Abstract
Heparan sulfates (HS) bind a diversity of protein ligands on the cell surface and in the extracellular matrix and thus can modulate cell signaling. The state of sulfation in glucosamines and uronic acids within the chains strongly influences their binding. We have previously cloned and characterized two human extracellular endoglucosamine 6-sulfatases, HSulf-1 and HSulf-2, which selectively liberate the 6-O sulfate groups on glucosamines present in N, 6-O, and 2-O trisulfated disaccharides of intact HS and heparins. These enzymes serve important roles in development and are upregulated in a number of cancers. To determine whether the Sulfs act on the trisulfated disaccharides that exist on the cell surface, we expressed HSulfs in cultured cells and performed a flow cytometric analysis with the RB4CD12, an anti-HS antibody that recognizes N- and O-sulfated HS saccharides. The endogenously expressed level of the cell surface RB4CD12 epitope was greatly diminished in CHO, HEK293, and HeLa cells transfected with HSulf-1 or HSulf-2 cDNA. In correspondence with the RB4CD12 finding, the N, 6-O, and 2-O trisulfated disaccharides of the HS isolated from the cell surface/extracellular matrix were dramatically reduced in the Sulf-expressed HEK293 cells. We then developed an ELISA and confirmed that the RB4CD12 epitope in immobilized heparin was degraded by purified recombinant HSulf-1 and HSulf-2, and conditioned medium (CM) of MCF-7 breast carcinoma cells, which contain a native form of HSulf-2. Furthermore, HSulf-1 and HSulf-2 exerted activity against the epitope expressed on microvessels of mouse brains. Both HSulf activities were potently inhibited by PI-88, a sulfated heparin mimetic with anti-cancer activities. These findings provide new strategies for monitoring the extracellular remodeling of HS by Sulfs during normal and pathophysiological processes.
Keywords: confocal microscopy, extracellular sulfatase, flow cytometry, heparan sulfate, PI-88
Introduction
Heparan sulfate proteoglycans (HSPGs) are cell surface or extracellular matrix components, which consist of core protein to which one or more glycosaminoglycan chains are covalently attached (Bernfield et al. 1999; Esko and Lindahl 2001). Heparan sulfate (HS) chains and heparins (structural analogs of HS chains) comprise repeating disaccharide units of glucuronic/iduronic acid (GlcA/IdoA) and glucosamine (GlcN) that are modified through a set of deacetylation, epimerization, and sulfation reactions (Gallagher 2001). The N-, 3-O, and 6-O positions of glucosamine and the 2-O position of the uronic acid residues in the HS disaccharide units are potentially substituted by sulfate groups by a group of Golgi-resident HS sulfotransferases (Habuchi et al. 2004). These synthetic reactions along the HS chains are spatially and temporally regulated, conferring upon the chains structural diversity, which underlie important roles in pathological and biological processes (Lin 2004; Parish 2006; Bishop et al. 2007). HS contain highly sulfated domains, “S-domains,” and partially sulfated or non-sulfated domains, which are transitional (Gallagher 2001; Powell et al. 2004). S-Domains are the most common units in heparin. Within the S-domains, a trisulfated disaccharide structure [-IdoA(2-OSO3)-GlcNSO3(6-OSO3)-] occurs. This structure is considered to be a key element in molecular interactions between HS/heparin and many protein ligands, including growth factors and chemokines (Esko and Selleck 2002; Kreuger et al. 2006). We have previously identified and cloned a human extracellular endosulfatase (HSulf-1), an ortholog of QSulf-1 (Dhoot et al. 2001). We also described a closely related protein, designated HSulf-2 (Morimoto-Tomita et al. 2002). Both Sulfs are posttranslationally modified with N-linked glycans (Morimoto-Tomita et al. 2002; Ambasta et al. 2007) and formylglycines (Cosma et al. 2003; Dierks et al. 2003), processed by furin endoproteases and can be secreted (Morimoto-Tomita et al. 2002; Tang and Rosen 2009). The Sulfs remove 6-O sulfates on glucosamine residues in the trisulfated disaccharides of heparin (Morimoto-Tomita et al. 2002; Saad et al. 2005) and heparan sulfate (Ai et al. 2003; Viviano et al. 2004). Sulf-2 mobilizes heparin-bound vascular endothelial growth factor (VEGF), FGF-1, and SDF-1 (Uchimura et al. 2006a). The enzyme is proangiogenic in the chick chorioallantoic membrane assay, presumably through its ability to reverse the association between angiogenic factors and heparin/HSPGs (Morimoto-Tomita et al. 2005). Studies of quail and Xenopus embryos (Dhoot et al. 2001; Freeman et al. 2008) and of Sulf-deficient mice have demonstrated developmental roles for the Sulfs (Lamanna et al. 2006; Ai et al. 2007; Holst et al. 2007; Lum et al. 2007). Furthermore, increasing evidence implicates the Sulfs in cancer, in some cases augmenting cancer cell growth and in others inhibiting it (Lai et al. 2004, 2008; Dai et al. 2005; Narita et al. 2007; Nawroth et al. 2007).
Antibodies against HS have been established as useful tools to evaluate the expression and localization of HS in cultures and tissues. The 10E4 monoclonal anti-heparan sulfate antibody (David et al. 1992) has been widely used to detect HS in biological and pathological sets. Another monoclonal anti-heparan sulfate antibody, HepSS-1, has also been characterized (van den Born et al. 2005). The HS epitopes of recently developed phage display antibodies have been defined using derivatives of HS and heparins (van Kuppevelt et al. 1998). One of them, RB4CD12, recognizes N- and O-sulfated saccharides of HS/heparin (Jenniskens et al. 2000; Dennissen et al. 2002). Its recognition epitope is proposed to be [-GlcNSO3(6-OSO3)-IdoA(2-OSO3)-GlcNSO3(6-OSO3)-], a trisulfated disaccharide-containing HS oligosaccharide (Jenniskens et al. 2002). In the present study, we have employed RB4CD12 to demonstrate that HSulfs exhibit 6-O-endoglucosamine activities against HS on the cell surface of transfected cells and in the extracellular matrix (ECM) of mouse brain microvessels, as well as on immobilized heparin in an ELISA. We then use RB4CD12 to show that both Sulfs are potently inhibited by PI-88, an antiangiogenic and antimetastatic agent.
