Abstract
Large numbers of guanine-rich sequences with potential to form G-quadruplexes have been identified in genomes of various organisms. Such sequences are constrained at both ends by long DNA duplex with a complementary strand in close proximity to compete for duplex formation. G-quadruplex/duplex competition in long double-stranded DNA has rarely been studied. In this work, we used DMS footprinting and gel electrophoresis to study G-quadruplex formation in long double-stranded DNA derived from human genome under both dilute and molecular crowding condition created by PEG. G-quadruplex formation was observed in the process of RNA transcription and after heat denaturation/renaturation under molecular crowding condition. Our results showed that the heat denaturation/renaturation treatment followed by gel electrophoresis could provide a simple method to quantitatively access the ability of G-quadruplex formation in long double-stranded DNA. The effect of K+ and PEG concentration was investigated and we found that stable G-quadruplexes could only form under the crowding condition with PEG at concentrations near the physiological concentration of biomass in living cells. This observation reveals a physical basis for the formation of stable G-quadruplexes in genome and supports its presence under the in vivo molecular crowding condition.
INTRODUCTION
Nucleic acids with multiple runs of guanine-rich (G-rich) motifs can fold back to form a four-stranded intramolecular G-quadruplex structure in the presence of mono mental ions, for instance, K+ or Na+ (1,2). Such G-quadruplex sequences have been found to present in many essential regions of human genome, such as telomeres (3), promoter of oncogenes (4), immunoglobulin switch (5) and insulin regulatory (6) regions. The prevalence of such sequences is demonstrated by many recent bioinformatic analyses that have revealed up to several hundred thousands of putative quadruplex sequences in the genome of human and other species (7–15). Although direct evidence is still lacking, the existence of G-quadruplex structure in genome is supported by several observations. G-quadruplex formation in plasmid has been suggested to occur during the intracellular transcription of its G-rich region (16). Mutation in the G-rich region upstream of the C-MYC promoter to reduce G-quadruplex formation resulted in increase in basal transcriptional activity of the promoter (17). In contrast, stabilization of G-quadruplexes by chemical compounds has been shown to downregulate the promoter activities of the C-MYC (17) and KRAS (18) gene in model plasmids and the expression of the native C-MYC gene (19). The presence of proteins that interact with G-quadruplexes suggest that such structures are functional elements in biological processes (20). It is now believed that G-quadruplexes play important role in regulating gene expression and thus constitute valuable therapeutic targets against cancer and other diseases (21–28).
In genomic DNA, all the quadruplex forming sequences, except for the telomere DNA, are located at internal positions of long double-stranded DNA (dsDNA). They are constrained at both ends by long DNA duplex with a complementary strand in close proximity to compete for duplex formation. So far, studies on G-quadruplex/duplex competition have been carried out almost exclusively using the core sequences of both the G- and C-rich strand (29–35). In these situations, the two reactants are all free molecules and the G-quadruplex/duplex competition strongly depends on their concentration because of the intermolecular nature of the duplex formation. In addition, the presence of flanking duplex in genome should reinforce the formation of duplex. Therefore it is difficult to obtain information from separate core sequences that can truly reflect the G-quadruplex/duplex competition in genomic DNA.
To better understand the quadruplex/duplex competition in genomic DNA, we prepared long dsDNA from human genome carrying G-quadruplex-forming sequences with flanking duplex at both sides and studied G-quadruplex formation under both dilute and molecular crowding conditions during the process of in vitro transcription and heat denaturation/renaturation. Our data revealed that molecular crowding creates an essential environment for stable G-quadruplex to form in dsDNA. We also explored how G-quadruplex formation was affected by the concentration of K+ and PEG. While K+ only affected the competition at millimolar level that is far below the physiological concentration of this cation, lasting G-quadruplex could form at PEG concentrations near the physiological concentration of biomass in living cells. This fact strongly suggests that stable G-quadruplex can form and coexist with duplex in genome inside cells under the in vivo crowding condition. Our results also show that gel electrophoresis can provide a useful tool to quantify G-quadruplex formation in long dsDNA.
MATERIALS AND METHODS
Preparation of dsDNA
Genomic DNA was isolated from HeLa cells as described (36). dsDNA (dsDNA) carrying a T7 promoter was prepared by overlap extension polymerase chain reaction (OE-PCR) (Table 1). Other dsDNA was prepared by direct amplification of specific region of genomic DNA containing G-quadruplex sequence using polymerase chain reaction (PCR). Primers (Table 2) for PCR were purchased from Invitrogen (Shanghai, China). For some experiments, one primer was labeled at the 5′-end with a fluorescein (FAM) for quantitation. PCR was conducted in a total volume of 50 µl containing 75 mM Tris–HCl, pH 8.8, 20 mM (NH4)2SO4, 0.01% Tween-20, 250 µM dNTP, 0.4 µM primer, 1.5 mM MgCl2, 5% (v/v) DMSO, 2 U Taq Polymerase (Fermantas, MBI) and 170 ng genomic DNA as template. Thermal cycling was carried out on a Biometro thermal cycler with initial denaturation at 94°C for 3 minutes, followed by addition of Taq Polymerase and subsequent 30 cycles of 94°C for 30 s, 58°C for 30 s and 72°C for 30 s. PCR product was purified using the TIANgel Midi Purification Kit (Tiangen, China).
Table 1.
Sequence of dsDNAs used in Figures 1–4
| Gene | Sequencea |
|---|---|
| C-MYC | 5′-GGCTTCGGAGTCCCCTGCACTATGACTCCTGA CAATtaatacgctcactataggGTGGGGAGGGTGGGGAA GAGCTATGATGCGTTCGATCACTCCATGTGAT CCTACACTCGCCGCAGGCTGG-3′ |
| NRAS | 5′-GGCTTCGGAGTCCCCTGCACTATGACTCCTGA CAATtaatacgactcactataggGAGGGGCGGGTCTGGG AAGAGCTATGATGCGTTCGATCACTCCATGTG ATCCTACACTCGCCGCAGGCTGG-3′ |
| Control | 5′-GGCTTCGGAGTCCCCTGCACTATGACTCCTGA CAATtaatacgactcactataggGAAGAGTCAGAGTGGGG AAGAGCTATGATGCGTTCGATCACTCCATGTG ATCCTACACTCGCCGCAGGCTGG-3′ |
aThe lower case region indicates the promoter sequence for T7 RNA polymerase, italic region indicates the range of transcription and underlined region the G-quadruplex-forming sequence.
Table 2.
