance
| Total input | Keyword enhancement time |
|---|---|
| 100 abstracts | 3 s |
| 100 genes | 2 min |
This table can be used to estimate the time required for a Martini analysis, assuming linear scaling with total input size. For example, to perform a keyword enhancement using two sets of 500 genes (=total input of 1000 genes) takes ∼20 min, i.e. 10 times longer than for 100 genes. The estimates given here are for genes with nine Medline abstracts (i.e. the median number for human genes). Scaling can be highly non-linear, e.g. including well-studied genes can take much longer. However, in practice the actual time taken is often less than the time estimated from this table.
Cell-cycle dataset
We next tested the keyword enhancement feature of Martini on a set of 600 human cell-cycle-regulated genes (20). The human cell cycle is relatively well-studied and understood, and many of the genes in this data set are well-characterized (98% are linked to Medline abstracts describing their function and 86% have GO annotations levels 3–9 in the GO ontology). Thus, we may expect not only that methods such as Martini should perform well with these data, but also that this set may be a good benchmark, since it should be straightforward to assess the accuracy of the resulting keywords and GO terms.
Each of these 600 genes has been assigned to a specific time point within the cell cycle at which the gene is maximally expressed (20). These time points are given as a percentage of cell-cycle progress rather than hours since the cycle duration varies between growth conditions. To construct pairs of gene sets, we used a sliding window spanning 10% of the cell cycle, and we compared all genes within the window with the remaining cell-cycle genes. Sliding the window in 1% steps, we generated 100 Martini keyword analyses spanning the entire cell cycle.
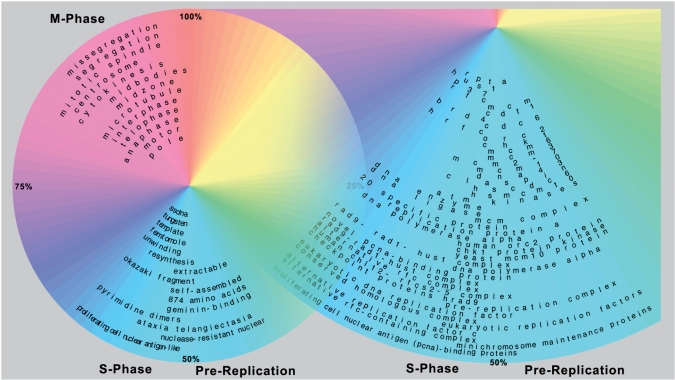
In Figure 2, these results are arranged in a cyclic layout (see ‘Methods’ section), where each keyword has been placed to show the exact region of the cell cycle where the keyword is significantly over-represented. The keywords cluster into three distinct groups: (i) a pre-replication phase (late G1, corresponding to cell-cycle progress from 41 to 52% in Figure 2) defined by keywords that describe the initiation of DNA replication and the checkpoints that can prevent initiation from taking place; (ii) S-phase (53–63%), defined by keywords that describe the proteins, complexes and processes associated with the replication machinery; (iii) M-phase (79–100%), which has no keywords for proteins or complexes, but has keywords that describe the cell division sub-processes. In G1 and G2 phase (1–40% and 64–78%, respectively) no enhanced keywords are seen, consistent with the generally-accepted belief that relatively few processes are specific to these ‘gap’ phases.
Figure 2.
Keywords found by Martini from cell-cycle genes. The figure shows all keywords found by Martini using 600 cell-cycle-regulated genes that have been experimentally assigned to specific time points within the human cell cycle. Percentage numbering indicates cell cycle progress, with cell division occurring at 100% or 12 o’clock. The arc spanned by each keyword shows the exact region where it is statistically significant. The radius of each keyword is determined by word length. The left-portion of the figure shows keywords that describe biological processes and functions—these keywords cluster into three distinct phases: M-, S- and a pre-replication phase. The right-portion of the figure is a close-up of the pre-replication and S-phase regions showing keywords that specify genes, proteins or complexes. The keywords shown, and their positions show a surprisingly accurate and precise match to the sub-phases, processes, and entities known to occur in the cell cycle (see also Table 3).
Assessed qualitatively, Figure 2 shows a surprisingly accurate and precise match to the events and entities known to occur at different stages of the cell cycle. Of the 72 total keywords found by Martini, we considered 67 to be ‘true positives’, i.e. to occur at the correct position in the cell cycle. The remaining five keywords—‘874 Amino Acids’, ‘Extractable’, ‘Femtomole’, ‘Tungsten’, ‘20 specific protein’—we would classified as ‘uninformative’ rather than ‘false positives’, since these keywords do not imply incorrect processes or entities.
To quantitate the accuracy and precision of the keywords and terms, we divided the 600 genes into four groups corresponding to the classic phases G1 (cell cycle progress from 1 to 40% in Figure 2, giving 113 genes), S (41–63%, 154 genes), G2 (64–78%, 82 genes) and M (79–100%, 251 genes). These gene sets were then used to perform a much simpler four-step analysis, shown in Table 2, where we compared the genes in each phase with those in the other three phases (e.g. G1 versus S+G2+M, etc.). For each of the tools, we then manually classified each term found as either true positive, false positive or uninformative using the following criteria: True positives are keywords that have definitely been assigned to the correct cell-cycle phase, i.e. they match to processes or entities known to occur specifically within that phase. False positives are keywords that match to cell-cycle processes, but have definitely been assigned to the incorrect phase, e.g. FatiGO finds the term ‘M phase’ associated with G1 genes. Since the dataset was defined as genes specific to the mitotic cell cycle, we considered any meiosis-specific keywords to be false positives. Finally, uninformative keywords are those that are not clearly right (true positive) and not clearly wrong (false positive).
Table 2.