Results
Cell surface expression of the RB4CD12 epitope is diminished by transfection of HSulf cDNA
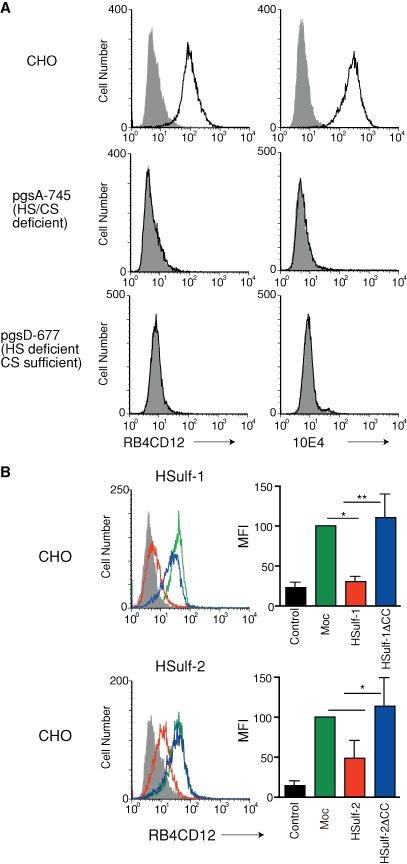
RB4CD12 is a single-chain variable fragment (scFv) antibody selected for reactivity to skeletal muscle heparin/HS glycosaminoglycans utilizing a phage display system (Jenniskens et al. 2000). RB4CD12 recognizes highly sulfated HS/heparin saccharide structures present in S-domains of HS and heparin. Antibody binding depends on all three sulfate modifications (Dennissen et al. 2002). The 10E4 mAb is widely used as a reporter for HS expression in cells and tissues. The 10E4 epitope is known to involve HS oligosaccharides that contain GlcNSO3 and GlcNAc residues with O-sulfations dispensable (van den Born et al. 2005). We found by flow cytometry that the RB4CD12 and 10E4 epitopes were expressed on the cell surface of wild-type CHO cells (Figure 1A). To confirm that the RB4CD12 epitope on CHO cells was dependent on HS structures and not on chondroitin sulfate (CS), we employed mutant CHO cells in which the biosynthesis of glycosaminoglycans was disrupted. The pgsA-745 cell line is deficient in both HS and CS synthesis (Esko et al. 1985). The pgsD-677 cell line is HS deficient but CS sufficient. Staining with RB4CD12 antibody was not observed in either pgsA-745 or pgsD-677 cells (Figure 1A) but was present in wild-type cells (Jenniskens et al. 2000). 10E4 staining of these cells showed the same pattern (Figure 1A).
Fig. 1.
Cell surface expression of the RB4CD12 heparan sulfate epitope in wild-type, heparan sulfate-deficient, and HSulf-transfected CHO cells. (A) Wild-type CHO cells (CHO), CHO deficient in glycosaminoglycan xylosylation initiation (pgsA-745), or CHO deficient in heparan sulfate chain polymerization (pgsD-677) were analyzed by flow cytometry with RB4CD12 scFv or 10E4. (B) Flow cytometry of CHO cells stably transfected with pcDNA3.1 empty vector alone (Mock); cDNA of HSulf-1 (HSulf-1), HSulf-1ΔCC (HSulf-1ΔCC), HSulf-2 (HSulf-2), or HSulf-2ΔCC (HSulf-2ΔCC) were analyzed for staining with RB4CD12. These data are representative of three independent determinations. Mean fluorescence intensity (MFI) relative to “Mock” of three independent determinations is shown. MPB49, a non-HS binding scFv antibody, was used as “control”. **, P < 0.001; *, P < 0.01; #, P < 0.05.
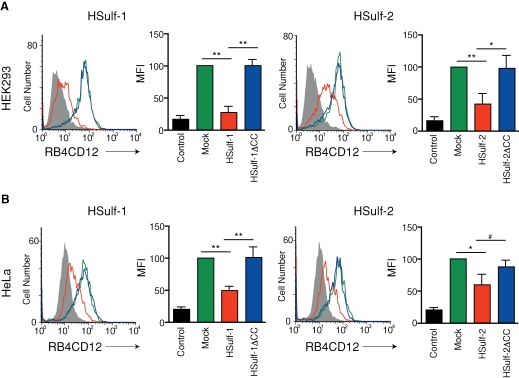
As reviewed above, HSulf-1 and HSulf-2 remove 6-O-sulfates from heparin/HS chains (Morimoto-Tomita et al. 2002; Ai et al. 2003; Viviano et al. 2004). Analysis of HS chains in cells derived from Sulf null mice is also consistent with this enzyme specificity (Lamanna et al. 2006, 2008; Ai et al. 2007). To determine whether the cell surface expression level of the sulfated saccharide structure was reduced by the HSulfs, we established CHO cells stably transfected with cDNA encoding HSulf-1 or HSulf-2 and performed flow cytometry with RB4CD12. A non-HS control staining (MPB49 antibody) was employed to show the basal level of the staining signal. HSulf-1 diminished the cell surface expression to 25% of the level seen in the unaffected mock control, as quantified by mean fluorescence intensity (MFI) (Figure 1B). HSulf-2 reduced expression to 50% of the background level (Figure 1B). CHO cells expressing inactive forms of enzymes (HSulf-1ΔCC and HSulf-2ΔCC) retained RB4CD12 staining equivalent to that of the mock transfectant (Figure 1B). We also examined the effects of the Sulfs on the expression of the RB4CD12 epitope in transiently transfected HEK 293 and HeLa cells. Transfection with either HSulf-1 or HSulf-2 cDNA significantly reduced the RB4CD12 epitope on the surface of these cells (to 25% in HEK293 with HSulf-1, 40% in HEK293 with HSulf-2, 50% in HeLa with HSulf-1, 60% in HeLa with HSulf-2), consistent with the results obtained with CHO stable transfectants (Figure 2A and B). Cells transfected with HSulf-1ΔCC or HSulf-2ΔCC plasmids sustained the epitope at the control level (Figure 2A and B). We also stained Sulf-transfected HEK293 and HeLa cells with the HepSS-1 antibody (van den Born et al. 2005). HSulf-1 expression significantly augmented staining with the antibody in HEK293 (3.2-fold) and HeLa cells (1.2-fold) (Supplementary Figure 1A). HSulf-2 increased the level of HepSS-1 staining on both cell types (1.5-fold in HEK293, 1.2-fold in HeLa cells) (Supplementary Figure 1B). Expression of inactive forms of HSulf-1 and HSulf-2 (HSulf-1ΔCC and HSulf-2ΔCC) did not alter staining by HepSS-1 (Supplementary Figure 1A and B). We examined the structure of HS isolated from the cell surface/ECM of Sulf-transfected HEK293 cells. The disaccharide composition of the isolated HS was analyzed. Transfection with either HSulf-1 or HSulf-2 cDNA substantially reduced the trisulfated disaccharide units of the HS (to 19% and 42% of the “Mock” control level with HSulf-1 and HSulf-2, respectively) (Table I). A moderate reduction was observed in total 6-O-sulfation of the isolated HS (to 75% and 85% of the “Mock” control level with HSulf-1 and HSulf-2, respectively) (Table I).
