Abstract
The white gene, which is responsible for eye pigmentation, is widely used to study position effects in Drosophila. As a result of insertion of P-element vectors containing mini-white without enhancers into random chromosomal sites, flies with different eye color phenotypes appear, which is usually explained by the influence of positive/negative regulatory elements located around the insertion site. We found that, in more than 70% of cases when mini-white expression was subject to positive position effects, deletion of the white promoter had no effect on eye pigmentation; in these cases, the transposon was inserted into the transcribed regions of genes. Therefore, transcription through the mini-white gene could be responsible for high levels of its expression in most of chromosomal sites. Consistently with this conclusion, transcriptional terminators proved to be efficient in protecting mini-white expression from positive position effects. On the other hand, the best characterized Drosophila gypsy insulator was poorly effective in terminating transcription and, as a consequence, only partially protected mini-white expression from these effects. Thus, to ensure maximum protection of a transgene from position effects, a perfect boundary/insulator element should combine three activities: to block enhancers, to provide a barrier between active and repressed chromatin, and to terminate transcription.
INTRODUCTION
Enhancer-mediated activation is a fundamental mechanism of gene activation in eukaryotes (1,2). Enhancers can act over large distances to activate transcription of a certain gene, regardless of their orientation and position relative to the promoter and without affecting adjacent genes. According to recent data, enhancers interact directly with tagged genes by looping out the intervening sequences (3–6). The assumed ability of enhancers to stimulate unrelated promoters provided a basis for the model suggesting the existence of a specific class of regulatory elements that form independent transcriptional domains and preclude undesirable interactions between enhancers and promoters (7).
The sequences referred to as insulators due to their ability to prevent activation or repression signals from passing across them to a promoter have been found in different organisms (8–13). Insulators are defined by two properties: these nucleoprotein complexes can block enhancer action on a promoter when interposed between them and can protect transgenes they flank from chromosomal position effects.
Over many years, the white gene has been widely used as a model system for analyzing the enhancer-blocking and boundary activities of insulators in Drosophila (14–20). The reasons for this are several. The white gene, being well characterized molecularly, is not essential for fly viability (21–24). A tissue-specific enhancer is responsible for white activation in the eyes (25), and the level of eye pigmentation is a sensitive indicator of the amount of white transcription. In test systems, the mini-white gene of the CaSpeR vector has usually been used (26). This gene contains ∼300 bp of 5′ and 630 bp of 3′ flanking DNA and has the greater part of the first intron deleted. Transformants carrying the mini-white gene show a range of eye coloration from pale yellow to red, depending on the position of mini-white insertion into the genome (21,23,24). To explain the high sensitivity of the mini-white gene to chromosomal position effects, it has been speculated that the white promoter can function as an enhancer trap, meaning that enhancers located either 5′ or 3′ of the transposon are able to stimulate transcription of the mini-white gene.
Using the mini-white model, it was shown that insulators could protect white expression from the influence of nearby enhancers (14,16,18). With regard to the proposed property of insulators to protect a transgene promoter from unspecific functional interactions with neighboring enhancers, we re-examined the ability of the mini-white gene to be activated by non-specific enhancers in random genomic positions and the functioning of the gypsy insulator as a boundary element.
MATERIALS AND METHODS
Plasmids and constructs
Promoterless gene mini-white was PCR-amplified with primers 5′-ctccaagcggtttacgcc-3′ and 5′-cagccgaatgaattctagttcc-3′ from the pCaSpeRΔ700 plasmid (27), digested with EcoRI, and subcloned into the P-elements-containing pCT plasmid digested with SmaI–EcoRI (pCTdW). The promoter of the white gene (Prw) was obtained from pCaSpeR2 digested with Eco47III and AflII; TASC was PCR-amplified with primers 5′-gttaccgacaaacgacagtccac-3′ and 5′-cggcgtgtgctacttgtcttagg-3′ from genomic DNA; TSV40 was obtained from pUASt plasmid digested with BamHI and XbaI; a 340-bp fragment containing the Su(Hw)-binding region (Gy) was PCR-amplified from the gypsy retrotransposon. An 8-kb fragment containing the yellow gene and the cDNA yellow clone were kindly provided by P. Geyer. A 3-kb SalI–BamHI fragment containing the yellow regulatory region (yr) was subcloned into pGEM7 cleaved with BamHI–XhoI (yr plasmid). A 5-kb BamHI–BglII fragment containing the yellow coding region (yc) was subcloned into pCaSpeR2 (C2-yc). The Eco47III–ScaI fragment of the yellow coding region (fragment from −893 to +4774 bp relative to the yellow transcription start site) was cloned into pCTdW digested with EcoRI (pCTdWY).
(Prw)dW, (TASC)dW and (Gy)dW
Fragments Prw, TASC and Gy were flanked by loxP sites and inserted into the pCTdWY plasmid digested with XbaI.
YΔ−893 and (UAS)YΔ−893
The yellow enhancers were deleted from the yr plasmid by digestion with Eco47III and NcoI. The resulting DNA fragment yrΔ was cloned into the yc plasmid (YΔ-893). The UAS promoter digested with BamHI and HpaI from the pUASt plasmid was cloned between the loxP sites (UAS). The resulting DNA fragment was inserted into yr digested with Eco47III and NcoI [yrΔ(UAS)]. The yrΔ(UAS) fragment was cloned into the yc plasmid.
(Ee)(TASC)W and (Ee)(TSV40)W
The white eye enhancer (Ee; fragment from −1465 to −1084 bp relative to the white transcription start site (24) flanked by frt sites was inserted into pCaSPeR2 digested with XbaI [(Ee)W]. Thereafter, TASC and TSV40 fragments flanked by loxP sites were inserted between the eye enhancer and white promoter at the HpaI restriction site.
UAS(TASC)dW, UAS(TSV40)dW and UAS(Gy)dW
The UAS promoter digested with BamHI and HpaI from the pUASt plasmid was inserted into pSK–lacZ digested with BamHI and EcoRV. A 2-kb UAS–lz fragment digested with BamHI and HincII was then subcloned into pCTdWY digested with XbaI. Fragments TASC, TSV40 and Gy flanked by loxP sites were cloned into pCTUAS–lz–dWY between the UAS promoter and the white coding region at the XhoI restriction site.