PCR primers used for preparing dsDNAs used in Figures 5–7
| Gene | Upstream primer (5′-3′) | Downstream primer (5′-3′) | dsDNA (bp)a | Ref. |
|---|---|---|---|---|
| AAVS1 | GGTAGACAGGGCTGGGGTG | CTCTCCTGCCCCTTCCTACA | 62 + 30 + 73 | (58) |
| ASS1 | AGGTGGCTGTGAACGCTGA | GACCGGGGACACGTGGC | 32 + 21 + 36 | (58) |
| BCL2 | GGGGCCACGGAGAGCG | GTCGGGCTGTGCAGAGAATG | 62 + 23 + 50 | (59) |
| C-KIT | GGCATTAACACGTCGAAAGA | TCCCTCTGCGCGCCGGC | 56 + 20 + 34 | (60) |
| C-MYC | TGGGCGGAGATTAGCGAGAG | CCTAGAGCTAGAGTGCTCGGC | 80 + 30 + 39 | (61) |
| CSTB | GGCTTCGGAGTCCCCTGC | CCAGCCTGCGGCGAGTG | 51 + 21 + 40 | (62) |
| EPO | TCTGCATGTGTGCGTGCG | CCGGCGAGCCTCAACC | 51 + 30 + 39 | (58) |
| GUK1 | GGCGGGTCGTGATGTTAG | ACCGCAGGGGCGTTCA | 62 + 22 + 24 | (13) |
| hTEL | GGCTTCGGAGTCCCCTGC | CCAGCCTGCGGCGAGTG | 39 + 21 + 36 | |
| ITGB1 | CGCGGCAGCACTTAAAGC | CCTCCTGGACTAGCCTGGAAT | 62 + 23 + 51 | (58) |
| KRAS | AGCTATCGATGCGTTCCG | GAGCACTCCTTCTCCCCG | 62 + 27 + 35 | (63) |
| LRE2 | GAGATCACATGGACACAGGAAGGG | CTGCACCCACTAATGTGTCATCTA | 50 + 23 + 49 | (58) |
| NRAS | GGCGAAAGAATGGAAGCG | GGCCTCCGAACCACGAGT | 83 + 18 + 34 | (64) |
| PSMA4 | ACCGCTCACCGAATAACCG | CGAGGGGCACGGGTTCTA | 55 + 22 + 57 | (13) |
| UTX | TTAAGTGGAGCCACGGCTGAC | CTGAGGGGATTCGTTGGAGAC | 78 + 16 + 93 | (13) |
aThe three numbers indicate, from left to right, the size of the duplex flanking the core G-rich sequence at the 5′ side, the core G-rich sequence and the duplex flanking the core G-rich sequence at the 3′ side, respectively.
In vitro transcription
Transcription was carried out using 0.6 pmol FAM-labeled dsDNA in a total volume of 29 µl (or its multiples) at 37°C for 1 h in transcription buffer containing 150 mM KCl, 20 U T7 RNA Polymerase (Fermentas, MBI), 0.5 mM GTP and TTP (for C-MYC) or 0.5 mM ATP and GTP (for NRAS and control), in the absence or presence of 40% (m/v) PEG 200. The reaction was stopped by addition of 1/29 vol of 0.5 M EDTA followed by treatment with 0.6 mg/ml Proteinase K (Fermentas, MBI) at 37°C for 1 h.
Heat denaturation/renaturation
dsDNA was made in 10 mM Tris–HCl (pH 7.4) buffer containing 1 mM EDTA and the indicated concentration of KCl (or LiCl) and PEG 200, heated at 95°C for 5 min and then cooled down to room temperature at a rate of 0.02°C per second.
Examination of G-quadruplex formation by dimethyl sulfate footprinting
Ten picomoles of FAM-labeled dsDNA in 200 µl volume derived from samples that had undergone transcription or heat denaturation/renaturation were mixed with 4 µl of 10% (v/v) dimethyl sulfate (DMS) in ethanol and incubated for 6 min at room temperature. The reaction was stopped by addition of 200 µl stop buffer (0.6 M NaOAc, 0.1 M β-mercaptoethanol, 20 µg sperm DNA). After phenol/chloroform extraction and ethanol precipitation, the DNA was dissolved in 50 µl water. The NTPs in the samples that had undergone transcription were removed by the desalt column mini Quick Spin Oligo Column (Roche, Germany). Thereafter, 50 µl 20% (v/v) piperidine in water was added and the samples were heated at 90°C for 30 min, followed by phenol/chloroform extraction and ethanol precipitation. The precipitated DNA was dissolved in 50% (v/v) deionized formamide in water, denatured at 95°C for 5 min and resolved on a denaturing 12% polyacrylamide gel.
Examination of G-quadruplex formation by native gel electrophoresis
DNA samples were loaded on 8% polyacrylamide gel containing 150 mM KCl, 40% (w/v) PEG 200 and electrophoresed at 4°C, 8 V/cm, in 1X TBE buffer containing 150 mM KCl. For the DNA samples amplified with unlabeled primers, the gel was stained with ethidium bromide (EB) and recorded on a ChemiImager 5500 (Alpha Innotech, San Leandro, CA, USA). For those amplified using FAM-labeled primers, the gel was scanned on a Typhoon phosphor imager (Amersham Biosciences, Sweden) and quantitated with the software ImageQuant 5.2. In some experiments, the DNA samples were incubated with 15 µM T4 Gene 32 single-stranded DNA-binding protein (SSB) (NEB, USA) on ice for 1 h before electrophoresis.
Examination of G-quadruplex formation by single-stranded DNA endonuclease
Ten picomoles of FAM-labeled dsDNA in 90 µl 10 mM Tris–HCl (pH 7.4) buffer containing 150 mM K+ solution and 40% (w/v) PEG 200 were treated as described in the heat denaturation/renaturation section. Thereafter, 10 µl of 20 U mung bean nuclease (MBN) or 50 U S1 nuclease (Takara, China) were added. After incubation at 37°C for 5 min, the reaction was subjected to phenol/chloroform extraction and ethanol precipitation. The precipitated DNA was dissolved in 50% (v/v) deionized formamide in water, denatured at 95°C for 5 min and resolved on a denaturing 12% polyacrylamide gel.
Thermal melting profiling by fluorescence resonance energy transfer
Oligonucleotides, 5′-GGGAGGGGCGGGTCTGGG-3′ (F-NRAS-T), 5′-CCGCACGCACCGCTCG-CCTTTTGGCGAGCGGTGCGTGCGG-3′ (F-rNRAS-T), 5′-GGGTTAGGGTTAGGGTTAGGG-3′ (F-TEL-T), and 5′-CACACTCACCTCCACACTCCATTTTTGGAGTGTGGAGGTGAGT-GTG-3′ (F-rTEL-T), labeled at the 5′-end with a fluorescein (FAM) and the 3′-end with a tetramethylrhodamine (TAMRA), respectively, were purchased from Takara Biotech (Dalian, China). The loop region in F-rNRAS-T and F-rTEL-T is underlined. The Thermal melting analyses were carried out in 10 mM lithium cacodylate buffer (pH 7.4) containing either 150 mM KCl or 5 mM KCl/145 mM LiCl and the indicated concentration of PEG 200 as described (37,38) on a Rotor-Gene 2000 Real-time Cycler.