Cell-cycle keywords and GO terms
| Tool | G1 | S | G2 | M |
|---|---|---|---|---|
| Martini | – | True positive: (ATR-interacting protein) protein kinase complex9; Ataxia Telangiectasia9; ATR–ATRIP complex9; Claspin6; DNA polymerase alpha8; DNA replication4; eukaryotic DNA replication factor8; Fork8; Human CHL12/RFCs2-5 complex8; HUS19; novel PCNA-binding protein8; Processivities8; Proliferating cell nuclear antigen (PCNA)-binding proteins8; Replication protein A8; RFA1-T118; RP-A8; Single-stranded8; ssDNA8. Uninformative: 20 specific proteins; Double-stranded DNA; Template. | – | True positive: Anaphase15; APC/C15; Aster18; BUBR115; Centrosome18; Cytokinesis11; Interphase11; Kinetochore18; Metaphase14; Microtubule18; Midzone19; Mitotic11; Mitotic spindle18; Multinucleated16; Prometaphase13; Prophase12; Segregation15; Telophase16. Uninformative: Monopolar |
| Marmite | – | – | – | – |
| CoPub | False positive: Mitosis11; Nucleotide-excision repair5; Oogenesis. Uninformative: Adipocyte differentiation; Apoptosis; Budding; Cell adhesion; Cell cycle, death, differentiation, fate determination, fate commitment, shape; Decidualization; Dephosphorylation; Epidermal differentiation; Epithelial cell proliferation, epithelial cell differentiation; Gene silencing; Growth pattern; Histogenesis; Intracellular transport; Keratinocyte differentiation; Morphogenesis; mRNA stabilization; Nucleocytoplasmic transport; Oncogenesis; Organogenesis; Phosphorylation; Protein folding, import; Response to hypoxia; RNA interference; RNA processing; Senescence; Signal transduction; Synaptonemal complex formation, synaptogenesis; Transcription; Transduction; Translation. | True positive: Base-excision repair5; Chromatin silencing3, remodeling, modification, insulator sequence binding; DNA damage response5, metabolism2, methylation4, recombination4, repair5, replication4, synthesis2, unwinding5; Double-strand break repair5; Hyperphosphorylation3; Mismatch repair5; Nucleotide-excision repair5; Postreplication repair5; Recombination4; Recombinational repair5; Single-stranded DNA binding8; Strand displacement8; Telomere maintenance5. False positive: Division11; M phase11; Mitosis11; Meiosis chromosome segregation, meiosis; Meiosis sister chromatid cohesion, meiosis II; Meiotic recombination; Metaphase14.Uninformative: Apoptosis; ATP transport, regeneration, metabolism, hydrolysis, catabolism, biosynthesis; Budding; Cell cycle, arrest, checkpoint; Cell death, growth and/or maintenance, cell growth, proliferation; DNA amplification, fragmentation; Drug resistance; Gene conversion, expression AND epigenetic, silencing; Histone acetylation; Induction of apoptosis; Mating behavior, mating; mRNA splicing, splice site selection; Mutagenesis; Oncogenesis; Phosphorylation; Rb:E2F-1:DP-1 transcription factor; RNA interference; Senescence; Transcription, mitotic, initiation; Translation; Viral replication. | False Positive: Anaphase A and B15; Chromatin silencing3, remodeling, modification, insulator sequence binding; Chromosome condensation13, segregation15, movement; Division11; DNA damage response15, methylation4, repair5, replication4, synthesis2; M phase11; Meiosis sister chromatid cohesion, II spindle assembly, I spindle assembly, chromosome segregation; Metaphase14; Mismatch repair5; Mitosis11; Nucleotide-excision repair5; Pachytene; Prophase12; Recombination4; Spermatogenesis. Uninformative: Antigen presentation; Apoptosis; Autophosphorylation; Budding; Caspase activation; Cell cycle, arrest, checkpoint, death, invasion; Chemotaxis; Conjugation; DNA fragmentation; Dephosphorylation; Fragmentation; Gene silencing; Glutathione metabolism, conjugation reaction, catabolism, biosynthesis; Histone acetylation; Induction of apoptosis; Inflammatory response; mRNA transcription; RNA interference; Transcription, initiation. | True positive: Abscission16; Anaphase A and B15; Cell division11; Centrosome separation15, cycle; Chromosome condensation13, segregation15, movement13–15; Cytokinesis11; Division11; Envelope breakdown12; Exit from mitosis14; M phase11; Metaphase14; Metaphase–anaphase transition14; Microtubule18 processes; Mitosis11; Prometaphase13; Prophase12; Sister chromatid cohesion and separation15; Spindle elongation13, assembly13, stabilization14, checkpoint14; Telophase16. False positive: DNA replication and synthesis4; Embryonic development; Fertilization; Meiosis sister chromatid cohesion, II spindle assembly, I spindle assembly, chromosome segregation; Oocyte maturation; Oogenesis; Osteoblast differentiation; Pachytene; Spermatogenesis; ssDNA binding8. Uninformative: Adipocyte differentiation; Angiogenesis; Anti-apoptosis; Antiviral response; Apoptosis; Autophosphorylation; Budding; Caspase activation; Cell adhesion, cycle arrest, cycle checkpoint, death, differentiation, fate specification, fate determination, fate commitment, growth and-or maintenance, growth, invasion, migration, motility, polarity, proliferation, size; Cell–cell adhesion; Cytoskeleton; Dephosphorylation; DNA damage response, fragmentation, repair; Double-strand break repair; Epithelial cell proliferation, differentiation; Fibroblast proliferation; Fragmentation; G-protein signaling; G2 checkpoint; G2/M checkpoint, transition; Gastrulation; Gene expression, epigenetic, silencing; Growth; Histone acetylation, modification; Hyperphosphorylation; Induction of apoptosis; Lung development; Mammary gland development; Mevalonate transport, pathway; Migration; Morphogenesis; mRNA processing, splicing, splice site selection, transport; Myogenesis; Neurogenesis; Nucleocytoplasmic transport; Oncogenesis; Phosphorylation; Programmed cell death; Protein prenylation; RNA interference, splicing; Senescence; Signal transduction; T-cell differentiation, homeostasis; Transcription, mitotic, initiation; Transduction; Vasculogenesis; Viral replication. |