Fig. 2.
Cell surface expression of the RB4CD12 epitope in HSulf-transfected HEK293 and HeLa cells. HEK293 (A) and HeLa (B) cells transiently transfected with a pcDNA3.1 empty vector alone (Mock); cDNA of HSulf-1 (HSulf-1), HSulf-1ΔCC (HSulf-1ΔCC), HSulf-2 (HSulf-2), or HSulf-2ΔCC (HSulf-2ΔCC) were analyzed by flow cytometry with RB4CD12. These data are representative of at least three independent experiments. Mean fluorescence intensity (MFI) relative to “Mock” of three independent experiments is shown. MPB49, a non-HS binding scFv antibody, was used as “control”. **, P < 0.001; *, P < 0.01; #, P < 0.05.
Table I.
Disaccharide composition of extracellular HS of HSulf-transfected HEK 293 cells. Total extracellular HS was degraded with a mixture of heparinases I, II, and III. The products were subjected to reverse-phase ion pair chromatography with a post-column fluorescent labeling. The values are representative of two independent experiments
| Unsaturated disaccharidea | |||||||
|---|---|---|---|---|---|---|---|
| Transfectant | ΔDi-0S | ΔDi-NS | ΔDi-6S | ΔDi-(N,6)diS | ΔDi-(N,2)diS | ΔDi-(N,6,2)triS | Total 6-O-sulfationb |
| Mock | 49.7 (100) | 14.5 (100) | 13.5 (100) | 9.1 (100) | 9.2 (100) | 3.9 (100) | 26.5 (100) |
| HSulf-1 | 55.1 (110.7) | 15.8 (109.1) | 14.3 (105.7) | 4.8 (52.8) | 9.2 (100.3) | 0.8 (19.2) | 19.8 (74.7) |
| HSulf-1 ΔCC | 50.9 (102.3) | 14.5 (100) | 13.4 (99.5) | 9.3 (102.3) | 7.8 (84.2) | 4.1 (104.2) | 26.8 (101.1) |
| Hsulf-2 | 51.6 (103.6) | 15.7 (108.2) | 14.0 (103.4) | 6.9 (75.4) | 10.2 (111.1) | 1.6 (42.1) | 22.5 (84.9) |
| HSulf-2 ΔCC | 49.5 (99.6) | 15.2 (104.9) | 13.5 (99.9) | 9.2 (101.4) | 7.7 (83.5) | 4.8 (123.2) | 27.5 (103.8) |
aPercentage of total disaccharides (% of “Mock”).
bPercentage of total 6-O-sulfation (% of “Mock”) determined by the sum of the percentages of 6-O-sulfated disaccharide products.