RESULTS
Detection of G-quadruplex formation in long dsDNA by DMS footprinting
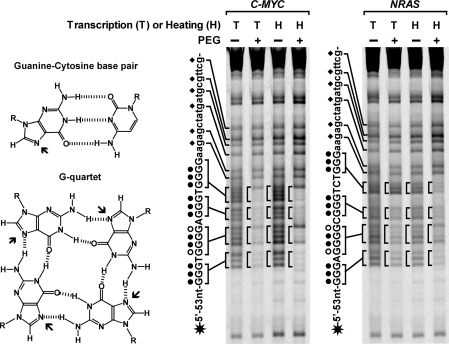
Using overlap PCR and genomic DNA from HeLa cells as template, we first constructed two dsDNAs carrying the core G-rich sequence from the C-MYC and NRAS gene, respectively. The G-rich core sequence was on the nontemplate strand and connected with a flanking promoter sequence for the T7 RNA polymerase at its 5′ side (Table 1). The two dsDNAs were subjected to heat denaturation/renaturation or transcription with T7 RNA polymerase. To mimic the intracellular environment, the treatments were carried out at neutral pH in 150 mM K+ solution containing 40% (w/v) PEG 200 or no PEG (Figure 1). G-quadruplex formation was then detected by the accessibility of the N7 of guanines to DMS. The N7 in DNA duplex is prone to methylation by DMS and subsequent cleavage with piperidine, but protected by the Hoogsteen bonding in the G-quartet of G-quadruplex structure. The two dsDNAs were labeled at the 5′-end of the G-rich strand with a fluorescent dye FAM. Cleavage fragments were resolved by denaturing gel electrophoresis. In Figure 1, distinct bands corresponding to the cleavage of the four runs of guanines marked with a circle in the core G-rich sequence can be clearly seen for the dsDNAs that were subjected to transcription or heat denaturation/renaturation in the absence of PEG. However, these bands become protected from cleavage when the treatment was carried out in the presence of 40% (w/v) PEG, indicating that in these DNAs G-quadruplex composed of three G-quartets formed during the process of RNA transcription and heat denaturation/renaturation. In contrast, the guanines in the flanking sequences were always similarly attacked no matter whether the samples were processed in the presence or absence of PEG indicating these sequences were in the duplex form.
Figure 1.
G-quadruplex formation in long dsDNA carrying G-quadruplex-forming sequence from the C-MYC, NRAS gene examined by DMS footprinting. The N7 (arrowed in the structures on the left side) is not protected from chemical attack in the G-C pair but is in the G-quartet. DNAs labeled at the 5′-end of the G-rich strand with a FAM (asterisk) were subjected either to RNA transcription (T) followed by protease treatment or heat denaturation/denaturation (H) in 150 mM K+ solution in the absence (–) or presence (+) of 40% (w/v) PEG 200 before footprinting. Filled circles beside the sequences indicate the guanines that are fully protected, open circles indicate the guanines that are partially protected in the G-quartets; diamonds indicate the unprotected guanines in the duplex region. To increase the resolution, some small bands were allowed to run out of the gel.
Detection and quantitation of G-quadruplex formation in long dsDNA by gel electrophoresis
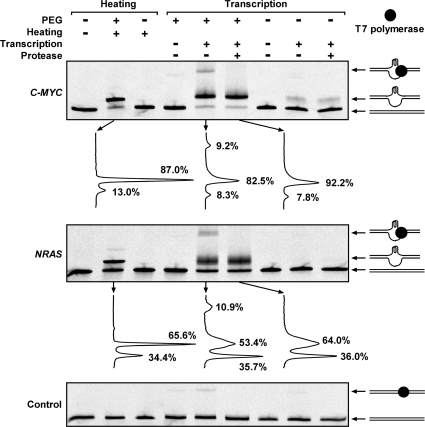
DMS footprinting is intuitive for identifying G-quadruplex structure, but difficult to quantify the amount of G-quadruplex formed. We anticipated that when the G-quadruplex forms, the structural change should alter the electrophoretic migration of a dsDNA. To test this possibility, the two dsDNAs and one control dsDNA without G-quadruplex-forming sequence was examined (Figure 2). The dsDNA with G-quadruplex-forming sequence from C-MYC or NRAS without prior heat treatment or heated in the absence of PEG appeared as a single band on the gel. A new major slow band emerged if they were heat treated in the presence of 40% (w/v) PEG 200. According to the DMS footprinting results, the new band should present the dsDNA containing G-quadruplex. In the two dsDNAs that had undergone transcription in the presence of PEG, two new slow bands appeared. The lower major band should be the G-quadruplex-containing dsDNA since it had a similar migration rate as the new band in the heat-treated dsDNA in PEG solution. The slowest band disappeared when the samples were treated with protease after transcription suggesting that it was a DNA/polymerase complex. The quantification by digital scan revealed that this band carried G-quadruplex since its disappearance intensified the G-quadruplex-containing band beneath it. When the transcription was carried out in the absence of PEG, a weak G-quadruplex-containing band was detected in the C-MYC DNA, but not in the NRAS DNA. This weak band did not appear in the heat-treated C-MYC DNA. A possible interpretation could be that the transcribed RNA bound to the C-rich template enhanced the competition of the C-MYC G-quadruplex against duplex. This enhancement was not seen in the NRAS DNA probably because the NRAS G-quadruplex was too weak to compete with duplex even with the transcribed RNA. For the control dsDNA, that did not contain G-quadruplex-forming sequence, very fainter bands were also detected in the samples treated with T7 polymerase. These bands all disappeared if the samples were treated with protease before electrophoresis, suggesting that they were DNA/polymerase complexes. Their amount was lower than that in the C-MYC and NRAS DNA. It is possible that formation of G-quadruplex facilitated the association of the polymerase with the DNAs. However, the quantity of such bands in the C-MYC and NRAS DNA did not correlate with the quantity of G-quadruplexes, suggesting the complex formation may also depend on sequence. By supplying only two NTPs, the transcription was allowed to proceed for 8 nt. This ensured homogeneous transcription product and sharp band for the DNA carrying G-quadruplex. When full-length transcription was allowed, the DNA band containing G-quadruplex became extremely smeary due to the heterogeneity in the progress of transcription (data not shown).
Figure 2.
G-quadruplex formation in long dsDNA carrying G-quadruplex-forming sequence from the C-MYC, NRAS gene examined by native gel electrophoresis. DNAs, including the control that does not form G-quadruplex, were labeled at the 5′-end of the G-rich strand with a FAM and subjected either to RNA transcription or heat denaturation/denaturation as described in Figure 1. The samples marked as transcription negative were treated in the same way as the transcription positive samples except that no NTP was supplied. The drawing at the right side shows schematic illustration of the DNA structure associated with the indicated DNA band. The graph beneath the C-MYC and NRAS gel is the digital scan of the indicated lane to quantitate the percentage of each DNA band.
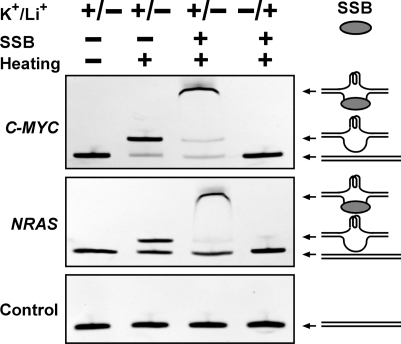
To verify that the DNA band immediately following the original duplex dsDNA band in gel electrophoresis carried G-quadruplex, electrophoretic mobility shift assay was conducted, in which the DNA was incubated with the SSB T4 gene 32 protein before electrophoresis (Figure 3). When the G-rich region in a dsDNA turns into G-quadruplex, its complementary C-rich region will be liberated into single-stranded form that will be recognized by the SSB. Indeed, the new band was shifted to a higher position in the gel, indicating the formation of a DNA/SSB complex and the presence of single-stranded DNA. G-quadruplex is stabilized by K+ or Na+, but not by Li+ (39–44). When the K+ was substituted by Li+, the slower migrating bands disappeared.