| FatiGO | False positive: M phase of mitotic cell cycle11. Uninformative: Cell cycle; Intracellular organelle (membrane-bound); Ribonucleoprotein complex. | True positive: DNA metabolic process2, repair5, replication4; DNA-dependent DNA replication4; Response to DNA damage stimulus5; Nucleobase, nucleoside, nucleotide and nucleic acid metabolic process2; Regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process2. False positive: Cell division11 Uninformative: Biopolymer metabolic process; cell communication, surface receptor linked signal transduction; cellular metabolic process; DNA binding; Hydrolase activity; intracellular membrane-bound organelle; macromolecule metabolic process; membrane-bound organelle; mitotic cell cycle; nucleic acid binding; primary metabolic process; Regulation of cellular metabolic process; regulation of metabolic process; response to endogenous stimulus; response to stress; RNA metabolic process. | – | True positive: Cell division11; M phase of mitotic cell cycle11; microtubule18-based process; mitosis11; regulation of mitosis11. False positive: DNA metabolic process2, repair5, replication4; Nucleobase, nucleoside, nucleotide and nucleic acid metabolic process2. Uninformative: Biopolymer metabolic process; cell cycle, phase, process; cellular metabolic process; cytokine production; cytoskeleton, organization and biogenesis; macromolecule metabolic process; microtubule cytoskeleton organization and biogenesis; nucleic acid binding; primary metabolic process; response to DNA damage stimulus; response to endogenous stimulus; response to stress. |
| ProfCom | Uninformative: Hydrolase activity. | – | Uninformative: Hydrolase activity. | – |
The output of different tools applied to four gene sets, corresponding to the cell-cycle phases G1, S, G2 and M. Each output term was categorized as ‘true positive’, ‘false positive’ or ‘uninformative’. Superscripts indicate matches to the cell-cycle benchmark (Table 3). Qualitatively, Martini gave the best performance.
CoPub cannot compare two lists, and the results shown were generated effectively by comparing each of the four gene subsets against the background of all other human genes. As expected, CoPub gives less precise results with more false positives. In fact due to space limitations in Table 2, we show only ‘biological processes’ from CoPub; including the other CoPub categories (‘drug’, ‘pathway’, ‘disease’ and ‘liver pathology’) gives nearly twice as many keywords with a similar pattern of true and false positives.
Some of the keywords we classified as uninformative could arguably be regarded as false positives. For example, CoPub finds ‘G2 checkpoint’ and ‘G2/M checkpoint’ associated with M-phase genes, however, since these terms describe a process happening between two phases, in this simple four-state analysis, we considered such terms to be neither clearly right or wrong. Similarly, the Rb:E2F-1:DP-1 transcription factor found by FatiGO belongs to the switch from G1 to S phase. Terms such as ‘cell cycle’, ‘cell cycle checkpoint’ and ‘hydrolase’ are not incorrect, but since they refer to processes throughout the entire cycle, it is also not correct to assign them to a single cell-cycle phase. Another borderline case is ‘DNA damage’, which is a key feature of S-phase, but is also present in other phases, hence we regarded it as a true positive if it occurs in S-phase, but as uninformative for other phases. CoPub also finds terms such as ‘lung development’ that appear to be incorrect, given how the gene set was defined, however since this term does not clearly match to any specific cell-cycle process, we categorized it as ‘uninformative’.
To calculate a recall score, we created a benchmark or ‘score card’ that defines 20 main phases, sub-processes and key components in the human cell cycle (Table 3). Each true positive in Table 2 was then mapped onto one row of Table 3, allowing us to count non-redundant true positives (tp), and also to count false negatives (fn, i.e. rows in Table 3 for which a tool has no matching keywords). The recall was then calculated as tp/(tp + fn), and the precision calculated as tp/(tp + fp), where fp stands for the number of false positives in Table 3. Note that the number of false positives has no clear limit, hence the precision score used here is an estimate of the ‘true’ precision.
Table 3.