The RB4CD12 epitope on heparin is degraded by recombinant HSulfs
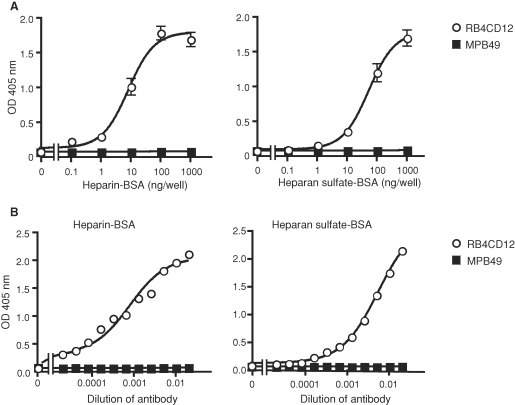
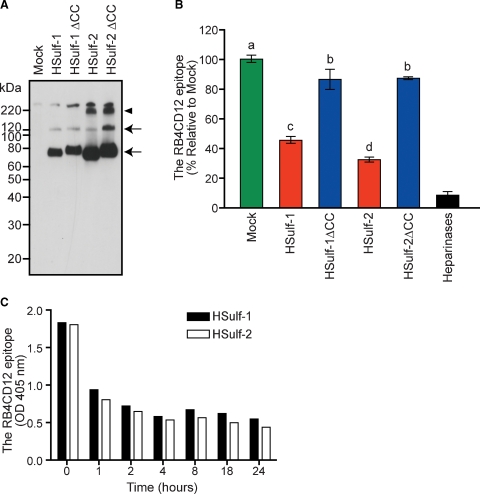
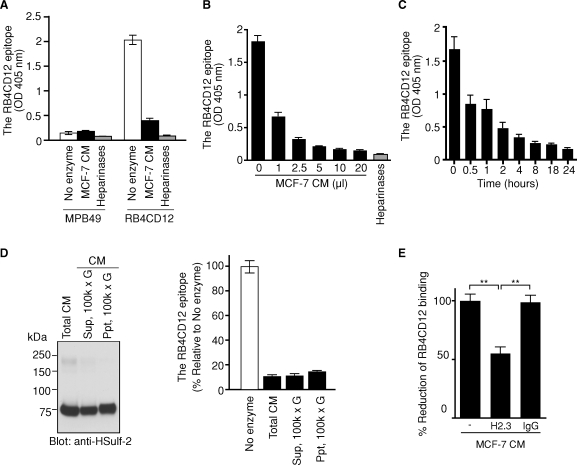
We then asked whether HSulf-1 and HSulf-2 could reduce the RB4CD12 epitope in a cell-free ELISA. The RB4CD12 antibody recognized heparin-BSA (>1 ng/well) and heparan sulfate-BSA (>10 ng/well) immobilized onto plastic wells (Figure 3A) in a concentration-dependent manner (Figure 3B). Amino terminal FLAG-tagged, carboxy terminal His-tagged HSulf-1 and HSulf-2, and corresponding inactive mutants (HSulf-1ΔCC and HSulf-2ΔCC) were purified from the conditioned medium (CM) of transfected HEK293 cells on Ni-NTA beads (Figure 4A). Immunoblotting with an anti-FLAG antibody revealed two specific protein bands in HSulf-1 and HSulf-1ΔCC (∼135 and ∼75 kDa) and three specific bands in HSulf-2 and HSulf-2ΔCC (∼240, ∼135, and ∼75 kDa) (Figure 4A). It has recently been reported that the 75 kD, ∼135 kD, and ∼240 kD bands correspond to the N-terminal subunit, the heterodimer of N- and C-terminal subunits, and a higher order oligomer, respectively (Tang and Rosen 2009). RB4CD12 binding to heparin-BSA was substantially reduced by pre-treatment of the heparin-BSA with the purified HSulf-1 (reduced to 45% of control) or HSulf-2 (reduced to 35% of control), (Figure 4B). HSulf-1ΔCC and HSulf-2ΔCC had only minor effects on RB4CD12 binding. The effects of the inactive Sulfs in the RB4CD12 assays might be attributable to a competing heparin-binding activity of Sulf protein due to its basic hydrophilic domain (Morimoto-Tomita et al. 2002; Ai et al. 2006; Frese et al. 2009). A mixture of heparinases was used as a positive control for digestion of immobilized heparin and was found to reduce the RB4CD12 recognition by >90% (Figure 4B). Both HSulf-1 and HSulf-2 reduced binding of RB4CD12 to heparin-BSA in a time-dependent manner (Figure 4C).
Fig. 3.
ELISA for the RB4CD12 epitope with immobilized heparin and heparan sulfate. (A) RB4CD12 was tested for binding to varying amounts of immobilized heparin-BSA or heparan sulfate-BSA by ELISA. (B) Reaction of serially diluted RB4CD12 to immobilized heparin-BSA (10 ng/well) or heparan sulfate-BSA (100 ng/well). MPB49 is a non-HS binding phage display antibody used as a control. Data in (B) are representative of two independent trials.
Fig. 4.
Expression of recombinant HSulf-1 and HSulf-2 and their effects on the RB4CD12 epitope in immobilized heparin. (A) Recombinant HSulfs (HSulf-1 and HSulf-2) and their enzymatically inactive forms (HSulf-1ΔCC and HSulf-2ΔCC) tagged with N-terminal FLAG and C-terminal His were produced in HEK293 cells. The proteins were purified on Ni-NTA beads and subjected to immunoblotting for the FLAG tag. The “Mock” control was based on testing bead-bound materials from vector control transfected HEK293 cells. Specific bands detected in HSulf-1 and HSulf-1ΔCC (∼130 and ∼75 kDa, arrows) and in HSulf-2 and HSulf-2ΔCC (∼240, ∼130 and ∼75 kDa, arrows and an arrowhead) are denoted. Note that a slight shift of bands in HSulf-1ΔCC and HSulf-2ΔCC was also seen in our previous work (Uchimura et al. 2006a). (B) The purified Sulfs were tested for their effects on immobilized heparin-BSA in the RB4CD12 epitope ELISA. The Sulfs were prepared from 0.5 mL of CM collected from the transfected HEK293 cells. A mixture of bacterial heparinases I, II, and III was used as a control for the epitope degradation (heparinases). Different letters among treatments indicate significant differences (P < 0.05). (C) The Sulfs purified from 0.5 mL of the CM were tested as a function of reaction time in the RB4CD12 ELISA.
The RB4CD12 epitope on heparin is degraded by MCF-7 conditioned medium
We have previously shown that MCF-7 cells, a human breast carcinoma cell line, produce Sulf-2 and secretes the protein into CM as an active form (Morimoto-Tomita et al. 2005; Uchimura et al. 2006a). To determine if a native form of Sulf exhibited the same activity as the recombinant enzyme, we employed the MCF-7 CM. Pretreatment of immobilized heparin-BSA with a fixed concentration of MCF-7 CM produced a >80% reduction in RB4CD12 binding (Figure 5A). The activity was both concentration- and time-dependent (Figure 5B and C). After centrifugation of MCF-7 CM at 100,000 × g for 1 h, HSulf-2 protein was detected in both the supernatant and the precipitate (Figure 5D). Both fractions exhibited activity (Figure 5D). By scanning the blot in Figure 5D, the Sulf activities relative to HSulf-2 proteins in both fractions were determined. The specific activities in the supernatant and the precipitate were comparable (data not shown). Preclearing the CM with the H2.3 anti-HSulf2 antibody reduced the activity to 55%, while control IgG did not, confirming that the reduction in RB4CD12 binding was due to HSulf-2 (Figure 5E). Thus, a significant portion of secreted Sulf-2 meets a stringent test of solubility and retains its enzymatic activity.