Figure 3.
G-quadruplex formation in dsDNA derived from the C-MYC, NRAS gene examined by gel electrophoretic mobility shift. DNAs, including the control that does not form G-quadruplex, were subjected to heat denaturation/renaturation in 150 mM K+ or Li+ solution containing 40% (w/v) PEG 200, then followed by incubation in the presence or absence of single-stranded DNA binding protein (SSB) before native gel electrophoresis. The gel was stained with ethidium bromide (EB). The drawing at the right side shows schematic illustration of the structure associated with the indicated DNA band.
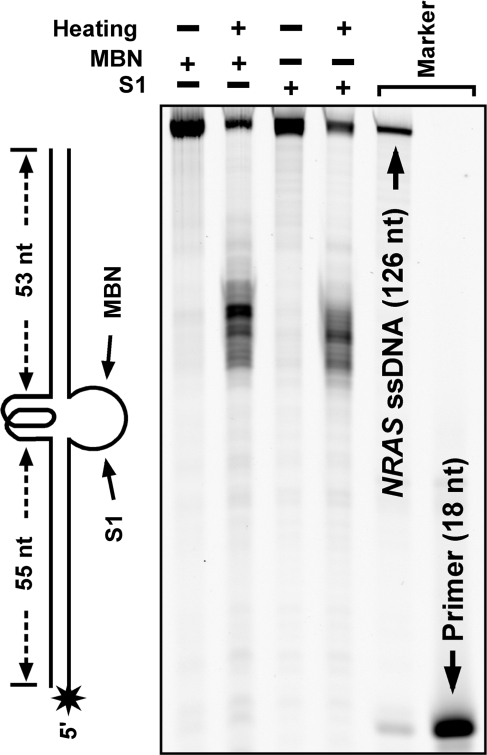
The incomplete transcription of merely 8 nt resulted in only a local and partial separation of the two DNA strands. However, in the heat denaturation/renaturation treatment, the two strands of the dsDNAs were completely dissociated before they re-annealed. There was a possibility that the slow migrating band immediately following the original dsDNA could be the DNA that remained in single-stranded form as a result of G-quadruplex formation. To test this possibility, we treated the NRAS dsDNA samples carrying a fluorescent dye FAM at the 5′-end of the C-rich strand before electrophoresis with MBN and S1 nuclease, respectively, that specifically degrades single-stranded nucleic acids. If the slow migrating band was single-stranded DNA, it should be fully degraded. However, the results shown in Figure 4 indicate that the cleavage only occurred in the middle of the DNA. This fact further verifies that the band immediately following the original dsDNA duplex band was dsDNA, carrying a G-quadruplex in the middle of the sequence. This result also confirms that the sequences flanking the core G-rich region were in duplex form. Very occasionally, this major band could be followed by an extremely faint band in the heat-treated samples, but not in the transcription samples (e.g. the 2nd lane of Figure 2 for NRAS). This faint band could be single-stranded DNA and such bands could usually be suppressed by slower cooling rate.
Figure 4.
G-quadruplex formation in long dsDNA carrying G-quadruplex-forming sequence from the NRAS gene examined by hydrolysis with mung bean nuclease (MBN) and S1 nuclease. DNA labeled at the 5′-end of the C-rich strand with a FAM (asterisk) was subjected to heat denaturation/renaturation in 150 mM K+, 40% (w/v) PEG 200 solution, then followed by hydrolysis with the indicated nuclease before denaturing gel electrophoresis. The single-stranded region opposite to the G-quadruplex is susceptible to the attack of the two nucleases.
G-quadruplex/duplex competition in long dsDNAs and the effect of K+ and PEG concentration
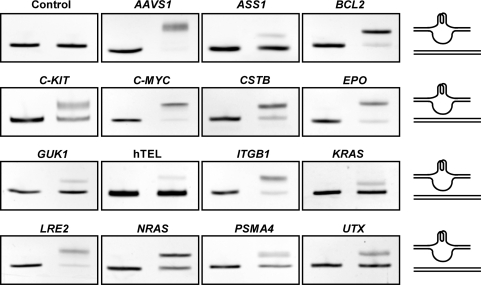
The results presented in the above section demonstrate that the gel electrophoresis can provide a simple method to identify and quantify G-quadruplex formation in long dsDNA. Using this method, we next studied the competition between G-quadruplex and duplex formation and how it is affected by K+ and PEG concentration in 15 long dsDNAs derived from human genome by PCR (Table 2). In these dsDNAs, the G-rich sequences were flanked by non-G-rich sequences at both ends. The core telomere sequence (T2AG3)4 was also made into double-stranded form flanked by non-G-rich sequences at both ends by overlap PCR as a reference since it is the most extensively studied G-quadruplex sequence. G-quadruplex formation was induced by the heat denaturation/renaturation treatment since it is a pure physical process and is expected to more faithfully reflect the competition between the two DNA structures themselves without potential influence from protein binding in transcription. Figure 5 shows the gels of these dsDNAs stained by EB. G-quadruplex formation varied significantly in the 15 dsDNAs as judged from the ratio of the two DNA bands in the heat-treated sample in each gel. For example, the majority of the AAVS1 and BCL2 DNA formed G-quadruplex, while the GUK1, ASS1 and telomere DNA were largely in duplex form.
Figure 5.
G-quadruplex formation in 16 long dsDNAs derived from human genome examined by native gel electrophoresis. DNAs in 150 mM K+, 40% (w/v) PEG 200 solution were subjected to heat denaturation/renaturation (right lane) or were not heat-treated (left lane) before electrophoresis. The gel was stained with ethidium bromide (EB). The drawing at the right side shows schematic illustration of the structure associated with the corresponding DNA band.
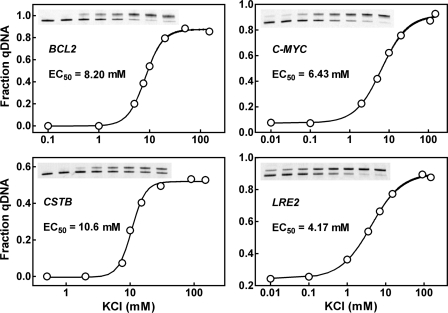
Since G-quadruplex is poorly stained by EB (45), a quantitative evaluation of G-quadruplex formation as a fraction of total DNA is difficult. Therefore, the rest analyses were carried out using dsDNAs amplified with fluorescently labeled primers. Animal cells contain high concentration of K+ (150 mM) intracellularly (46). So far, most of the G-quadruplexes have been found to form at submicromolar or micromolar level of K+ when single-stranded oligonucleotide was examined (32,43,47–49). To investigate the effect of K+ on dsDNA, G-quadruplex formation in four dsDNAs were examined as a function of K+ concentration. The data in Figure 6 show that the K+ requirement for G-quadruplex formation is roughly 10 times higher for dsDNA than for single-stranded G-rich strand. For instance, the C-MYC sequence has an EC50 of 6.43 mM K+ for dsDNA (Figure 6) but between 0.01 and 0.1 mM for the single-stranded form (47). All the four dsDNAs reached half transition at the neighborhood of 10 mM K+, which is much lower than the physiological concentration of K+. This fact suggests that G-quadruplex formation should be insensitive to fluctuation in K+ concentration inside cells.