(TASC)W and (TSV40)W
TASC and TSV40 fragments flanked by loxP sites were inserted into the pCaSPeR2 plasmid digested with HpaI.
(Gy)W(Gy)
One gypsy insulator (Gy) flanked by frt sites was inserted into the pCaSPeRΔ700 plasmid digested with EcoRI [W(Gy)]. The second gypsy insulator (Gy) flanked by loxP sites was inserted into the W(Gy) plasmid digested with XbaI.
Y(TASC)W
The Eco47III–ScaI fragment of the yellow coding region was cloned into (TASC)W digested with XbaI.
Generation and analysis of transgenic lines
The construct and P25.7wc plasmid were injected into yacw1118 preblastoderm embryos (28). The resultant flies were crossed with yacw1118 flies, and the transgenic progeny were identified by their eye color or bristle pigmentation. Chromosome localization of various transgene insertions was determined by crossing the transformants with the yacw1118 balancer stock containing dominant markers, In(2RL), CyO for chromosome 2 and In(3LR)TM3, Sb for chromosome 3.
To determine the levels of yellow and white expressions, we visually estimated the degree of pigmentation in the abdominal cuticle and wing blades (yellow) and in the eyes (white) of 3–5-day-old males developing at 25°C. For yellow, a five-grade scale was used, with grade 1 corresponding to the total loss of yellow expression and grade 5 corresponding to wild-type pigmentation. Identical data were obtained for the wing and body pigmentation in all experiments. On the nine-grade scale for white, bright red (R) and white (W) eyes corresponded to the wild type and the total loss of white expression, respectively. Intermediate levels of eye pigmentation, in the order of decreasing gene expression, were brownish red (BrR), brown (Br), dark orange (dOr), orange (Or), dark yellow (dY), yellow (Y) and pale yellow (pY). The pigmentation scores were independently determined by two investigators. These scores (every unit representing one line) were entered into the corresponding table and used to assess changes in gene expression.
The lines with DNA fragment excisions were obtained by crossing the transposon-bearing flies with the Flp (w1118; S2CyO, hsFLP, ISA/Sco;+) or Cre (yw; CyO, P[w+,cre]/Sco;+) recombinase-expressing lines (29,30). The Cre recombinase induces 100% excisions in the next generation. The high level of FLP recombinase (almost 90% efficiency) was produced by daily heat shock treatment for 2 h during the first three days after hatching. All excisions were confirmed by PCR analysis with the pairs of primers flanking the −893 insertion site (5′-atccagttgattttcagggacca-3′ and 5′-ttggcaggtgattttgagcatac-3′) relative to the yellow transcription start site and the insertion site upstream from the white gene promoter (5′-gattaacccttagcatgtccg-3′ and 5′-tttcacactttcccctgc-3′).
To induce GAL4 expression, we used the modified yw1118; P[w−, tubGAL4]117/TM3,Sb line (Bloomington Center #5138), in which the marker mini-white gene was deleted as described (31).
Construct insertion sites in transgenic lines were determined with inverse PCR technique. Genomic DNA extracted from transgenic flies was treated with RsaI or MboI endonuclease. The cleaved DNA was ligated and PCR-amplified with primers 5′-aagattcgcagtggaaggctgcac-3′ and 5′-tccgcacacaacctttcctctcaac-3′ (after RsaI cleavage) or 5′-cccttagcatgtccgtggggtttg-3′ and 5′-cgctgtctcactcagactcaatacgacac-3′ (after MboI cleavage). The PCR products were sequenced, and the coordinates and directions of insertions were determined with the Flybase R5.13 database.
RESULTS
Stimulation of the mini-white gene in transgenic lines is determined in most cases by transcription through the transposon
As shown previously, transformants carrying the mini-white gene frequently showed eye pigmentation in the range of orange to red, which indicated elevation of mini-white expression above the basal level (pale yellow to dark yellow eyes) (21,23,24). There are two explanations of the mini-white stimulation in some genomic positions. On the one hand, mini-white expression can be stimulated (or repressed) by surrounding regulatory elements such as enhancers or silencers or due to local chromatin organization. On the other hand, transposon insertion in the transcribed region can lead to an increase in the amount of mRNA products transcribed through the white coding region.
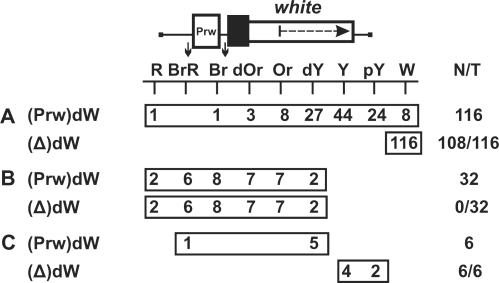
To decide between these models, we flanked the white promoter from −110 to + 276 by sites for Cre recombinase (loxP) and reinserted it into the promoterless mini-white gene [(Prw)dW, Figure 1]. For selecting transgenic lines, the marker yellow gene (responsible for cuticle and bristle pigmentation) was inserted on the 3′ side of the mini-white gene. The Wari insulator (27) located on the 3′ side of the mini-white gene was deleted from the construct. We obtained 154 independent transgenic lines carrying a single copy of the (Prw)dW construct (Figure 1). Flies in these lines had eye phenotypes ranging from pale yellow to red, which confirmed previous observations that mini-white expression is sensitive to position effects.
Figure 1.
Stimulation of the mini-white gene in transgenic lines. Levels of eye pigmentation in transgenic lines in which the deletion of the white promoter (A) resulted in the white color of the eyes, (B) had no effect on eye pigmentation, or (C) partially reduced it. The white gene is shown as a white rectangle with the arrow indicating the direction of transcription; the white square marked ‘Prw’ represents the white promoter. The black box shows the promoter deletion in mini-white. Downward arrows indicate cleavage sites for Cre recombinase; in construct names, the corresponding excisable element is parenthesized. The ‘white’ columns show the numbers of transgenic lines with the white eye pigmentation levels. N is the number of lines in which flies acquired a new w phenotype relative to the initial lines. T is the total number of lines examined for each particular construct.