Cell-cycle benchmark and score-card
| Cell-cycle phases, processes and components |
Martini | Marmite | CoPub | FatiGO | ProfCom | |
|---|---|---|---|---|---|---|
| Synonyms/related genes | ||||||
| 1. G1-Phase | Gap 1 | |||||
| 2. S-Phase | DNA metabolism, synthesis; synthesis phase | 1 | 1 | |||
| Sub-processes: | 1 | |||||
| (i) Replication initiation | Chromatin silencing; Hyperphosphorylation | |||||
| (ii) DNA replication | DNA methylation, synthesis, recombination | 1 | 1 | 1 | ||
| (iii) DNA repair | Base-excision repair; DNA damage response, unwinding; double-strand break repair; mismatch repair; nucleotide-excision repair; post-replication repair; Telomere maintenance | 1 | 1 | |||
| Key components: | ||||||
| (i) Origin of replication complex | Claspin; ORC | |||||
| (ii) Mini-chromosome maintenance complex | MCM2-7 | |||||
| (iii) Replication fork | CHL12; DNA ligase; DNA polymerase; DNA replication factor; Helicase; holoenzyme; lagging strand; leading strand; Okazaki fragments; PCNA; PCNA-binding protein; pre-replication complex; primase; processivity; replication protein A (RP-A); replication factor C (RFC); single-stranded DNA; ssDNA-binding proteins; Strand displacement; Topoisomerase. | 1 | 1 | |||
| 3. DNA repair proteins | Ataxia Telangiectasia mutated gene (ATM), ATR, ATR-interacting proteins; ATRIP; CHK1 kinase, CHK2 kinase; HUS1 | 1 | ||||
| 4. G2-Phase | Gap 2 | |||||
| 5. M-Phase | Cell division; ‘Karyokinesis and cytokinesis’; Mitosis; Mitotic division; ‘Not interphase’ | 1 | 1 | 1 | ||
| Sub-processes: | 1 | 1 | ||||
| (i) Prophase | Envelope breakdown. | |||||
| (ii) Prometaphase | Chromosome condensation; spindle assembly; spindle elongation | 1 | 1 | |||
| (iii) Metaphase | BubR1; chromosome alignment; hyperphosphorylation; metaphase–anaphase transition; mitotic checkpoint; mitotic exit checkpoint; mitotic spindle checkpoint; spindle stabilization | 1 | 1 | |||
| (iv) Anaphase | APC/C; centrosome separation; chromatid separation; chromosome segregation; sister chromatid separation | 1 | 1 | |||
| (v) Telophase | Multinuclear | 1 | 1 | |||
| (vi) Cytokinesis | Abscission | 1 | ||||
| Key components: | 1 | 1 | 1 | |||
| (i) Mitotic spindle | Aster; centrosomes; centriole pair; Kinetochore; microtubules; mitotic center | |||||
| (ii) Metaphase plate | Midzone | 1 | ||||
| (iii) Cleavage furrow | Contractile ring; non-muscle myosin II+actin filaments | |||||
| True Positives (non-redundant) | 12 | 0 | 12 | 5 | 0 | |
| False Negatives | 8 | 20 | 8 | 15 | 20 | |
| False Positives | 0 | 0 | 58 | 6 | 0 | |
| Recall | 60% | 0% | 60% | 25% | 0% | |
| Precision | 100% | – | 17% | 46% | – | |
This table lists 20 key features of the human cell cycle, and uses this feature list as a benchmark to compare the performance of different tools based on their output (Table 2).
Of the five tools tested against this benchmark, Martini clearly gave the best performance, with 60% recall and 100% precision. CoPub found many more keywords and had similarly good recall (60%), but only 17% precision (i.e. many false positives). FatiGO also found more keywords than Martini, but had lower recall (25%) and lower precision (45%). Marmite found zero terms in all of the phases, while ProfCom found only the single term ‘hydrolase activity’ that we judged to be uninformative.
Melanoma dataset
We next tested keyword enhancement using two gene sets, one associated with primary melanoma and another with metastatic melanoma (4). In contrast to the very specific comparisons of cell-cycle phases in Table 2, comparing these two types of melanoma corresponds to asking a more general question. We considered the melanoma dataset to be not a good candidate as a benchmark, unlike the cell-cycle dataset, but probably a more realistic or typical case.
Table 4 compares the output of FatiGO, Marmite, Martini and ProfCom with these data. We manually classified each keyword found as either mitosis-related, uninformative, or ‘not mitosis-related’ using the following criteria (different to the cell-cycle criteria). Mitosis-related keywords have a clear relation to the major mitosis-specific processes. Since mitotic cell division is what we would expect to see associated with metastatic cancer, we considered these keywords to be true positives. Uninformative keywords were either too generic (e.g. ‘assemblies’ or ‘biogenesis’), or related to experimental techniques (e.g. ‘co-immunoprecipitation’), or related to model organisms (‘cerevisiae’ or ‘sporulation’). Any remaining keywords were classified as Not mitosis-related. Keywords in this final category are the most interesting as their connection to melanoma and metastasis is, in many cases, not immediately obvious. In contrast to Arabidopsis and the human cell-cycle, where many of us have extensive experience, we had little previous experience with the melanoma literature, and hence we were less confident in deciding true and false positives.
Table 4.
Keywords for metastatic versus primary melanoma
| Tool | Keywords/GO terms |
|---|---|
| Martini | Mitosis-related: Anaphase; APC/C; Arrest; Ataxia telangiectasia; BUBR1; Camptothecin; CDC20; CDH1; CDK2; CDK2 kinase; CDT1; Cell cycle; Cell division; Centrosome; Checkpoint; Chromatid; Claspin; Cohesin; Cyclin B; Cyclin-dependent; Cyclin-dependent kinases; Cyclosome; Cytogenetic; Cytokinesis; DNA damage; DNA repair; DNA replication; DNA-binding protein; Double-strand break; Double-stranded DNA; E2F transcription factor; E2F4; EGF; Elongation; Elongation factor; Fork; G2-phase; Guanine nucleotide; H2B; Helicase, Initiation factor; High-sensitive reverse transcription-nested polymerase chain; Histone; Histones; Intermediate filament; Interphase; Junctional; Kinase; Kinases; Kinetics; Kinetochore; Ligases; Ligated; M-phase; MAD1; MAD2; Metaphase; Microtubule; Midbodies; Midzone; Minichromosome; Misalignment; Missegregation; Mitotic; Mitotic spindle; Monopolar; Multipolar; Non-phosphorylatable; Nuclear antigen; Nuclear pore; Nuclear protein; Nucleolar; Nucleus; Oligoribonucleotides; PCNA; Phosphopeptides; Phosphoprotein; Phosphoproteins; Phosphorylation; Pol; Polo-like kinase 1; Polypeptide chain elongation factor 1 alpha; Posttranslational; Prometaphase; Prophase; Proteasome; Protein kinase; RAD17; rDNA; Replication; Replication protein A; Ribonucleotide; Ribosomal gene; Ribosome; Ring-shaped; RNA polymerase; RNA polymerase I subunit; RP-A; S-phase; Segregation; Small nuclear ribonucleoprotein; Small nuclear ribonucleoprotein particle; spindle associated proteins; ssDNA; Telophase; Topoisomerase; Topoisomerase I; Ubiquitin; ubiquitin ligases; Ubiquitins |