Figure 6.
G-quadruplex formation in dsDNAs as a function of K+ concentration. DNAs were derived from BCL2, C-MYC, CSTB and LRE2 by PCR using fluorescent primers, made in buffer containing 40% (w/v) PEG 200 and various concentration of K+ and subjected to heat denaturation/renaturation before native gel electrophoresis. G-quadruplex formation was quantitated as fraction of total DNA.
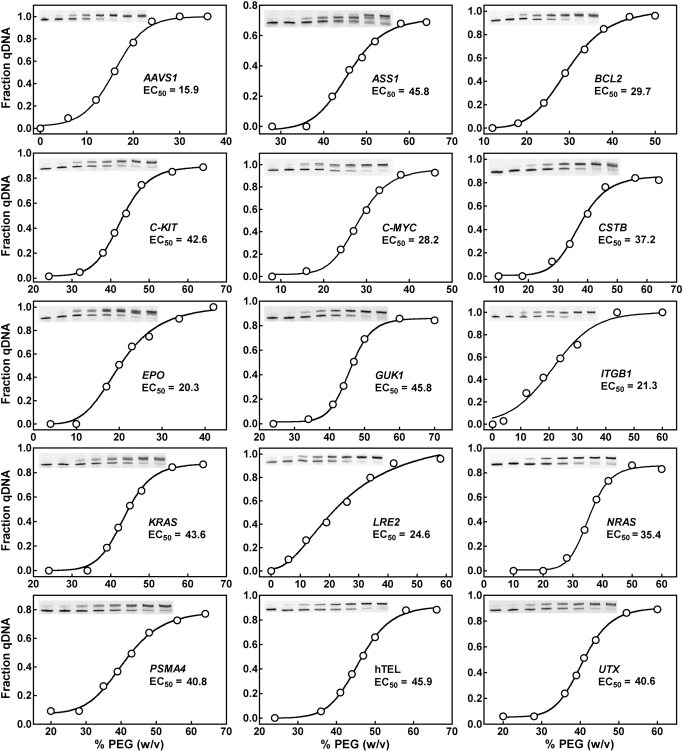
With regard to the molecular crowding condition, we examined the 15 dsDNAs for the dependence of G-quadruplex formation on PEG concentration (Figure 7). No G-quadruplex was detected in all these DNAs in the absence of PEG. Among the DNAs, the AAVS1 showed the lowest EC50 of 15.9% PEG and hTEL the highest EC50 of 45.9% PEG for G-quadruplex formation. The slope of the dose–response curves also displayed large difference. For example, the G-quadruplex formation in the LRE2 was affected over a broad range of PEG concentration, while that in the GUK1 and NRAS was responsive within a narrower range. In contrast to the case of K+, the effective concentrations of PEG for the dsDNAs to reach half transition (EC50) are all close to the physiological concentration of biomolecules (30–40%, w/v) in living cells (50,51). For the ASS1 gene, two new bands were induced by PEG (Figure 7). This could indicate the presence of more than one G-quadruplex structure.
Figure 7.
G-quadruplex formation in dsDNAs as a function of PEG 200 concentration. DNAs were derived from human genome by PCR using fluorescent primers, made in 150 mM K+ solution containing various concentration (w/v) of PEG 200 and subjected to heat denaturation/renaturation before native gel electrophoresis. G-quadruplex formation was quantitated as fraction of total DNA.
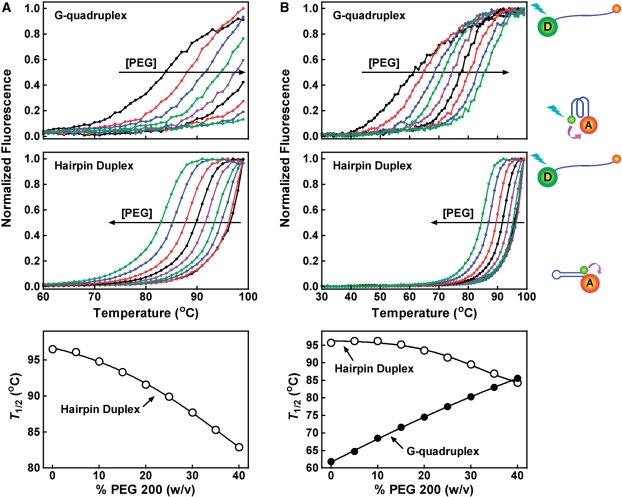
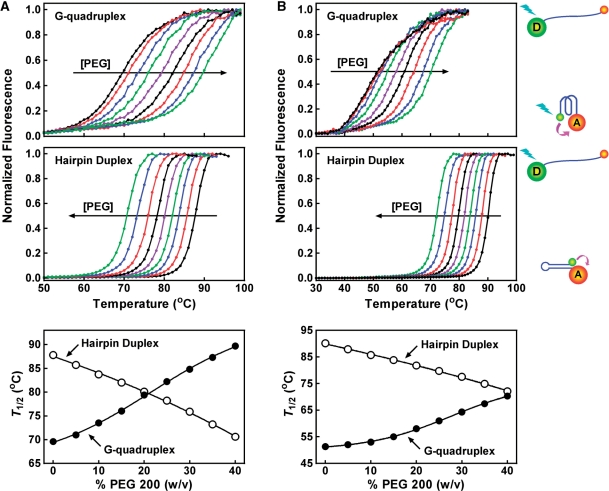
Several studies have reported that molecular crowding can stabilize G-quadruplex and destabilize duplex structure (32,52–57). We analyzed the effect of PEG 200 at various concentrations on the thermal stability of two G-quadruplexes (F-NRAS-T, F-TEL-T) and their corresponding hairpin duplexes (F-rNRAS-T, F-rTEL-T) in which the sequences were randomized to prevent G-quadruplex formation (Figures 8 and 9). These oligonucleotides were labeled at their 5′-end with a fluorescein (FAM) as donor and 3′-end a tetramethylrhodamine (TAMRA) as acceptor. The thermal denaturation of the structures moved the two fluorophores apart and decreased the fluorescence resonance energy transfer (FRET) between them, thus leading to an increase in donor fluorescence. As judged from the T1/2 that represents the temperature for the donor fluorescence to reach midvalue between the minimal and maximal emission, PEG 200 enhanced the stability of the G-quadruplexes and reduced the stability of the duplexes in a concentration-dependent manner. In 150 mM K+ solution, PEG stabilized the F-NRAS-T G-quadruplex at all concentrations, although the G-quadruplex was too stable to be fully denatured. In contrast, the corresponding hairpin duplex was destabilized (Figure 8A). By decreasing the K+ concentration to 5 mM to lower the stability of the F-NRAS-T G-quadruplex, a simultaneous stabilization of G-quadruplex and destabilization of duplex by PEG was observed (Figure 8B). The same effect was also observed for the human telomere sequence F-TEL-T and F-rTEL-T (Figure 9). Therefore, the opposite effect of PEG 200 on the stabilities of the G-quadruplexes and duplexes favored the competition of G-quadruplex to duplex structure and provides an explanation for the formation of G-quadruplex in PEG solution.