Next, we deleted the white promoter by inducing recombination between the loxP sites. In 116 transgenic lines (Figure 1A), the deletion of the promoter resulted in the white color of the eyes, indicating the loss of mini-white expression. In 95 out of these 116 transgenic lines, the eye color in flies ranged from pale yellow to dark yellow, corresponding to the basal level of the mini-white expression driven by the promoter alone. In 11% (13 out of 116) of the transgenic lines, flies had darker eye pigmentation, which could be indicative of mini-white activation by a regulatory element located outside the transposon.
In 38 out of 154 transgenic lines, the deletion of the white promoter either had no effect on mini-white expression (32 lines, Figure 1B) or partially reduced it (six lines, Figure 1C). In 30 out of 32 transgenic lines in which the pigmentation level remained unchanged after the deletion of the promoter, the eye color in flies was in the range from orange to red, indicating that mini-white expression was above the basal level.
To gain an insight into the nature of mini-white activation at different genomic positions, we determined its chromosomal insertion sites in 23 transgenic lines. Table 1 shows positions of the mini-white gene relative to neighboring genes in these lines.
Table 1.
Sites of (Prw)dW construct insertion in transgenic lines
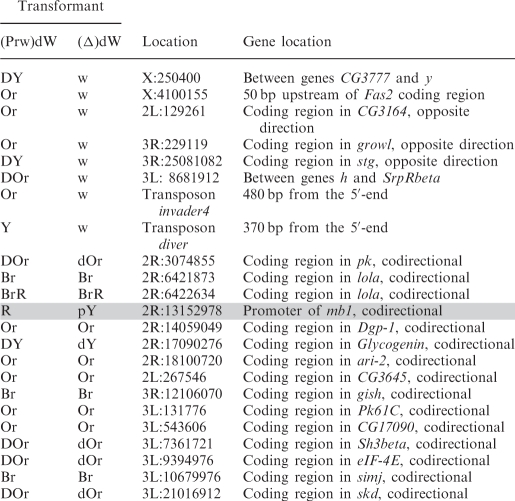 |
In eight transgenic lines in which flies acquired white eyes after the deletion of the promoter, the transposon was inserted either into intergenic regions (five lines) or into genes oriented opposite to the direction of transcription (three lines).
In contrast, all transgenic lines displaying promoter-independent mini-white expression were generated by insertions into genes whose transcription direction coincided with that of the mini-white gene. According to available data (NCBI GEO), some of these genes are specifically active in the eye imaginal discs or are expressed throughout development in all tissues. Thus, mini-white expression could be due to transcription driven by the promoter of a tagged gene. Since the deletion of the white promoter did not alter mini-white expression (except for the line marked grey in Table 1), it seems likely that transcription through the mini-white gene resulted in inactivation of the white promoter, as was previously shown for Drosophila genes Ubx, Abd-A (32,33) and dihydrofolate reductase (34).
Taken together, these results suggest that only in 13 out of 154 (8%) transgenic lines could the high level of mini-white expression be due to stimulation of the white promoter by neighboring enhancers or transcriptionally active chromatin.
The enhancerless yellow gene is rarely activated by surrounding regulatory elements
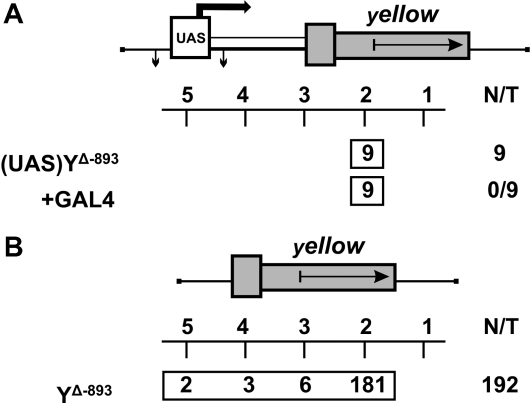
We extended our study to one more tissue specific gene, yellow, that is required for larval and adult cuticle pigmentation. The spatiotemporal pattern of its expression is controlled by at least five independent, tissue-specific transcriptional enhancers (35,36). As shown previously, the intensity of cuticle pigmentation correlates with the level of yellow gene expression (37). At first, we tested whether transcription from upstream promoters can stimulate yellow expression, as in the case of the mini-white gene. The UAS promoter flanked by lox sites was cloned at –893 relative to the yellow transcription start sites (Figure 2A). The yellow wing and body enhancers were deleted from the construct. We obtained nine independent transgenic lines, each carrying a single copy of the construct. To express the GAL4 protein, we used the transgenic line carrying the GAL4 gene under control of the ubiquitous tubulin promoter (tubGAL4). In transgenic lines carrying the enhancerless yellow gene fused with GAL4-binding sites, tubGAL4 stimulates yellow transcription in all cuticle structures (data not shown). At the same time, induction of the UAS promoter by GAL4 expression did not change wing and body pigmentation in transgenic flies, indicating that transcripts generated from the upstream promoter failed to produce the functional Yellow protein. Thus, in contrast to the situation with the mini-white gene, an increase in yellow expression could be attributed only to stimulation of the promoter by regulatory elements located near the site of the yellow transgene insertion.
Figure 2.
Stimulation of the enhancerless yellow gene in transgenic lines. (A) Testing the ability of the UAS promoter located at –893 to produce the functional Yellow protein. The UAS promoter is represented by the white rectangle marked ‘UAS’. The ‘yellow’ columns show the numbers of transgenic lines with different levels of body and wing pigmentation. ‘+GAL4’ indicates that yellow phenotypes in transgenic lines were examined after induction of GAL4 expression. (B) Testing the frequency of yellow stimulation in random genomic positions. For other designations, see Figure 1.
Next, we used the construct containing the yellow sequences from −893 to +5204, including those with the bristle enhancer located in the intron. On the whole, we obtained 192 transgenic lines, each carrying a single insertion. Among them, only 11 lines showed a weak (six lines) or strong (five lines) increase in yellow expression (Figure 2B). Thus, in only a minor part of the transgenic lines was the yellow promoter activated by an enhancer or chromatin located outside the construct. These results confirm our main conclusion that enhancer–promoter interactions are specific and that incorrect stimulation of a promoter by a wrong enhancer is a relatively rare event.