Fig. 5.
The effect of MCF-7 conditioned medium (CM) on the RB4CD12 epitope in heparin. (A) The effect of MCF-7 conditioned medium (10 μL, overnight incubation time) on the RB4CD12 epitope in immobilized heparin-BSA. (B and C) The activity of MCF-7 CM on the RB4CD12 epitope as a function of CM volume (1 h incubation) and reaction time (1 μL of CM). MPB49, a non-HS binding scFv antibody, was used as a control for RB4CD12. A mixture of bacterial heparinases I, II, and III was used as a positive control to produce epitope degradation (heparinases). (D) HSulf-2 was analyzed by immunoblotting in unfractionated MCF-7 CM (5 μg protein, “total CM”), the supernatant fraction (5 μg protein, “Sup, 100 k × g”) and the precipitated fraction (1.5 μg protein, “Ppt, 100 k × g”) of the CM after a centrifugation at 100,000 × g for 1 h. HSulf-2 protein was detected by H2.3 anti-HSulf2 antibody (Morimoto-Tomita et al. 2005). The activity of each fraction (with the same amount of proteins as above) was analyzed by the RB4CD12 ELISA (overnight incubation). (E) Inhibition of the activity in MCF-7 CM by the H2.3 anti-HSulf2 antibody. Two micrograms of H2.3 antibody or rabbit IgG was incubated with MCF-7 CM (1 μL, 4 h incubation). **, P < 0.001.
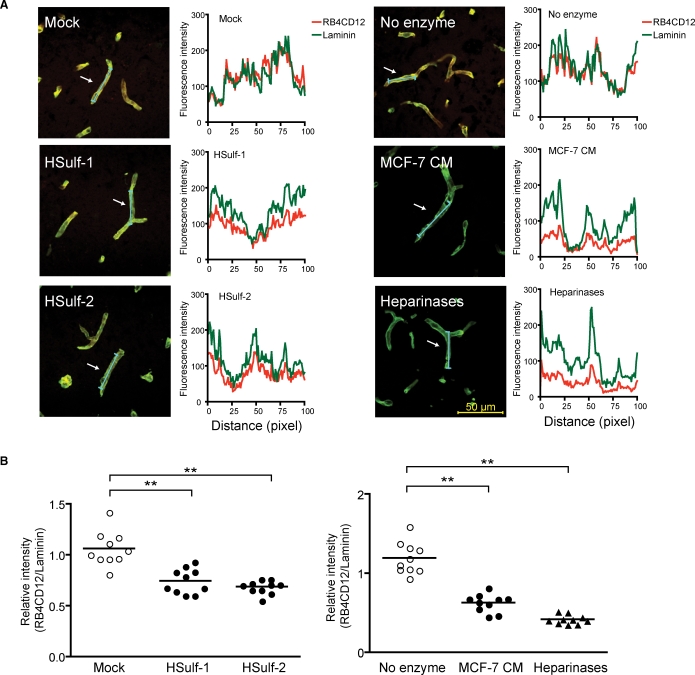
HSulfs degrade the RB4CD12 epitope in tissue
To determine whether the HSulfs could degrade the RB4CD12 epitope in tissues, we treated cryostat-cut sections of mouse brains with HSulfs. RB4CD12 stained microvessels in sections of mouse brain (Figure 6A). The staining co-localized with laminin, a marker of basement membranes (Figure 6A). No specific staining was observed when RB4CD12 substituted with MPB49 (data not shown). Treatment of sections with purified HSulf-1, purified HSulf-2, or MCF-7 CM produced significant reductions in RB4CD12 staining of microvessels. Similarly, treatment with heparinases also diminished staining. For quantification, the intensity of RB4CD12 staining was normalized to that of laminin (see Material and methods). HSulf-1 and HSulf-2 reduced RB4CD12 staining to 70% and 65%, respectively, of the “Mock” treatments (Figure 6B). MCF-7 CM and a mix of heparinases reduced RB4CD12 staining to 52% and 35%, respectively, of the “no enzyme” treatments (Figure 6B).
Fig. 6.
Sulf degradation of the RB4CD12 epitope in the basement membrane of mouse brain microvessels. (A) Cryostat-cut sections of mouse brains were incubated overnight with recombinant HSulf-1 (0.4 μg) and HSulf-2 (0.4 μg) prepared from CM of transfected HEK293 cells (HSulf-1, HSulf-2), buffer only (no enzyme), or CM of MCF-7 human breast cancer cells (MCF-7 CM). The Ni-NTA resin-bound materials that were prepared from HEK293 cells transfected with the empty vector were eluted and used (Mock). A mix of bacterial heparinases (Heparinases) served as a positive control. RB4CD12 binding was visualized by a Cy3-conjugated anti-VSV tag antibody with a confocal laser scanning microscopy (red). Basement membranes of vessels were stained by an anti-laminin antibody in conjunction with a Cy2-conjugated secondary antibody (green). The signal intensities along the line markers (blue lines) indicated by arrows in the images were measured for RB4CD12 and laminin staining as described in Material and methods. Representative images and measurements of 10 randomly selected vessels are shown. (B) The relative intensity of RB4CD12 to laminin is shown, n = 10 vessels per treatment. Data are from two experiments with five vessels analyzed in distinct brain specimens from two donors for each treatment. Each symbol represents one vessel image; small horizontal lines indicate the mean. Magnification used 630×. **, P < 0.001.