| Uninformative: Assembled; Assemblies; Binaries; Biogenesis; Cerevisiae; Clade; Classification; Co-immunoprecipitation; Connection; Coordination; Cross-validation; Daughter; Degradation; Depletion; Dispensable; Dynamics; Empirical; End; Essential; Eukaryote; Eukaryotic; Eukaryotic cell; False-positive; Fraction; Global; Health; Hit; Human protein-protein; Immunodepleted; Imprinting; In-gel; Independent; Integrated; Invaluable; Lates; Layer; Leaves; Linings; Liquid chromatography-tandem mass spectrometry; Lysate; Machine; Mappings; Mass spectrometries; Matched; Mechanism; Microinjection; Motif; Motor; Multisubunit; Mus sp; Nascent; Nationals; Network; Nicotiana tabacum; Non-redundant; Oligoribonucleotide; Pairwise; Percent; Permit; Purified; Remain; Removed; Saccharomyces cerevisiae; Scale; Schizosaccharomyces; Schizosaccharomyces pombe; Spectral; Stable isotope; Stem-loop; Step; Stringent; Supplemental; Surprisingly; Systematics; Tobacco; Unlike; Unprecedented; Unsolved; Upper | |
| Not mitosis-related: 12 MAGE; 5′-phosphates; 941 pseudogenes; ATPase; aneuploidy; ATM kinases; Autism; Bone marrow cell; Bronchial; Calcium; Calcium-dependent; Chaperone; CHK1 protein kinase; Cornea; Cornified; Cystic fibrosis gene; Desmosome; Epidermal cell; Epithelial; Epithelial cell; Epithelium; Human gene MAGE-1; Guanine nucleotide exchange factors; Heat-shock; HSPs; Hair follicle; HeLa cell; HeLa cell nuclear; Involucrin; Keratinocyte; Lymphoid-specific; MAGE; MAGE-A; MAGE-A antigens; Melanoma antigen gene; Neurite; Novel protein-protein; Over 100 disease-associated proteins; Polyploid; Pre-rRNA; Protein complex; Proteome; PSMS; Psoriatics; Psoriasis; Pyrophosphatase; Quiescent; Rebinding; Regulatory; Repair; Reversible; single MAGE-A antigen; Site-specific; Specific kinase-substrate; Squamous epithelium; Stimuli; Stratified; Stratified epithelia; Subcellular; Subcomplex; Substrate; Subunit; Superfamilies; Suprabasal; Surface; Skin diseases; Starvation; Stratum corneum; Synthesis; Temporal; Topologies; Transmission; Unaligned; Unattached; Viral protein; Xeroderma pigmentosum | |
| Marmite | Not mitosis-related: Psoriasis |
| FatiGO | Mitosis-related: Chromosome, organization and biogenesis; DNA metabolic process; E2F; M phase of mitotic cell cycle; Cell cycle, phase, process; Cell division; Chromosome segregation; DNA recombination, repair, replication; HNF1; Intermediate filament cytoskeleton; MEF-2; Mitosis, cell cycle; Nucleobase, nucleoside, nucleotide and nucleic acid metabolic process; Rb:E2F-1:DP-1; Response to DNA damage stimulus |
| Not mitosis-related: Anatomical structure development, morphogenesis; Apical junction complex; Apicolateral plasma membrane; ATP binding; Biopolymer metabolic process; Cation binding; Cell adhesion, junction, part; Cellular component organization and biogenesis, metabolic process; Cornified envelope; Cytosol; Defense response; Ectoderm development; Epidermis development; Extracellular region part, space; Integral to membrane, plasma membrane; Intercellular junction; Intracellular, membrane-bound organelle; Intrinsic to membrane; Macromolecule metabolic process; Membrane-bound organelle; Multicellular organismal development; Non-membrane-bound organelle; Nuclear part; Nucleic acid binding; Nucleotide binding; Nucleus; Organ development; Organelle lumen; Plasma membrane; Primary metabolic process; Protein folding; Purine nucleotide binding; Response to endogenous stimulus; RNA binding, localization, processing; System development; Tissue development | |
| ProfCom | — |
In this table, different tools have been used to compare a set of genes associated with primary melanoma, and a second set of genes associated with metastatic melanoma. Each keyword or GO term found has been classified as either mitosis-related, uninformative, or ‘not mitosis-related’. Compared with the other tools, Martini found more keywords, more-specific keywords, but also more uninformative keywords.
Martini found 264 significantly-enhanced keywords, a much larger number than the other methods (Table 4). Of the keywords found by Martini, 109 were mitosis-related and 79 were uninformative. This left 76 keywords assigned as ‘not mitosis-related’; for each of these we manually checked the literature for evidence of a connection to melanoma or metastasis. For some keywords, this connection was straightforward, e.g. skin, cornea, lymphoid, HeLa cells, desmosome, intermediate filaments, involucrin, calcium, as well as several skin diseases. For other keywords, the connection was less obvious, but was supported by the literature: e.g. polyploidy (24), cornification and bone marrow cells (25), heat-shock/chaperone proteins (26), cystic fibrosis (27), ATM kinases (28), CHK1 (29), neurites (30). Perhaps the most interesting keywords found by Martini are the names of several of the MAGE (melanoma-associated genes) gene family. These genes are normally expressed only in developing sperm, where they play a role in meiotic cell division. However, these genes are also expressed in melanoma (31,32).
FatiGO found 4 transcription factors and 47 GO terms, of which 36 were classified as not mitosis-related (Table 4). As with Martini, all the terms in the ‘not mitosis-related’ category seemed to have a general connection to melanoma or metastasis, hence none were obviously false positives. Interestingly, FatiGO does not find the link to spermatogensis.
Comparing Martini and FatiGO qualitatively, both seemed to have similar precision with this dataset, i.e. all terms and keywords found were either uninformative or, as best as we could judge true positives, correctly indicating a connection to melanoma or metastasis. Martini found many more keywords, more-specific keywords and also more uninformative keywords. Martini also found many processes related to melanoma and metastasis that were not found by FatiGO. Thus, we conclude that Martini had qualitatively a somewhat higher recall, however, unlike the human cell cycle, we cannot quantify this since we do not have the background to construct a benchmark covering all the major processes and components involved. Marmite and ProfCom did not perform well with this dataset, finding almost no terms (Table 4).