Transcriptional terminators can protect mini-white expression from activating chromosomal position effects
If the high level of the mini-white expression in most genomic positions is accounted for by transcription through the gene, transcriptional terminators should function like boundaries. To test this assumption, we chose the well-studied SV40 terminator (designated TSV40) and the terminator named TASC that was previously identified in the regulatory region of the Achaete–Scute gene complex (O.Maksimenko and P.Georgiev, unpublished data).
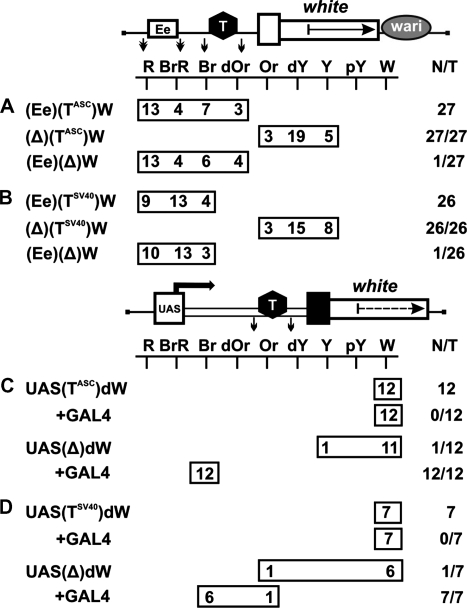
To verify their potential effect on the interaction between the eye enhancer and the white promoter, we made constructs in which these terminators flanked by lox sites were inserted between the eye enhancer flanked by frt sites and the white promoter (Figure 3A and B). As in common enhancer-blocking assay with the mini-white gene, the Wari insulator (27) was left intact in these constructs. The flies of all resultant transgenic lines displayed strong eye pigmentation that remained unchanged upon deletion of the terminators but decreased considerably upon deletion of the eye enhancer. Therefore, the terminators failed to influence enhancer–promoter communication.
Figure 3.
Functional activity of selected terminators in the eyes. Testing the enhancer-blocking activity (A) of the terminator found in the regulatory region of the Achaete–Scute gene complex (TASC) and (B) of the SV40 terminator (TSV40). A terminator (T) is shown as a black hexagon; in construct names, terminators are indicated in superscript. The eye enhancer is represented by the rectangle marked ‘Ee’. The Wari insulator is shown as a gray oval marked ‘wari’. Downward arrows indicate cleavage sites for Cre or FLP recombinase. Testing the activity of (C) TASC and (D) TSV40 terminators in the eyes. ‘+GAL4’ indicates that eye phenotypes in transgenic lines were examined after induction of GAL4 expression. In this case, N is the number of lines in which flies acquired a new w phenotype upon induction of GAL4. For other designations, see Figures 1 and 2.
To test the above terminators for the ability to arrest transcription elongation in the eyes, we used a model system that contained the UAS promoter, 2-kb spacer from the lacZ gene, and the promoterless mini-white gene with deleted Wari insulator (Figure 3C and D). The terminators flanked by lox sites were inserted into the spacer. The flies of all resultant transgenic lines had white eyes, with eye pigmentation remaining unchanged upon induction of the UAS promoter by crossing with the tubGAL4 line. In derivative transgenic lines obtained by deleting the terminators, induction of the UAS promoter by GAL4 expression resulted in the brown eye color of transgenic flies, which was indicative of strong mini-white activation. These results confirm that the mini-white gene is effectively translated when containing an additional sequence of at least 2 kb at the 5′-end and that the terminators are able to effectively terminate transcription in the eyes in all transgenic lines tested.
Next, we examined whether the terminators can function as boundaries and effectively protect mini-white expression from the positive effects of surrounding sequences. The terminators flanked by loxP sites were inserted in front of the mini-white gene (Figure 4A and B). These constructs contained the Wari insulator (27) on the 3′ side of the mini-white gene.
Figure 4.
Testing (A) TASC and (B) TSV40 terminators for the ability to protect mini-white expression from positive position effects. (C) Testing TASC for the negative influence on mini-white expression. The yellow gene is shown as a gray rectangle with the arrow indicating the direction of transcription. (D) Testing TASC for the ability to terminate endogenous transcription at different genomic sites. For other designations, see Figures 1–3.
As a result, we obtained 59 lines carrying TASC (Figure 4A) and 22 lines carrying TSV40 (Figure 4B). In 75 transgenic lines, flies had eye color ranging from pale yellow to dark yellow, which corresponded to the basal level of mini-white expression. A stronger eye pigmentation was observed in only six transgenic lines flies, confirming our previous observation that activation of the white promoter by a neighboring enhancer or/and chromatin is a rare event. Deletion of the terminators resulted in a higher level of eye pigmentation in 32 out of 81 transgenic lines, providing evidence that the mini-white gene is often stimulated by transcription initiated upstream of this gene.
To test TASC for the negative influence on mini-white expression, we made the construct Y(TASC)W in which the yellow gene with the regulatory region was inserted upstream of the mini-white gene in (TASC)W (Figure 4C). In such a transgene, the yellow gene functioned as a buffer protecting the mini-white gene from the position effects of sequences located upstream of the transposon insertion site. In 16 transgenic lines, the eye color of flies ranged from yellow to dark yellow, with pigmentation level remaining unchanged upon the deletion of TASC. Thus, it appears unlikely that TASC directly affects the mini-white expression.
To obtain additional evidence that TASC can terminate endogenous transcription, thereby protecting mini-white expression, we used the (TASC)dW construct containing the promoterless white gene (Figure 4D). The flies of 68 resultant transgenic lines had white eyes; after deletion of TASC, the flies of 14 lines acquired the eye color ranging from pale yellow to orange. Thus, TASC could effectively terminate endogenous transcription at different genomic sites.
Taken together, these results show that most of position effects on mini-white expression are caused by transcription through the transgene. Therefore, any regulatory element capable of terminating transcription can protect mini-white expression, acting like a boundary.