PI-88 inhibits the Sulfs
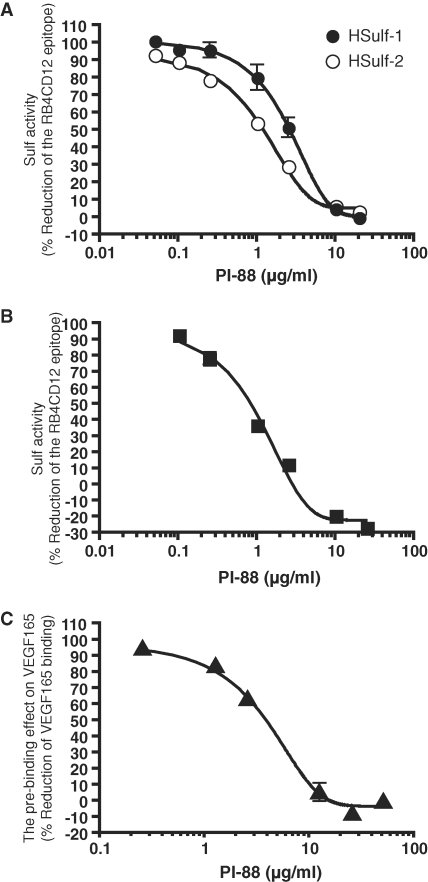
PI-88 is a phosphomannopentaose polysulfate, which was developed as an inhibitor of heparanase, an endoglycosidase which cleaves HS chains (Khachigian and Parish 2004). Since PI-88 was designed as a heparin mimetic, we wanted to know whether it might inhibit the Sulfs. We first tested its activity in the heparin-BSA ELISA described above. Reduction of the RB4CD12 epitope produced by HSulf-1, HSulf-2 (Figure 7A), or MCF-7 CM (Figure 7B) was efficiently inhibited by PI-88 in a dose-dependent manner. We previously showed that treatment of heparin-BSA with MCF-7 CM reduced the subsequent binding of VEGF165 (Uchimura et al. 2006a). When PI-88 was mixed with MCF-7 CM, its inhibitory activity in this assay was blunted (Figure 7C). IC50 and LogIC50 values of PI-88 in these various assays are summarized in Table II.
Fig. 7.
Inhibition of HSulf-1 and HSulf-2 activities by PI-88. Purified recombinant Sulfs (HSulf-1, 0.4 μg and HSulf-2, 0.4 μg) (A) and MCF-7 CM (10 μL) were used (B). PI-88 was tested at varying concentrations for its ability to inhibit the action of the Sulfs and the CM on the RB4CD12 epitope of immobilized heparin by ELISA as described in Material and methods. (C) PI-88 at varying concentrations was tested for its ability to inhibit the action of MCF-7 CM (10 μL) to reduce the binding of VEGF165 to immobilized heparin-BSA in a solid phase binding assay (Uchimura et al. 2006a). PI-88 exhibited negligible activity when assays were performed without the CM (data not shown). Over the concentration of 0.1 μg/mL (both HSulf-1 and HSulf-2) (A), 0.25 μg/mL (MCF-7 CM) (B), and 1.25 μg/mL (MCF-7 CM) (C), the data are statistically significant (P < 0.05). The values were analyzed by one-way ANOVA with Dunnett's test against the value obtained without PI-88.
Table II.
Effects of PI-88 on Sulf activities
| IC50b | |||
|---|---|---|---|
| Enzyme | Analyzed effecta | (μg/mL) | LogIC50c |
| HSulf-1 | Degradation of RB4CD12 epitope | 2.6 | 2.5 ± 0.1 |
| HSulf-2 | Degradation of RB4CD12 epitope | 1.2 | 0.9 ± 0.1 |
| MCF-7 CM | Degradation of RB4CD12 epitope | 0.6 | 0.9 ± 0.2 |
| Pre-binding effect on VEGF165 | 3.6 | 2.9 ± 0.1 |
aThese effects were measured and shown in Figure 7.
bThe concentration of PI-88 that inhibited the Sulf enzymatic activity by 50%.
cMeans ± SE.
Discussion
A large body of evidence has established that the Sulfs can modulate the signaling activities of a variety of HS-binding factors (Wnts, FGFs, noggin, HGF, GDNF) during development (Ai et al. 2003, 2007; Viviano et al. 2004; Freeman et al. 2008) and tumor growth (Lai et al. 2004; Dai et al. 2005; Nawroth et al. 2007). Besides these establishments, a more direct approach for the detection of the glucosamine-6-endosulfatase activities of the Sulfs has been anticipated. Here using a phage display-derived antibody, RB4CD12, we have devised a general method, compatible with both flow cytometry and immunohistochemistry, to monitor the actions of Sulf-1 and Sulf-2 on the cell surface and the ECM. RB4CD12 recognizes highly sulfated domains within HS since it reacts with highly sulfated oligosaccharides with most likely dependence on the presence of 6-O-, N-, and 2-O- sulfate esters (Dennissen et al. 2002). The antibody prefers oligosaccharides containing both GlcNSO3(6-OSO3) and IdoA(2-OSO3) (Dennissen et al. 2002; Jenniskens et al. 2002). As predicted from the specificity of the antibody, stable (CHO) or transient (HEK293, HeLa) transfection of Sulfs reduced the cell surface expression of the RB4CD12 epitope. The reduction in the N-, 6-O-, and 2-O- trisulfated disaccharide units [-IdoA(2-OSO3)-GlcNSO3(6-OSO3)-] (by 58–81%) observed in cell surface/ECM HS of HSulf-transfected cells is consistent with the reduction of the RB4CD12 epitope at the cell surface. The moderate loss (15–25%) of overall HS 6-O-sulfation in the HS of HSulf-transfected cells suggested that RB4CD12 specifically recognizes the trisulfated disaccharide structure as a component of the HS oligosaccharide [-GlcNSO3(6-OSO3)-IdoA(2-OSO3)-GlcNSO3(6-OSO3)-]. The expected parallel increase in N- and 2-O-disulfated disaccharide units in cell surface/ECM HS of HSulf-transfected cells was not observed. The basis for this result remains to be explained. We found that Sulf transfection generally caused a moderate increase in cell surface staining by HepSS-1. The increase in staining may reflect newly formed product via the enzymatic action of the HSulfs. Another possibility is that the accessibility of the antibody to the epitope was increased by liberation of 6-O-sulfate groups from HS chains.