Figure 8.
Effect of PEG 200 on the thermal stability of NRAS G-quadruplex and hairpin duplex structure. Fluorescence melting assays were carried out in (A) 150 mM K+ or (B) 5 mM K+/145 mM Li+ solution containing 0, 5, 10, 15, 20, 25, 30, 35, 40% (w/v) of PEG 200, respectively. The horizontal arrow indicates the direction of increasing in PEG concentration.
Figure 9.
Effect of PEG 200 on the thermal stability of human telomere G-quadruplex and hairpin duplex structure. Fluorescence melting assays were carried out in (A) 150 mM K+ or (B) 5 mM K+/145 mM Li+ solution containing 0, 5, 10, 15, 20, 25, 30, 35, 40% (w/v) of PEG 200, respectively. The horizontal arrow indicates the direction of increasing in PEG concentration.
DISCUSSION
In this work, we observed G-quadruplex formation at the internal region of long dsDNA in the process of RNA transcription and heat denaturation/renaturation and studied how it could be affected by the concentration of K+ and molecular crowding. From the analysis of 15 dsDNAs carrying different core G-rich sequences from human genome, our data show that molecular crowding with PEG is essential for the formation of G-quadruplexes in these dsDNAs. Without molecular crowding, none of the G-rich sequences was able to sustain stable G-quadruplex after the heat denaturation/renaturation process. Under the molecular crowding condition, G-quadruplexes can not only form, but also maintain as stable structure in the presence of a complementary strand in close vicinity. The effects of molecular crowding on the two structures clearly explain our observations. The dependence of G-quadruplex formation on PEG concentration revealed that the effective concentrations for the PEG to promote G-quadruplex formation (EC50) in the dsDNAs are near the physiological concentration of biomolecules (30–40%, w/v) in living cells (50,51). This fact suggests that the molecular crowding condition inside living cells may provide a proper environment for stable G-quadruplexes to form in genome when small region of DNA double helix is denatured. This observation is of physiological importance because it provides a physical basis for the formation of stable G-quadruplexes under physiological condition and indicates the requirement of G-quadruplex unwinding activity if this structure has to be disrupted in certain biological processes. Local and transient dissociation of DNA duplex take place during many biological DNA-processing events, such as replication, transcription and promoter recognition. These events should provide chances for G-quadruplex to form in vivo.
Among the 15 dsDNAs, the one carrying the human telomere sequence showed the least capability to form G-quadruplex with an EC50 of 45.9% (Figure 7). At 40% PEG, about 15% of the telomere sequence in the dsDNA was in G-quadruplex form as calculated from the PEG does curve. This value is much smaller than that of the same telomere sequence placed at the ends of dsDNA where more than half of them formed G-quadruplex (45). This difference demonstrates that the presence of flanking duplex significantly suppresses G-quadruplex formation. Therefore, studies using long dsDNA can yield more accurate and physiologically relevant information than those using only the core sequence regarding the competition between G-quadruplex and duplex in genome.
Our work shows that formation of G-quadruplex retards dsDNA migration in gel electrophoresis. This property provides a reliable and simple method for analyzing G-quadruplex formation in long dsDNA. Precise quantitation can be easily obtained by the ratio of the DNA bands if the dsDNA is prepared using primer labeled with radioactive isotopes or fluorescent dyes. Approximate results can be obtained by staining with dyes, such as EB, without labeling of primer. For a same G-rich sequence, the extent of G-quadruplex formation in different biological processes may be different because of the different proteins and mechanisms involved. The heat denaturation/renaturation treatment followed by gel electrophoresis provides an easy and general assessment of the capability of G-quadruplex formation in long dsDNA. Because of the abundance of G-quadruplex sequences in genome, G-quadruplex stabilization by small molecules is emerging as a therapeutic strategy against cancer and other diseases (21–28). How a small molecule will promote G-quadruplex formation and stabilize G-quadruplex in genomic DNA would be better tested using long dsDNA as target. We expect that the heat treatment followed by gel electrophoresis used in this work can provide a useful tool to fulfill this purpose.
FUNDING
Grant numbers 2007CB507402 from MSTC; and 30670451, 90813031 and 20621502 from NSFC. Funding for open access charge: MSTC (Grant number 2007CB507402).
Conflict of interest statement. None declared.
REFERENCES
- 1.Gilbert DE, Feigon J. Multistranded DNA structures. Curr. Opin. Struct. Biol. 1999;9:305–314. doi: 10.1016/S0959-440X(99)80041-4. [DOI] [PubMed] [Google Scholar]
- 2.Simonsson T. G-quadruplex DNA structures–variations on a theme. Biol. Chem. 2001;382:621–628. doi: 10.1515/BC.2001.073. [DOI] [PubMed] [Google Scholar]
- 3.Blackburn EH. Structure and function of telomeres. Nature. 1991;350:569–573. doi: 10.1038/350569a0. [DOI] [PubMed] [Google Scholar]
- 4.Simonsson T, Pecinka P, Kubista M. DNA tetraplex formation in the control region of c-myc. Nucleic Acids Res. 1998;26:1167–1172. doi: 10.1093/nar/26.5.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sen D, Gilbert W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature. 1988;334:364–366. doi: 10.1038/334364a0. [DOI] [PubMed] [Google Scholar]
- 6.Hammond-Kosack MC, Kilpatrick MW, Docherty K. The human insulin gene-linked polymorphic region adopts a G-quartet structure in chromatin assembled in vitro. J. Mol. Endocrinol. 1993;10:121–126. doi: 10.1677/jme.0.0100121. [DOI] [PubMed] [Google Scholar]
- 7.Zhang R, Lin Y, Zhang CT. Greglist: a database listing potential G-quadruplex regulated genes. Nucleic Acids Res. 2008;36:D372–D376. doi: 10.1093/nar/gkm787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yadav VK, Abraham JK, Mani P, Kulshrestha R, Chowdhury S. QuadBase: genome-wide database of G4 DNA–occurrence and conservation in human, chimpanzee, mouse and rat promoters and 146 microbes. Nucleic Acids Res. 2008;36:D381–D385. doi: 10.1093/nar/gkm781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Verma A, Halder K, Halder R, Yadav VK, Rawal P, Thakur RK, Mohd F, Sharma A, Chowdhury S. Genome-wide computational and expression analyses reveal G-quadruplex DNA motifs as conserved cis-regulatory elements in human and related species. J. Med. Chem. 2008;51:5641–5649. doi: 10.1021/jm800448a. [DOI] [PubMed] [Google Scholar]