Flanking the mini-white gene by gypsy insulators only partially protects it from position effects
The best studied Drosophila insulator was found in the regulatory region of the gypsy retrotransposon (38,39). The authors of previous studies (14,15) observed that the gypsy insulator completely insulated mini-white expression. However, they performed experiments with the yellow gene inserted in the opposite orientation upstream of the mini-white gene, and its presence could strongly reduce the ability of transcripts to pass through the transgene to produce the functional White protein. Accordingly, we did not observe a high level of mini-white expression in any of Y(Δ)W transgenic lines carrying the construct in which the yellow gene was inserted upstream of the mini-white gene (Figure 4C). For this reason, we tested whether the gypsy insulator could protect mini-white expression from position effects.
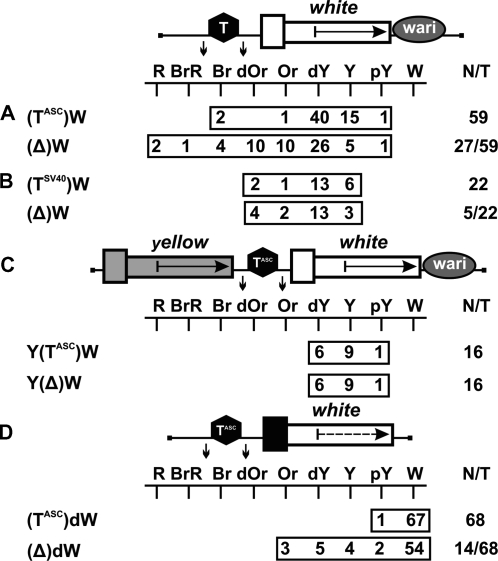
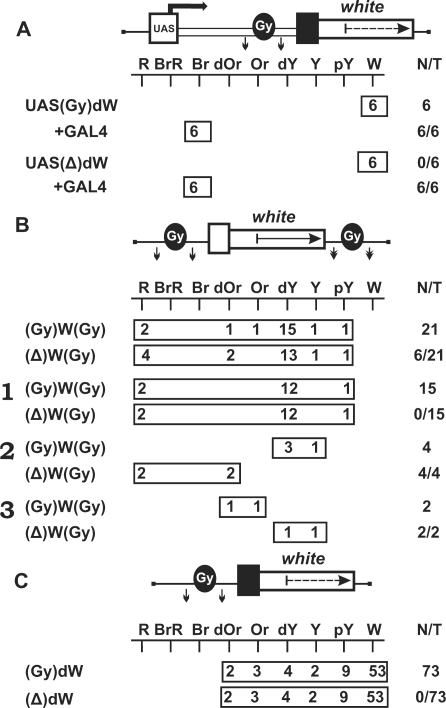
At first, we examined whether the gypsy insulator could terminate transcription in the eyes. The gypsy insulator flanked by lox sites was inserted in the direct orientation, as in the gypsy retrotransposon, in the spacer between the UAS promoter and the promoterless mini-white gene (Figure 5A). In all transgenic lines tested, flies had brown eyes after induction of the UAS promoter by GAL4 expression. Deletion of the insulator had no influence on eye pigmentation, suggesting that this insulator failed to terminate transcription in the eyes, despite the presence of AATAAA repeats in its sequence. The gypsy insulator in all subsequent constructs was inserted in the direct orientation.
Figure 5.
(A) Testing the gypsy insulator (Gy, black oval) for the ability to terminate transcription in the eyes. (B) Testing the gypsy insulator for the ability to protect mini-white expression from positive position effects. Numbers on the left (1–3) indicate groups of transgenic lines in which eye pigmentation (1) remained unchanged, (2) increased, or (3) decreased after the deletion of the insulator. (C) Testing the gypsy insulator for the ability to function as a transcriptional terminator in different genomic positions. For other designations, see Figures 1 and 2.
Next, we made the construct in which the mini-white gene was inserted between the loxP- and frt-flanked gypsy insulators (Figure 5B). In this construct, the Wari insulator (27) was deleted from the mini-white gene. We obtained a total of 21 independent lines, each containing a single copy of the construct in which the mini-white gene was flanked by the gypsy insulators. The eye color phenotypes of these transgenic lines varied from pale yellow to red, which contradicted the results obtained by Roseman et al. (14). Thus, the presence of the yellow gene proved to shield the mini-white gene from positive position effects. After the deletion of the gypsy insulator located upstream of the mini-white gene, the eye color phenotypes remained unchanged in 15 transgenic lines [Figure 5B (1)], with eye pigmentation increasing [Figure 5B (2)] or decreasing [Figure 5B (3)] in four and two transgenic lines, respectively.
We determined chromosomal insertion sites for the four transgenic lines in which flies had a red eye color after deletion of the gypsy insulator. The results showed that the mini-white gene was colinearly inserted in the coding regions of the CrebB-17A, mRpS9, Doa and ftz-f1 genes, which are strongly expressed in the eye imaginal discs (NCBI GEO) (Table 2). In two lines carrying insertions in the Doa and ftz-f1 genes, the white expression was considerably reduced in the presence of the gypsy insulator. In both lines, we found potential polyA sites immediately upstream of the insertion sites. In the other two lines in which gypsy had no influence on the white expression, we did not find any signal for transcription termination in the vicinity of the transposon insertion sites. As shown previously, the gypsy insulator can potentiate weak polyadenylation signals but fails to terminate transcription by itself (39–42). Thus, it appears that the gypsy insulator potentiated termination at weak polyA sites in the Doa and ftz-f1 genes but did not affect transcription in the CrebB-17A and mRpS9 genes.
Table 2.
Sites of (Gy)W(Gy) construct insertion in transgenic lines
| Transformant |
|||
|---|---|---|---|
| (Gy)W(Gy) | (Δ)W(Gy) | Location | Gene location |
| R | R | X:18265579 | Coding region in CrebB-17A, codirectional |
| R | R | 3R:3718330 | Coding region in mRpS9, codirectional |
| Y | R | 3R:18759108 | Coding region in ftz-f1, codirectional |
| dY | R | 3R:24714644 | Coding region in Doa, codirectional |
| dY | dOr | 2L:12507930 | Coding region in bun, opposite direction |
| dY | dOr | 3R:466875 | Coding region in CG9775, opposite direction |
| Or | Y | 2R:5936940 | Between CG18445 and CG2249 |
| dOr | dY | 3L:637935 | Between Reg-2 and ban |
The chromosomal insertion sites were also identified for transgenic lines in which eye pigmentation increased from dark yellow to dark orange after the deletion of the gypsy insulator (Figure 5B). In these cases, the transposon was mapped in the coding regions of genes oriented opposite to the transcription direction, suggesting that the gypsy insulator in these transgenic lines could block communication between a nearby enhancer and the white promoter.