It was possible that the effects we observed on the RB4CD12 epitope of transfected cells were due to primary action of the Sulfs but secondary signaling responses in the cells. We therefore carried experiments on heparin in a cell-free setting. The trisulfated disaccharide unit is a major structural unit (∼80% of total disaccharide units) in heparin. To show direct effects of the Sulfs on the RB4CD12 epitope, we established a solid-phase ELISA employing immobilized heparin as a substrate. As seen in Figure 4C, both HSulf-1 and HSulf-2 diminished the epitope in immobilized heparin at a saturation level by 70–75% binding, consistent with our previous demonstration that the Sulfs can degrade 80% of the total trisulfated disaccharide units in soluble heparins (Morimoto-Tomita et al. 2002). In the same ELISA, MCF-7 CM also exhibited strong degradation of the RB4CD12 epitope (by ∼80%) (Figure 5). The activity of the CM against the RB4CD12 epitope in immobilized heparin proceeded linearly up to 8 h. We attribute this activity to Sulf-2, as we have shown the specific inhibition of the activity by the H2.3 anti-HSulf2 antibody and have previously demonstrated that the endoglucosamine-6-sulfatase activity of MCF-7 CM resides in native Sulf-2 (Uchimura et al. 2006a). The Sulf activity and the HSulf-2 protein in MCF-7 CM were detected in both the supernatant and the precipitate after a centrifugation at 100,000 × g. The precipitate fraction contains the most of extracellular membrane vesicle components (Kim et al. 2002). Our data suggest that HSulf-2 might be able to associate with extracellular membrane vesicles, such as exosomes (Thery et al. 2002). We found that recombinant and native Sulfs reduced the RB4CD12 epitope on the basement membrane of brain microvessels. The degree of activity of the Sulfs could possibly be reflected by their enzyme stability and/or post-translational modifications (Zito et al. 2005). Besides verifying the activity of Sulfs on HS in a tissue context, this finding suggests the possible utility of this antibody as an immunohistochemical reporter of Sulf activity in situ.
PI-88 inhibits metastasis and angiogenesis during the growth and progression of solid tumors and is currently in clinical trials for several cancers. (Parish et al. 1999; Joyce et al. 2005). PI-88 may act through its inhibition of heparanase, an enzyme implicated in the remodeling of extracellular matrix by cancer cells (Joyce et al. 2005; Ilan et al. 2006; Vlodavsky et al. 2007). However, PI-88 also blocks a number of growth factor–HS interactions and may interfere with the signaling activities of these factors. We found that PI-88 also inhibited the Sulfs with IC50s in the range of 0.6–3.6 μg/mL (Table II), which are comparable to that for heparanase (2 μg/mL) (Parish et al. 1999). For several cancers, significant subsets of patients show upregulation of Sulf transcripts in tumors (Iacobuzio-Donahue et al. 2003; Li et al. 2005; Morimoto-Tomita et al. 2005; Nawroth et al. 2007; Lai et al. 2008). Furthermore, Sulf-2 is proangiogenic and can promote the growth of cancer cells in vitro and in vivo (Nawroth et al. 2007; Lai et al. 2008). Thus, the inhibitory effects of PI-88 on tumor progression might be attributable, at least in part, to inactivation of the Sulfs. The development of more selective Sulfs inhibitors should allow testing of this possibility. The assays described herein may provide the basis for screening chemical libraries for further examples of Sulf inhibitors.
Material and methods
Materials
The following materials were obtained commercially from the source indicated. Heparin conjugated with bovine serum albumin (Heparin-BSA), heparinases (I, II and III), monoclonal anti-VSV (vesicular stomatitis virus) glycoprotein-Cy3™ antibody, and monoclonal anti-FLAG® antibody were from Sigma (St. Louis, MO); CHO wild-type (CHO-K1), CHO mutants (pgsA-745, pgsD-677), and HeLa cells were from ATCC (Manassas, VA); HEK293 and MCF-7 cells were from Japan Health Science Research Resources Bank (Osaka, Japan); polyclonal rabbit anti-VSV-G antibody was from Bethyl Laboratories (Montgomery, TX); Cy™3-conjugated goat anti-Mouse IgM μ chain, alkaline phosphatase-conjugated polyclonal goat anti-rabbit IgG (H+L) and Cy™2-conjugated goat anti-Rabbit IgG (H+L) were from Jackson Immuno Research Laboratories (West Grove, PA); recombinant human VEGF-165 and polyclonal goat anti-human VEGF antibody were from R&D Systems (Minneapolis, MN); a biotinylated swine anti-goat IgG (H+L) antibody, a streptavidin conjugated with alkaline phosphatase, horseradish peroxidase-conjugated goat anti-mouse IgG1 antibody were from Caltag Laboratories (Burlingame, CA); 10E4 and HepSS-1 monoclonal anti-heparan sulfate antibodies were from Seikagaku (Tokyo, Japan). The RB4CD12 phage display-derived anti-heparan sulfate antibody (also known as HS3A8) was produced in a VSV-tag version and purified as described previously (Dennissen et al. 2002). DEAE-Sepharose™ was from GE Healthcare UK Ltd (Buckinghamshire, England). Heparan sulfate conjugated with BSA was prepared as described previously (Uchimura et al. 2006a). PI-88 (Parish et al. 1999) was provided by Progen Pharmaceutics Ltd.
Cell culture and transfection
CHO cells were cultured in Ham's F12 medium containing 10% fetal bovine serum (FBS) at 37°C in 5% CO2. HEK293 and HeLa cells were cultured in Dulbecco's Modified Eagle Medium containing 10% FBS. CHO, HEK293, and HeLa cells were transfected with pcDNA3.1 Myc/His-HSulf1, -HSulf2, -HSulf1ΔCC or -HSulf2ΔCC expression plasmid (Morimoto-Tomita et al. 2002) using the FuGENE® 6 transfection reagent according to the manufacturer's instructions (Roche Diagnostics, Basel, Switzerland). HSulf-1ΔCC and HSulf-2ΔCC are enzymatically inactive with two cysteines mutated to alanines (Morimoto-Tomita et al. 2002). Stable CHO transfectants were selected with 1 mg/mL G418 (Invitrogen). In transient transfections, the transfection efficiency was measured by counting the percentage of green-fluorescent cells in parallel experiments with an expression plasmid encoding green fluorescent protein. The efficiency was determined to be ∼60% in HEK293 and HeLa cells.