Ovarian cancer dataset
As a final test of keyword enhancement, we used FatiGO, Marmite, Martini and ProfCom to compare a set of 160 genes associated with clear-cell ovarian cancer (i.e. cells that are clear when viewed through a microscope), and a second set of 105 genes associated with non-clear-cell ovarian cancer. For this comparison, each of the tools found exactly zero significantly enhanced keywords or GO terms.
DISCUSSION
Which tool best predicts function?
In this work we have compared Martini with several other tools with similar functionality, and overall Martini performs favorably for the specific cases we tested. However, comparing such tools is complex and multifaceted. Many criteria need to be considered making it difficult to judge which tool is the ‘best’, for example, some end-users may prefer tools that offer advanced features and functionality, even though these tools may take longer to learn. Martini offers fewer advanced features than many other tools, since we designed it for end-users who require a simple, easy-to-use tool that gives quick insight into the functional differences between two gene lists.
Another important criterion is the range of organisms that a tool can be applied to. Most of the tools we could find that are comparable to Martini (e.g. FatiGO, Marmite and ProfCom) can work only with genes from human plus a small number of model organisms. Martini has a much greater range, since it can be used with any organism in the Entrez gene database. Furthermore, Martini even allows comparison of genes across different organisms, whereas almost all similar tools usually restrict comparisons to genes within the same organism.
Finally, accuracy is clearly a very important criterion for assessing tools such as Martini. Unfortunately, accuracy can be difficult to assess objectively and to quantify reliably. What is required is a set of reliable benchmarks tailored specifically for comparing two gene sets, ideally spanning a wide range of functions and organisms. In this article, we have taken a step in this direction by proposing one such benchmark (Table 3) that can be used with gene sets related to the human cell cycle (see ‘Results’ section). Since the human cell cycle is very well-studied, this benchmark probably represents a ‘best-case’, and the performance of such tools is likely to be worse for most other datasets (e.g. for the ovarian-cancer dataset, none of tools tested could find any keywords or GO terms). We designed this benchmark to cover only the 20 most important phases, sub-processes and components in the cell cycle; however, as tools improve it would eventually be useful to create a more detailed, fine-grained benchmark. Currently, the best performing tools reach only 60% recall (Table 3), indicating that there is considerable scope for improving such tools.
For the benchmark, we also included CoPub, even though this tool is not really similar to the others tested here, since it cannot compare two gene lists. Given this, CoPub performs rather well in the benchmark, with equal-best recall. The lower precision (i.e. more false positives) obtained by CoPub illustrates the benefit of the two-set approach.
Using the cell-cycle benchmark, Martini had markedly better performance compared with the other tools we tested (Table 3). In addition, assessed qualitatively, Martini also had better or equal accuracy for each of the other datasets presented here (Arabidopsis, melanoma and ovarian cancer). Taken together, these results suggest that Martini represents an advance in the state-of-the-art in automated comparison of gene sets. Once published, we await further feedback from end-users applying Martini to wider range of cases to see if this trend still holds.
Keywords versus GO terms
In our initial survey of tools for gene set analysis, we found that almost all such tools rely on GO terms, often exclusively (see ‘Introduction’ section), and only a small number of methods used keywords. This suggests a perception amongst many scientists in this field that GO terms are the preferred, more reliable source of functional annotation for genes. Indeed, when we shared the results presented here, many of our colleagues found it striking that the GO-based tools (ProfCom and FatiGO) performed in some cases much worse than a keyword-based tool such as Martini.
Summarizing the performance of the tools with the datasets we tested, we conclude that Martini performed best, followed by FatiGO, then Marmite and ProfCom, both performing similarly. Since FatiGO and ProfCom are both GO-based, and since Martini and Marmite are both based on similar keyword dictionaries, it is likely that the poorer performance of Marmite and ProfCom arises from the statistical methods used.
However, based on the performance difference between Martini and FatiGO, plus the relatively good performance of CoPub (Table 2), we conclude that keywords may be a richer source of functional annotations than GO terms. Since this runs contrary to the expectation of many scientists in the field, we decided to survey the density of gene annotations from GO terms versus keywords. As reported in the ‘Introduction’ section, in Entrez we found that the median numbers of GO terms and Medline citations per human gene are seven and nine, respectively. Using the AKS2 keyword dictionary, we get 32 keywords per abstract, and hence a median value of over 100 unique keywords per gene. For well-studied genes, the contrast between GO terms and keywords is even stronger, e.g. the Entrez entry for human p53 has 74 GO terms compared with 2527 Medline abstracts, which give rise to over 11 000 unique keywords using AKS2.
The appeal of GO terms likely derives from the use of a controlled vocabulary, as well as the fact that annotation of gene function using GO terms has been done rather systematically. In contrast, extracting keywords automatically from Medline abstracts could be expected to be time-consuming, noisy and error-prone. However, both Martini and CoPub demonstrate the feasibility of a keyword-based approach. Furthermore, in agreement with Küffner et al. (9), we find that keywords appear to give consistently better, and more specific results than GO terms.
Tips for using Martini
In this section, we discuss some practical tips and issues for end-users planning to use Martini to compare gene sets.
First, we would advise end-users not to have too high expectations when using any automated method to infer function. Like all such methods, Martini does not always produce good results. Martini depends entirely upon the underlying literature associated with the genes in the input sets: it may often occur that there is relatively little literature, or that the literature does not adequately describe the differences between the two genes sets. In the results, we presented one such case—the ovarian cancer dataset—where all of the tools tested produced exactly zero results.