In two transgenic lines, the deletion of the gypsy insulator resulted in decreasing mini-white expression. Transposon insertions in these lines were mapped to intergenic regions, suggesting that, in all likelihood, the above phenomenon is explained by the boundary activity of the gypsy insulator, which protected mini-white expression from the repressive chromatin structures.
To further test the ability of the gypsy insulator to function as a transcriptional terminator in different genomic positions, we inserted the lox-flanked gypsy insulator in front of the promoterless mini-white gene lacking the endogenous Wari insulator (Figure 5C). In 20 out of 73 transgenic lines carrying single copies of the construct, flies had eye pigmentation in the range from pale yellow to orange. The deletion of the gypsy insulator had no effect on eye pigmentation in any of the transgenic lines examined. These results confirm that, in most cases, the gypsy insulator fails to terminate transcription.
DISCUSSION
We have shown here that, in most genomic positions of the mini-white transgene, positive position effects are caused by transcription through the gene. It seems likely that the mini-white gene has an internal site for translation initiation, which allows the functional White protein to be produced from transcripts initiated from promoters located upstream of the white coding region.
The possibility that a heterologous enhancer is able to stimulate mini-white expression has been observed (but not proved) in only 8% of transgenic lines. Infrequent white stimulation by surrounding chromatin may be explained by inactivation of the white promoter in most of genomic sites due to transcription through the gene. In such a case, the ability of enhancers to stimulate the white promoter does not manifest itself in many transcriptionally active regions. However, stimulation of the white gene shielded by a transcription terminator still proved to be a relatively rare event: eye pigmentation was within an orange–brown range in only six out of 81 transgenic lines (7%). On the other hand, deletion of the terminators resulted in stimulation of white expression in 40% of transgenic lines. Thus, white expression in many transcriptionally active regions was not stimulated by surrounding chromatin or regulatory elements.
The results of our experiments with another tissue-specific gene, yellow, are similar: it was activated by surrounding regulatory elements in only ∼6% of transgenic lines. The yellow promoter is insensitive to transcription going through the gene (D. Chetverina, unpublished data). However, in contrast to the situation with white, the transcripts started upstream from the yellow promoter fail to produce the functional Yellow protein. Thus, we could not estimate the percentage of transcriptionally active regions in which the yellow promoter is insensitive to surrounding chromatin.
Taken together, these results indicate that tissue-specific promoters are infrequently activated by surrounding enhancers in genomic context at pupal–adult stages of Drosophila development. In embryos, conversely, extremely diverse, position-dependent expression patterns observed in various ‘enhancer trap’ experiments suggest that different endogenous enhancers can unspecifically activate a weak promoter located in the transposon (43–46). A probable explanation to these conflicting data is that specificity of enhancer–promoter interactions increases in the course of Drosophila development.
A major putative function of insulators is to protect integrated reporter genes from positive or negative effects of the surrounding chromatin (9–11). The best-characterized boundary elements such as Drosophila scs insulator (16) and vertebrate HS4 insulator (18) were initially identified by their ability to protect the white gene from positive effects of the surrounding chromatin, with the resultant eye pigmentation being consistently lighter. Different regions of the SF1 insulator were shown to be required for enhancer blocking in embryos and for shielding the mini-white gene from chromosomal position effects (47). MAR elements were also shown to protect white expression from the positive position effects (19,20).
Here, we have shown that transcriptional terminators can effectively shield mini-white expression from position effects. Thus, A/T-rich MAR elements, 1.8-kb scs, 1.2-kb HS4 and SF1 insulators appear to protect mini-white expression by terminating transcription through the transgene. In contrast, the gypsy insulator only partially protects mini-white expression from position effects. We cannot exclude, however, that the mini-white transgene flanked by the combination of gypsy and Wari insulators is better protected from position effects, as was shown in previous studies (14,15). The gypsy insulator is effective in blocking enhancer stimulation (39,48–51), PRE-mediated silencing (51,52) and heterochromatin repression (14,15). At the same time, the gypsy insulator fails to shield the transgene expression from the effects of transcription initiated in the vicinity of the transgene insertion sites.
In conclusion, our results show that the position effects generated by transcription through a transgene are most frequent and that transcriptional terminators, compared to the classical gypsy insulator, provide better protection from these effects.
FUNDING
Ministry of Science and Education of the Russian Federation (project no. 02.512.11.2252), the Molecular and Cell Biology Program of the Russian Academy of Sciences (to P.G.), the Russian Foundation for Basic Research (projects no. 09-04-01232 and 09-04-13880), MD-468.2009.4 (to O.M.). Funding for open access charge: Howard Hughes Medical Institute (to P.G.).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors are grateful to D. Chetverina and O. Zaytseva for their help with some experiments and sharing unpublished results and to N.A. Gorgolyuk for his help in preparing the manuscript.