- 10.Kikin O, Zappala Z, D'A;ntonio L, Bagga PS. GRSDB2 and GRS_UTRdb: databases of quadruplex forming G-rich sequences in pre-mRNAs and mRNAs. Nucleic Acids Res. 2008;36:D141–D148. doi: 10.1093/nar/gkm982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hershman SG, Chen Q, Lee JY, Kozak ML, Yue P, Wang LS, Johnson FB. Genomic distribution and functional analyses of potential G-quadruplex-forming sequences in Saccharomyces cerevisiae. Nucleic Acids Res. 2008;36:144–156. doi: 10.1093/nar/gkm986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eddy J, Maizels N. Conserved elements with potential to form polymorphic G-quadruplex structures in the first intron of human genes. Nucleic Acids Res. 2008;36:1321–1333. doi: 10.1093/nar/gkm1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huppert JL, Balasubramanian S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007;35:406–413. doi: 10.1093/nar/gkl1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Du Z, Kong P, Gao Y, Li N. Enrichment of G4 DNA motif in transcriptional regulatory region of chicken genome. Biochem. Biophys. Res. Commun. 2007;354:1067–1070. doi: 10.1016/j.bbrc.2007.01.093. [DOI] [PubMed] [Google Scholar]
- 15.Huppert JL, Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duquette ML, Handa P, Vincent JA, Taylor AF, Maizels N. Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA. Genes Dev. 2004;18:1618–1629. doi: 10.1101/gad.1200804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl Acad. Sci. USA. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cogoi S, Xodo LE. G-quadruplex formation within the promoter of the KRAS proto-oncogene and its effect on transcription. Nucleic Acids Res. 2006;34:2536–2549. doi: 10.1093/nar/gkl286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ou TM, Lu YJ, Zhang C, Huang ZS, Wang XD, Tan JH, Chen Y, Ma DL, Wong KY, Tang JC, et al. Stabilization of G-quadruplex DNA and down-regulation of oncogene c-myc by quindoline derivatives. J. Med. Chem. 2007;50:1465–1474. doi: 10.1021/jm0610088. [DOI] [PubMed] [Google Scholar]
- 20.Fry M. Tetraplex DNA and its interacting proteins. Front. Biosci. 2007;12:4336–4351. doi: 10.2741/2391. [DOI] [PubMed] [Google Scholar]
- 21.Ou TM, Lu YJ, Tan JH, Huang ZS, Wong KY, Gu LQ. G-quadruplexes: targets in anticancer drug design. ChemMedChem. 2008;3:690–713. doi: 10.1002/cmdc.200700300. [DOI] [PubMed] [Google Scholar]
- 22.Neidle S, Read MA. G-quadruplexes as therapeutic targets. Biopolymers. 2000;56:195–208. doi: 10.1002/1097-0282(2000)56:3<195::AID-BIP10009>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 23.Kerwin SM. G-Quadruplex DNA as a target for drug design. Curr. Pharm. Des. 2000;6:441–478. doi: 10.2174/1381612003400849. [DOI] [PubMed] [Google Scholar]
- 24.Hurley LH, Wheelhouse RT, Sun D, Kerwin SM, Salazar M, Fedoroff OY, Han FX, Han H, Izbicka E, Von Hoff DD. G-quadruplexes as targets for drug design. Pharmacol. Ther. 2000;85:141–158. doi: 10.1016/s0163-7258(99)00068-6. [DOI] [PubMed] [Google Scholar]
- 25.Han H, Hurley LH. G-quadruplex DNA: a potential target for anti-cancer drug design. Trends Pharmacol. Sci. 2000;21:136–142. doi: 10.1016/s0165-6147(00)01457-7. [DOI] [PubMed] [Google Scholar]
- 26.Mergny JL, Helene C. G-quadruplex DNA: a target for drug design. Nat. Med. 1998;4:1366–1367. doi: 10.1038/3949. [DOI] [PubMed] [Google Scholar]
- 27.Wong HM, Payet L, Huppert JL. Function and targeting of G-quadruplexes. Curr Opin Mol Ther. 2009;11:146–155. [PubMed] [Google Scholar]
- 28.Gatto B, Palumbo M, Sissi C. Nucleic acid aptamers based on the g-quadruplex structure: therapeutic and diagnostic potential. Curr. Med. Chem. 2009;16:1248–1265. doi: 10.2174/092986709787846640. [DOI] [PubMed] [Google Scholar]
- 29.Phan AT, Mergny JL. Human telomeric DNA: G-quadruplex, i-motif and Watson-Crick double helix. Nucleic Acids Res. 2002;30:4618–4625. doi: 10.1093/nar/gkf597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li W, Wu P, Ohmichi T, Sugimoto N. Characterization and thermodynamic properties of quadruplex/duplex competition. FEBS Lett. 2002;526:77–81. doi: 10.1016/s0014-5793(02)03118-6. [DOI] [PubMed] [Google Scholar]
- 31.Li W, Miyoshi D, Nakano S, Sugimoto N. Structural competition involving G-quadruplex DNA and its complement. Biochemistry. 2003;42:11736–11744. doi: 10.1021/bi034168j. [DOI] [PubMed] [Google Scholar]
- 32.Kan ZY, Yao Y, Wang P, Li XH, Hao YH, Tan Z. Molecular crowding induces telomere G-quadruplex formation under salt-deficient conditions and enhances its competition with duplex formation. Angew. Chem. Int. Ed. Engl. 2006;45:1629–1632. doi: 10.1002/anie.200502960. [DOI] [PubMed] [Google Scholar]
- 33.Kumar N, Maiti S. The effect of osmolytes and small molecule on Quadruplex-WC duplex equilibrium: a fluorescence resonance energy transfer study. Nucleic Acids Res. 2005;33:6723–6732. doi: 10.1093/nar/gki961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kumar N, Sahoo B, Varun KA, Maiti S. Effect of loop length variation on quadruplex-Watson Crick duplex competition. Nucleic Acids Res. 2008;36:4433–4442. doi: 10.1093/nar/gkn402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Risitano A, Fox KR. Stability of intramolecular DNA quadruplexes: comparison with DNA duplexes. Biochemistry. 2003;42:6507–6513. doi: 10.1021/bi026997v. [DOI] [PubMed] [Google Scholar]
- 36.Blin N, Stafford DW. A general method for isolation of high molecular weight DNA from eukaryotes. Nucleic Acids Res. 1976;3:2303–2308. doi: 10.1093/nar/3.9.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.De Cian A, Guittat L, Kaiser M, Sacca B, Amrane S, Bourdoncle A, Alberti P, Teulade-Fichou MP, Lacroix L, Mergny JL. Fluorescence-based melting assays for studying quadruplex ligands. Methods. 2007;42:183–195. doi: 10.1016/j.ymeth.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 38.Chen Z, Zheng KW, Hao YH, Tan Z. Reduced or diminished stabilization of the telomere G-quadruplex and inhibition of telomerase by small chemical ligands under molecular crowding condition. J. Am. Chem. Soc. 2009;131:10430–10438. doi: 10.1021/ja9010749. [DOI] [PubMed] [Google Scholar]