Secondly, our experience suggests that best results are often obtained by asking very specific questions, i.e. by comparing two closely related datasets. For example, in Table 2 we used CoPub to compared four sets of ∼150 genes, on average, with the background of the remaining ∼20 000+ human genes; this produced good recall but with many false positives, hence low precision. With Martini, we got better results by asking a more specific question, i.e. by comparing the ∼150 gene sets for each cell-cycle phase against the remaining ∼450 genes associated with the other phases (Table 2). In fact, as can be seen by comparing the Martini keywords in Table 2 with those in Figure 2, we got even better results (many more keywords and more specific keywords) with the same dataset, but asking an even more specific question, e.g. comparing on average ∼60 genes in each 10% sub-region of the cell cycle to the ∼540 genes in the remaining 90%.
Thirdly, in cases where only a single gene set is available, one strategy is to construct a second reference gene set by randomly selecting a subset of genes from the same organism. We suggest using a reference set that is several times larger than the experimental set, and choosing genes that each have greater than the median number of abstracts for that species: for human, this means genes with more than nine abstracts. We used a similar strategy for the Arabidopsis dataset, and in this case it produced good results. However, we stress again that there can be no guarantee of producing informative results with automated methods such as Martini.
Fourthly, an alternative strategy in the case of a single experimental set is to use a tool such as Anni (3) or TXTGate (2) that can interactively divide a single gene set into functionally related sub-clusters. These sub-clusters can then be compared using Martini.
Finally, to analyze the keywords produced by Martini, we recommend the strategy adopted for the melanoma dataset (Table 4) i.e. divide the keywords into three groups:
Keywords that are obvious, given the biological context.
Keywords that are uninformative (e.g. keywords such as ‘surprisingly’): Martini sometimes produces many of these (e.g. Table 4), partly due to its large keyword dictionary. Such generic keywords are usually more annoying than troublesome, and we plan to blacklist many of them in the future.
The remaining keywords are often the most interesting, and are most likely to give novel insight into the functional differences between the two gene sets.
In some cases, Martini produces a list of keywords that is very large. To help such cases, in the future, we plan that the output list of keywords will be automatically organized into similar biomedical concepts.
CONCLUSIONS
Martini is designed to be fast and easy-to-use, providing a quick first insight into the functional difference between two gene sets. Our results suggest that Martini offers a significant advance in the automated extraction of biological knowledge from sets of genes or abstracts. Currently, Martini focuses on finding differences between two input sets; in the near future we plan to add an option to search instead for commonalities between these sets, for example to find interactions involving genes from both sets. In addition, we plan to improve Martini by adding document classification, by enabling the input of single gene-lists, and by using sequence alignment tools to extend functional annotation to similar sequences.
FUNDING
European Molecular Biology Laboratory, by the European Union Framework Programme 6 grant ‘TAMAHUD’ (LSHC-CT-2007-037472, in part); Novo Nordisk Foundation Center for Protein Research. Funding for open access charge: EMBL internal budget.
Conflict of interest statement. None declared.
REFERENCES
- 1.Cohen AM, Hersh WR. A survey of current work in biomedical text mining. Brief Bioinform. 2005;6:57–71. doi: 10.1093/bib/6.1.57. [DOI] [PubMed] [Google Scholar]
- 2.Glenisson P, Coessens B, Van Vooren S, Mathys J, Moreau Y, De Moor B. TXTGate: profiling gene groups with text-based information. Genome Biol. 2004;5:R43. doi: 10.1186/gb-2004-5-6-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jelier R, Schuemie MJ, Veldhoven A, Dorssers LC, Jenster G, Kors JA. Anni 2.0: a multipurpose text-mining tool for the life sciences. Genome Biol. 2008;9:R96. doi: 10.1186/gb-2008-9-6-r96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Riker AI, Enkemann SA, Fodstad O, Liu S, Ren S, Morris C, Xi Y, Howell P, Metge B, Samant RS, et al. The gene expression profiles of primary and metastatic melanoma yields a transition point of tumor progression and metastasis. BMC Med. Genomics. 2008;1:13. doi: 10.1186/1755-8794-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Al-Shahrour F, Minguez P, Tarraga J, Medina I, Alloza E, Montaner D, Dopazo J. FatiGO+: a functional profiling tool for genomic data. Integration of functional annotation, regulatory motifs and interaction data with microarray experiments. Nucleic Acids Res. 2007;35:W91–W96. doi: 10.1093/nar/gkm260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Antonov AV, Schmidt T, Wang Y, Mewes HW. ProfCom: a web tool for profiling the complex functionality of gene groups identified from high-throughput data. Nucleic Acids Res. 2008;36:W347–W351. doi: 10.1093/nar/gkn239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R, Eilbeck K, Lewis S, Marshall B, Mungall C, et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32:D258–D261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khatri P, Draghici S. Ontological analysis of gene expression data: current tools, limitations, and open problems. Bioinformatics. 2005;21:3587–3595. doi: 10.1093/bioinformatics/bti565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Küffner R, Fundel K, Zimmer R. Expert knowledge without the expert: integrated analysis of gene expression and literature to derive active functional contexts. Bioinformatics. 2005;21:ii259–ii267. doi: 10.1093/bioinformatics/bti1143. [DOI] [PubMed] [Google Scholar]