REFERENCES
- 1.Ptashne M. Gene regulation by proteins acting nearby and at a distance. Nature. 1986;322:697–701. doi: 10.1038/322697a0. [DOI] [PubMed] [Google Scholar]
- 2.Dorsett D. Distant liaisons: Long-range enhancer−promoter interactions in Drosophila. Curr. Opin. Genet. Dev. 1999;9:505–514. doi: 10.1016/s0959-437x(99)00002-7. [DOI] [PubMed] [Google Scholar]
- 3.de Laat W, Grosveld F. Spatial organization of gene expression: The active chromatin hub. Chromosome Res. 2003;11:447–459. doi: 10.1023/a:1024922626726. [DOI] [PubMed] [Google Scholar]
- 4.West AG, Fraser P. Remote control of gene transcription. Hum. Mol. Genet. 2005;14:101–111. doi: 10.1093/hmg/ddi104. [DOI] [PubMed] [Google Scholar]
- 5.Fraser P. Transcriptional control thrown for a loop. Curr. Opin. Genet. Dev. 2006;16:490–495. doi: 10.1016/j.gde.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 6.Palstra RJ, de Laat W, Grosveld F. Beta-globin regulation and long-range interactions. Adv. Genet. 2008;61:107–142. doi: 10.1016/S0065-2660(07)00004-1. [DOI] [PubMed] [Google Scholar]
- 7.Udvardy A. Dividing the empire: Boundary chromatin elements delimit the territory of enhancers. EMBO J. 1999;18:1–8. doi: 10.1093/emboj/18.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuhn EJ, Geyer PK. Genomic insulators: connecting properties to mechanism. Curr. Opin. Cell Biol. 2003;15:259–265. doi: 10.1016/s0955-0674(03)00039-5. [DOI] [PubMed] [Google Scholar]
- 9.Capelson M, Corces VG. Boundary elements and nuclear organization. Biol. Cell. 2004;96:617–629. doi: 10.1016/j.biolcel.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 10.Brasset E, Vaury C. Insulators are fundamental components of the eukaryotic genomes. Heredity. 2005;94:571–576. doi: 10.1038/sj.hdy.6800669. [DOI] [PubMed] [Google Scholar]
- 11.Gaszner M, Felsenfeld G. Insulators: exploiting transcriptional and epigenetic mechanisms. Nature Rev. Genet. 2006;7:703–713. doi: 10.1038/nrg1925. [DOI] [PubMed] [Google Scholar]
- 12.Valenzuela L, Kamakaka RT. Chromatin insulators. Annu. Rev. Genet. 2006;40:107–138. doi: 10.1146/annurev.genet.39.073003.113546. [DOI] [PubMed] [Google Scholar]
- 13.Wallace JA, Felsenfeld G. We gather together: Insulators and genome organization. Curr. Opin. Genet. Dev. 2007;17:400–407. doi: 10.1016/j.gde.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roseman RR, Pirrotta V, Geyer PK. The su(Hw) protein insulates expression of the Drosophila melanogaster white gene from chromosomal position-effects. EMBO J. 1993;12:435–442. doi: 10.1002/j.1460-2075.1993.tb05675.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roseman RR, Swan JM, Geyer PK. A Drosophila insulator protein facilitates dosage compensation of the X chromosome mini-white gene located at autosomal insertion sites. Development. 1995;121:3573–3582. doi: 10.1242/dev.121.11.3573. [DOI] [PubMed] [Google Scholar]
- 16.Kellum R, Schedl P. A position-effect assay for boundaries of higher order chromosomal domains. Cell. 1991;64:941–950. doi: 10.1016/0092-8674(91)90318-s. [DOI] [PubMed] [Google Scholar]
- 17.Brasset E, Bantignies F, Court F, Cheresiz S, Conte C, Vaury C. Idefix insulator activity can be modulated by nearby regulatory elements. Nucleic Acids Res. 2007;35:2661–2670. doi: 10.1093/nar/gkm140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chung JH, Whiteley M, Felsenfeld G. A 5′ element of the chicken beta-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell. 1993;74:505–514. doi: 10.1016/0092-8674(93)80052-g. [DOI] [PubMed] [Google Scholar]
- 19.Namciu SJ, Blochlinger KB, Fournier REK. Human matrix attachment regions insulate transgene expression from chromosomal position effects in Drosophila melanogaster. Mol. Cell Biol. 1998;18:2382–2391. doi: 10.1128/mcb.18.4.2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Namciu SJ, Fournier REK. Human matrix attachment regions are necessary for the establishment but not the maintenance of transgene insulation in Drosophila melanogaster. Mol. Cell Biol. 2004;24:10236–10245. doi: 10.1128/MCB.24.23.10236-10245.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hazelrigg T, Levis R, Rubin GM. Transformation of white locus DNA in Drosophila: Dosage compensation, zeste interaction, and position effects. Cell. 1984;36:469–481. doi: 10.1016/0092-8674(84)90240-x. [DOI] [PubMed] [Google Scholar]
- 22.Levis R, O'H;are K, Rubin GM. Effects of transposable element insertions on RNA encoded by the white gene of Drosophila. Cell. 1984;38:471–481. doi: 10.1016/0092-8674(84)90502-6. [DOI] [PubMed] [Google Scholar]
- 23.Levis R, Hazelrigg T, Rubin GM. Separable cis-acting control elements for expression of the white gene of Drosophila. EMBO J. 1985;4:3489–3499. doi: 10.1002/j.1460-2075.1985.tb04108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pirrotta V, Steller H, Bozzetti M. Multiple upstream regulatory elements control the expression of the Drosophila white gene. EMBO J. 1985;4:3501–3508. doi: 10.1002/j.1460-2075.1985.tb04109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qian S, Varjavand B, Pirrotta V. Molecular analysis of the zeste–white interaction reveals a promoter-proximal element essential for distant enhancer–promoter communication. Genetics. 1992;131:79–90. doi: 10.1093/genetics/131.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pirrotta V. Vectors for P-mediated transformation in Drosophila. Biotechnology. 1988;10:437–456. doi: 10.1016/b978-0-409-90042-2.50028-3. [DOI] [PubMed] [Google Scholar]
- 27.Chetverina D, Savitskaya E, Maksimenko O, Melnikova L, Zaytseva O, Parshikov A, Galkin AV, Georgiev P. Red flag on the white reporter: a versatile insulator abuts the white gene in Drosophila and is omnipresent in mini-white constructs. Nucleic Acids Res. 2008;36:929–937. doi: 10.1093/nar/gkm992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karess RE, Rubin GM. Analysis of P transposable element functions in Drosophila. Cell. 1984;38:135–146. doi: 10.1016/0092-8674(84)90534-8. [DOI] [PubMed] [Google Scholar]