- 39.Sen D, Gilbert W. A sodium-potassium switch in the formation of four-stranded G4-DNA. Nature. 1990;344:410–414. doi: 10.1038/344410a0. [DOI] [PubMed] [Google Scholar]
- 40.Williamson JR. G-quartet structures in telomeric DNA. Annu. Rev. Biophys. Biomol. Struct. 1994;23:703–730. doi: 10.1146/annurev.bb.23.060194.003415. [DOI] [PubMed] [Google Scholar]
- 41.Mergny JL, Phan AT, Lacroix L. Following G-quartet formation by UV-spectroscopy. FEBS Lett. 1998;435:74–78. doi: 10.1016/s0014-5793(98)01043-6. [DOI] [PubMed] [Google Scholar]
- 42.Zhao Y, Kan ZY, Zeng ZX, Hao YH, Chen H, Tan Z. Determining the folding and unfolding rate constants of nucleic acids by biosensor. Application to telomere G-quadruplex. J. Am. Chem. Soc. 2004;126:13255–13264. doi: 10.1021/ja048398c. [DOI] [PubMed] [Google Scholar]
- 43.Yao Y, Wang Q, Hao YH, Tan Z. An exonuclease I hydrolysis assay for evaluating G-quadruplex stabilization by small molecules. Nucleic Acids Res. 2007;35:e68. doi: 10.1093/nar/gkm194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhuang XY, Tang J, Hao YH, Tan Z. Fast detection of quadruplex structure in DNA by the intrinsic fluorescence of a single-stranded DNA binding protein. J. Mol. Recognit. 2007;20:386–391. doi: 10.1002/jmr.847. [DOI] [PubMed] [Google Scholar]
- 45.Kan ZY, Lin Y, Wang F, Zhuang XY, Zhao Y, Pang DW, Hao YH, Tan Z. G-quadruplex formation in human telomeric (TTAGGG)4 sequence with complementary strand in close vicinity under molecularly crowded condition. Nucleic Acids Res. 2007;35:3646–3653. doi: 10.1093/nar/gkm203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P, editors. Molecular Biology of the Cell. 4th edn. New York, NY: Garland Science; 2002. [Google Scholar]
- 47.Simonsson T, Sjoback R. DNA tetraplex formation studied with fluorescence resonance energy transfer. J. Biol. Chem. 1999;274:17379–17383. doi: 10.1074/jbc.274.24.17379. [DOI] [PubMed] [Google Scholar]
- 48.Ueyama H, Takagi M, Takenaka S. A novel potassium sensing in aqueous media with a synthetic oligonucleotide derivative. Fluorescence resonance energy transfer associated with Guanine quartet-potassium ion complex formation. J. Am. Chem. Soc. 2002;124:14286–14287. doi: 10.1021/ja026892f. [DOI] [PubMed] [Google Scholar]
- 49.Gray RD, Chaires JB. Kinetics and mechanism of K+- and Na+-induced folding of models of human telomeric DNA into G-quadruplex structures. Nucleic Acids Res. 2008;36:4191–4203. doi: 10.1093/nar/gkn379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zimmerman SB, Trach SO. Estimation of macromolecule concentrations and excluded volume effects for the cytoplasm of Escherichia coli. J. Mol. Biol. 1991;222:599–620. doi: 10.1016/0022-2836(91)90499-v. [DOI] [PubMed] [Google Scholar]
- 51.Zimmerman SB, Minton AP. Macromolecular crowding: biochemical, biophysical, and physiological consequences. Annu. Rev. Biophys. Biomol. Struct. 1993;22:27–65. doi: 10.1146/annurev.bb.22.060193.000331. [DOI] [PubMed] [Google Scholar]
- 52.Miyoshi D, Matsumura S, Nakano S, Sugimoto N. Duplex dissociation of telomere DNAs induced by molecular crowding. J. Am. Chem. Soc. 2004;126:165–169. doi: 10.1021/ja036721q. [DOI] [PubMed] [Google Scholar]
- 53.Miyoshi D, Karimata H, Sugimoto N. Hydration regulates thermodynamics of G-quadruplex formation under molecular crowding conditions. J. Am. Chem. Soc. 2006;128:7957–7963. doi: 10.1021/ja061267m. [DOI] [PubMed] [Google Scholar]
- 54.Miyoshi D, Karimata H, Sugimoto N. Hydration regulates the thermodynamic stability of DNA structures under molecular crowding conditions. Nucleosides Nucleotides Nucleic Acids. 2007;26:589–595. doi: 10.1080/15257770701490282. [DOI] [PubMed] [Google Scholar]
- 55.Xue Y, Kan ZY, Wang Q, Yao Y, Liu J, Hao YH, Tan Z. Human telomeric DNA forms parallel-stranded intramolecular G-quadruplex in K+ solution under molecular crowding condition. J. Am. Chem. Soc. 2007;129:11185–11191. doi: 10.1021/ja0730462. [DOI] [PubMed] [Google Scholar]
- 56.Nakano S, Karimata H, Ohmichi T, Kawakami J, Sugimoto N. The effect of molecular crowding with nucleotide length and cosolute structure on DNA duplex stability. J. Am. Chem. Soc. 2004;126:14330–14331. doi: 10.1021/ja0463029. [DOI] [PubMed] [Google Scholar]
- 57.Miyoshi D, Nakamura K, Tateishi-Karimata H, Ohmichi T, Sugimoto N. Hydration of Watson-Crick base pairs and dehydration of Hoogsteen base pairs inducing structural polymorphism under molecular crowding conditions. J. Am. Chem. Soc. 2009;131:3522–3531. doi: 10.1021/ja805972a. [DOI] [PubMed] [Google Scholar]
- 58.Weitzmann MN, Woodford KJ, Usdin K. The development and use of a DNA polymerase arrest assay for the evaluation of parameters affecting intrastrand tetraplex formation. J. Biol. Chem. 1996;271:20958–20964. doi: 10.1074/jbc.271.34.20958. [DOI] [PubMed] [Google Scholar]
- 59.Dai J, Dexheimer TS, Chen D, Carver M, Ambrus A, Jones RA, Yang D. An intramolecular G-quadruplex structure with mixed parallel/antiparallel G-strands formed in the human BCL-2 promoter region in solution. J. Am. Chem. Soc. 2006;128:1096–1098. doi: 10.1021/ja055636a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fernando H, Reszka AP, Huppert J, Ladame S, Rankin S, Venkitaraman AR, Neidle S, Balasubramanian S. A conserved quadruplex motif located in a transcription activation site of the human c-kit oncogene. Biochemistry. 2006;45:7854–7860. doi: 10.1021/bi0601510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Phan AT, Modi YS, Patel DJ. Propeller-type parallel-stranded G-quadruplexes in the human c-myc promoter. J. Am. Chem. Soc. 2004;126:8710–8716. doi: 10.1021/ja048805k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Saha T, Usdin K. Tetraplex formation by the progressive myoclonus epilepsy type-1 repeat: implications for instability in the repeat expansion diseases. FEBS Lett. 2001;491:184–187. doi: 10.1016/s0014-5793(01)02190-1. [DOI] [PubMed] [Google Scholar]
- 63.Cogoi S, Quadrifoglio F, Xodo LE. G-rich oligonucleotide inhibits the binding of a nuclear protein to the Ki-ras promoter and strongly reduces cell growth in human carcinoma pancreatic cells. Biochemistry. 2004;43:2512–2523. doi: 10.1021/bi035754f. [DOI] [PubMed] [Google Scholar]
- 64.Kumari S, Bugaut A, Huppert JL, Balasubramanian S. An RNA G-quadruplex in the 5′ UTR of the NRAS proto-oncogene modulates translation. Nat. Chem. Biol. 2007;3:218–221. doi: 10.1038/nchembio864. [DOI] [PMC free article] [PubMed] [Google Scholar]