- 10.Chaussabel D, Sher A. Mining microarray expression data by literature profiling. Genome Biol. 2002;3:RESEARCH0055. doi: 10.1186/gb-2002-3-10-research0055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chagoyen M, Carmona-Saez P, Shatkay H, Carazo JM, Pascual-Montano A. Discovering semantic features in the literature: a foundation for building functional associations. BMC Bioinformatics. 2006;7:41. doi: 10.1186/1471-2105-7-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jelier R, Jenster G, Dorssers LC, Wouters BJ, Hendriksen PJ, Mons B, Delwel R, Kors JA. Text-derived concept profiles support assessment of DNA microarray data for acute myeloid leukemia and for androgen receptor stimulation. BMC Bioinformatics. 2007;8:14. doi: 10.1186/1471-2105-8-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Al-Shahrour F, Carbonell J, Minguez P, Goetz S, Conesa A, Tarraga J, Medina I, Alloza E, Montaner D, Dopazo J. Babelomics: advanced functional profiling of transcriptomics, proteomics and genomics experiments. Nucleic Acids Res. 2008;36:W341–W346. doi: 10.1093/nar/gkn318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frijters R, Heupers B, van Beek P, Bouwhuis M, van Schaik R, de Vlieg J, Polman J, Alkema W. CoPub: a literature-based keyword enrichment tool for microarray data analysis. Nucleic Acids Res. 2008;36:W406–W410. doi: 10.1093/nar/gkn215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Etzold T, Argos P. SRS—an indexing and retrieval tool for flat file data libraries. Comput. Appl. Biosci. 1993;9:49–57. doi: 10.1093/bioinformatics/9.1.49. [DOI] [PubMed] [Google Scholar]
- 16.Fisher RA. On the interpretation of χ2 from contingency tables, and the calculation of P. J. Roy. Stat. Soc. 1922;85:87–94. [Google Scholar]
- 17.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Roy. Stat. Soc. 1995;57:289–300. [Google Scholar]
- 18.Osier MV, Zhao H, Cheung KH. Handling multiple testing while interpreting microarrays with the Gene Ontology Database. BMC Bioinformatics. 2004;5:124. doi: 10.1186/1471-2105-5-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Swarbreck D, Wilks C, Lamesch P, Berardini TZ, Garcia-Hernandez M, Foerster H, Li D, Meyer T, Muller R, Ploetz L, et al. The Arabidopsis Information Resource (TAIR): gene structure and function annotation. Nucleic Acids Res. 2008;36:D1009–D1014. doi: 10.1093/nar/gkm965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jensen LJ, Jensen TS, de Lichtenberg U, Brunak S, Bork P. Co-evolution of transcriptional and post-translational cell-cycle regulation. Nature. 2006;443:594–597. doi: 10.1038/nature05186. [DOI] [PubMed] [Google Scholar]
- 21.Whitfield ML, Sherlock G, Saldanha AJ, Murray JI, Ball CA, Alexander KE, Matese JC, Perou CM, Hurt MM, Brown PO, et al. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol. Biol. Cell. 2002;13:1977–2000. doi: 10.1091/mbc.02-02-0030.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zorn KK, Bonome T, Gangi L, Chandramouli GV, Awtrey CS, Gardner GJ, Barrett JC, Boyd J, Birrer MJ. Gene expression profiles of serous, endometrioid, and clear cell subtypes of ovarian and endometrial cancer. Clin. Cancer Res. 2005;11:6422–6430. doi: 10.1158/1078-0432.CCR-05-0508. [DOI] [PubMed] [Google Scholar]
- 23.Wolfram Research I. Mathematica Edition: Version 7.0. Champaign, Illinois: Wolfram Research, Inc.; 2008. [Google Scholar]
- 24.Whang-Peng J, Chretien P, Knutsen T. Polyploidy in malignant melanoma. Cancer. 1970;25:1216–1223. doi: 10.1002/1097-0142(197005)25:5<1216::aid-cncr2820250529>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 25.Perez M, Migliaccio S, Taranta A, Festuccia C, Orru L, Brama M, Bologna M, Faraggiana T, Baron R, Teti A. Melanoma cells stimulate osteoclastogenesis, c-Src expression and osteoblast cytokines. Eur. J. Cancer. 2001;37:629–640. doi: 10.1016/s0959-8049(00)00436-6. [DOI] [PubMed] [Google Scholar]
- 26.di Pietro A, Tosti G, Ferrucci PF, Testori A. Oncophage: step to the future for vaccine therapy in melanoma. Expert Opin. Biol. Ther. 2008;8:1973–1984. doi: 10.1517/14712590802517970. [DOI] [PubMed] [Google Scholar]
- 27.Warren N, Holmes JA, al-Jader L, West RR, Lewis DC, Padua RA. Frequency of carriers of cystic fibrosis gene among patients with myeloid malignancy and melanoma. British Med. J. 1991;302:760–761. doi: 10.1136/bmj.302.6779.760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee JH, Paull TT. Activation and regulation of ATM kinase activity in response to DNA double-strand breaks. Oncogene. 2007;26:7741–7748. doi: 10.1038/sj.onc.1210872. [DOI] [PubMed] [Google Scholar]
- 29.Tse AN, Carvajal R, Schwartz GK. Targeting checkpoint kinase 1 in cancer therapeutics. Clin. Cancer Res. 2007;13:1955–1960. doi: 10.1158/1078-0432.CCR-06-2793. [DOI] [PubMed] [Google Scholar]
- 30.Sarkar D, Boukerche H, Su ZZ, Fisher PB. mda-9/Syntenin: more than just a simple adapter protein when it comes to cancer metastasis. Cancer Res. 2008;68:3087–3093. doi: 10.1158/0008-5472.CAN-07-6210. [DOI] [PubMed] [Google Scholar]
- 31.De Plaen E, Arden K, Traversari C, Gaforio J, Szikora J, De Smet C, Brasseur F, van der Bruggen P, Lethé B, Lurquin C, et al. Structure, chromosomal localization, and expression of 12 genes of the MAGE family. Immunogenetics. 1994;40:360–369. doi: 10.1007/BF01246677. [DOI] [PubMed] [Google Scholar]
- 32.Simpson AJ, Caballero OL, Jungbluth A, Chen YT, Old LJ. Cancer/testis antigens, gametogenesis and cancer. Nat. Rev. Cancer. 2005;5:615–625. doi: 10.1038/nrc1669. [DOI] [PubMed] [Google Scholar]