- 29.Golic KG, Lindquist S. The FLP recombinase of yeast catalyzes site-specific recombination in the Drosophila genome. Cell. 1989;59:499–509. doi: 10.1016/0092-8674(89)90033-0. [DOI] [PubMed] [Google Scholar]
- 30.Siegal ML, Hartl DL. Application of Cre/loxP in Drosophila. Site-specific recombination and transgene co-placement. Methods Mol. Biol. 2000;136:487–495. doi: 10.1385/1-59259-065-9:487. [DOI] [PubMed] [Google Scholar]
- 31.Kyrchanova O, Toshchakov S, Parshikov A, Georgiev P. Study of the functional interaction between Mcp insulators from the Drosophila bithorax complex: Effects of insulator pairing on enhancer–promoter communication. Mol. Cell Biol. 2007;27:3035–3043. doi: 10.1128/MCB.02203-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petruk S, Sedkov Y, Riley KM, Hodgson J, Schweisguth F, Hirose S, Jaynes JB, Brock HW, Mazo A. Transcriptional elongation of non-coding bxd RNAs promoted by the Trithorax TAC1 complex represses Ubx by a transcriptional interference mechanism. Cell. 2006;127:1209–1221. doi: 10.1016/j.cell.2006.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Petruk S, Sedkov Y, Brock HW, Mazo A. A model for initiation of mosaic HOX gene expression patterns by non-coding RNAs in early embryos. RNA Biol. 2007;4:1–6. doi: 10.4161/rna.4.1.4300. [DOI] [PubMed] [Google Scholar]
- 34.Martianov I, Ramadass A, Barros AS, Chow N, Akoulitchev A. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature. 2007;445:666–670. doi: 10.1038/nature05519. [DOI] [PubMed] [Google Scholar]
- 35.Geyer PK, Corces VG. Separate regulatory elements are responsible for the complex pattern of tissue-specific and developmental transcription of the yellow locus in Drosophila melanogaster. Genes Dev. 1987;1:996–1004. doi: 10.1101/gad.1.9.996. [DOI] [PubMed] [Google Scholar]
- 36.Martin M, Meng YB, Chia W. Regulatory elements involved in the tissue-specific expression of the yellow gene of Drosophila. Mol. Gen. Genet. 1989;218:118–126. doi: 10.1007/BF00330574. [DOI] [PubMed] [Google Scholar]
- 37.Moris JR, Petrov DA, Lee AM, Wu CT. Enhancer choice in cis and in trans in Drosophila melanogaster: role of the promoter. Genetics. 2004;167:1739–1747. doi: 10.1534/genetics.104.026955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Holdridge C, Dorsett D. Repression of hsp70 heat shock gene transcription by the suppressor of hairy-wing protein of Drosophila melanogaster. Mol. Cell Biol. 1991;11:1894–1900. doi: 10.1128/mcb.11.4.1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Geyer PK, Corces VG. DNA position-specific repression of transcription by a Drosophila zinc finger protein. Genes Dev. 1992;6:1865–1873. doi: 10.1101/gad.6.10.1865. [DOI] [PubMed] [Google Scholar]
- 40.Dorsett D, Viglianti GA, Rutledge BJ, Meselson M. Alteration of hsp82 gene expression by the gypsy transposon and suppressor genes in Drosophila melanogaster. Genes Dev. 1989;3:454–468. doi: 10.1101/gad.3.4.454. [DOI] [PubMed] [Google Scholar]
- 41.Dorsett D. Potentiation of a polyadenylation site by a downstream protein–DNA interaction. Proc. Natl Acad. Sci. USA. 1990;87:4373–4377. doi: 10.1073/pnas.87.11.4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lankenau DH, Corces VG, Engels WR. Comparison of targeted-gene replacement frequencies in Drosophila melanogaster at the forked and white loci. Mol. Cell Biol. 1996;16:3535–3544. doi: 10.1128/mcb.16.7.3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bellen HJ, O’Kane CJ, Wilson C, Grossniklaus U, Pearson RK, Gehring WJ. P-element mediated enhancer detection: a versatile method to study development in Drosophila. Genes Dev. 1989;3:1288–1300. doi: 10.1101/gad.3.9.1288. [DOI] [PubMed] [Google Scholar]
- 44.Bier E, Vaessin H, Shepherd S, Lee K, McCall K, Barbel S, Ackerman L, Carretto R, Uemura T, Grell E, et al. Searching for pattern and mutation in the Drosophila genome with a P-lacZ vector. Genes Dev. 1989;3:1273–1287. doi: 10.1101/gad.3.9.1273. [DOI] [PubMed] [Google Scholar]
- 45.Butler JEF, Kadonaga JT. Enhancer-promoter specificity mediated by DPE or TATA core promoter motifs. Genes Dev. 2001;15:2515–2519. doi: 10.1101/gad.924301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.O’Kane CJ, Gehring WJ. Detection in situ of genomic regulatory elements in Drosophila. Proc. Natl Acad. Sci. USA. 1987;84:9123–9127. doi: 10.1073/pnas.84.24.9123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Majumder P, Roy S, Belozerov VE, Bosu D, Puppali M, Cai HN. Diverse transcription influences can be insulated by the Drosophila SF1 chromatin boundary. Nucleic Acids Res. 2009;37:4227–4233. doi: 10.1093/nar/gkp362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cai H, Levine M. Modulation of enhancer−promoter interactions by insulators in the Drosophila embryo. Nature. 1995;376:533–536. doi: 10.1038/376533a0. [DOI] [PubMed] [Google Scholar]
- 49.Cai HN, Levine M. The gypsy insulator can function as a promoter-specific silencer in the Drosophila embryo. EMBO J. 1997;16:1732–1741. doi: 10.1093/emboj/16.7.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scott KS, Geyer PK. Effects of the su(Hw) insulator protein on the expression of the divergently transcribed Drosophila yolk protein genes. EMBO J. 1995;14:6258–6267. doi: 10.1002/j.1460-2075.1995.tb00316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scott KS, Taubman AD, Geyer PK. Enhancer blocking by the Drosophila gypsy insulator depends upon insulator anatomy and enhancer strength. Genetics. 1999;153:787–798. doi: 10.1093/genetics/153.2.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sigrist CJ, Pirrotta V. Chromatin insulator elements block the silencing of a target gene by the Drosophila polycomb response element (PRE) but allow trans interactions between PREs on different chromosomes. Genetics. 1997;147:209–221. doi: 10.1093/genetics/147.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mallin DR, Myung JS, Patton JS, Geyer PK. Polycomb group repression is blocked by the Drosophila suppressor of Hairy-wing [su(Hw)] insulator. Genetics. 1998;148:331–339. doi: 10.1093/genetics/148.1.331. [DOI] [PMC free article] [PubMed] [Google Scholar]