Preparation of HSulfs
N-terminal-FLAG-, C-terminal-His-tagged versions of HSulf1, HSulf2, HSulf1ΔCC, or HSulf2ΔCC in the pSecTag plasmid (Uchimura et al. 2006a) were prepared. HEK293 cells were transiently transfected with FuGENE® 6 and then grown in OptiMEM (Invitrogen) at 37°C in 5% CO2 for 48 h. Ten milliliters of conditioned medium (CM) was collected and mixed with 100 μL of a Ni-NTA agarose resin (Pro-Bond™ resin, Invitrogen) followed by rotation at 4°C for 4 h. After incorporation into a chromatography column (Poly-Prep® column, Bio-Rad), the resin was washed with the buffer containing 50 mM HEPES, pH 7.5, and 0.05% Tween 20 (20 times the volume of the resin). The resin-bound proteins were eluted with the buffer containing 50 mM HEPES, pH 7.5, 10 mM MgCl2, and 100 mM imidazole (double volume of the resin). The “Mock” control was prepared from CM of HEK293 cells transfected with an empty vector. Native HSulf-2 was obtained from CM of cultured MCF-7 cells, a human breast carcinoma cell line (Morimoto-Tomita et al. 2005). The cells were grown in RPMI-1640 containing 10% FBS in a 150 cm2 flask. Cells (80% confluent) were rinsed with phosphate-buffered saline (PBS) and OptiMEM, and then incubated in 25 mL of OptiMEM at 37°C in 5% CO2 for 72 h. The CM was collected and then 100-fold concentrated on a Centricon-30 (Millipore). Subsequently, the concentrated CM was dialyzed into 50 mM HEPES, pH 7.5. The CM was fractionated into a supernatant (soluble fraction) and a precipitate (containing membrane fragments) by centrifugation (100,000 × g for 1 h at 4°C). The supernatant was 100-fold concentrated as above. The precipitate was dissolved in the buffer containing 50 mM HEPES, pH 7.5, and 0.05% Tween-20.
Cell surface expression of heparan sulfate epitopes
Adherent cells were detached from culture flasks by incubating in 0.02% EDTA/PBS (Sigma) at 37°C for 10 min and then dissociated into monodispersed cells. Cells (1 × 105) were collected and suspended in 100 μL 1× FACS buffer (1% BSA and 0.1% NaN3 in PBS, filtered by a 0.22 μm filter unit) containing an Fc Blocker (eBioscience) and incubated at 4°C for 15 min. Then, 10E4 (10 μg/mL), HepSS-1 (20 μg/mL), or RB4CD12 (1:50 dilution) were added to the suspension. Parallel staining was also done with mouse IgM (eBioscience, San Diego, CA), as isotype-matched control for 10E4 and HepSS-1, or with MPB49 (1:50 dilution), a non-HS binding phage display antibody, as a control for RB4CD12. After incubation at 4°C for 30 min, cells were washed twice with the 1× FACS buffer. Cells were then incubated in 100 μL of 1× FACS buffer containing Cy3-conjugated goat anti-Mouse IgM (3 μg/mL, for 10E4 and HepSS-1) or Cy3-conjugated anti-VSV (4 μg/mL, for RB4CD12 and MPB49) at 4°C for 30 min. Subsequently, cells were washed as above, fixed in 2% paraformaldehyde in PBS and then transferred into a 5 mL polystyrene round-bottom tube capped with a cell-strainer cap (BD, Franklin Lakes, NJ). Antibody-bound cells were then analyzed by FACS using a FACSCalibur flow cytometer, CellQuest software (BD), and FlowJo software (Tree Star, Ashland, OR).
Preparation and structural analysis of HS
HSulf-transfected adherent cells (2 × 106 cells) cultured in OptiMEM were trypsinized for 15 min. The supernatants containing cell surface/ECM HS were collected. The HS was purified by a DEAE-Sepharose column chromatography as described (Habuchi et al. 2007). The disaccharide compositions of the HS were determined by reverse-phase ion pair chromatography with a post-column fluorescent labeling as described previously (Habuchi et al. 2007).
ELISA for Sulf activities and PI-88 inhibition
To immobilize heparin or heparan sulfate, 100 ng/mL of heparin-BSA or 1 μg/mL of heparan sulfate-BSA in PBS was added to the wells (100 μL/well) of a 96-well plate (Immulon 2HB, Dynex Laboratories). The plate was placed at 4°C overnight. The wells were washed three times with PBS containing 0.1% Tween-20 (PBS-T) and then blocked with 3% BSA (Sigma) in PBS containing 0.01% NaN3 at RT for 2 h. The wells were washed as above and incubated with bead-bound or purified FLAG-His-tagged enzymes or concentrated MCF-7 CM in 100 μL of a reaction mixture containing 50 mM of HEPES, pH 7.5, 10 mM of MgCl2 at 37°C overnight. To test for inhibitory effects of PI-88 on Sulfs, various amounts of PI-88 were incubated together with the reaction mixture. The wells were washed three times with PBS-T and then incubated with 100 μL/well of primary antibody RB4CD12 (1:750 diluted by 0.1% BSA in PBS) at RT for 1 h. The wells were washed as above and incubated with 100 μL/well of secondary rabbit anti-VSV antibody (1 μg/mL in 0.1% BSA in PBS) at RT for 45 min. Then, the wells were washed and incubated with 100 μL/well of alkali phosphatase-conjugated goat anti-rabbit IgG (0.3 μg/mL in 0.1% BSA in PBS) at RT for 45 min. The wells were washed as above and incubated with PNPP (Pierce) at RT for 5–10 min. OD 405 nm was read on a microplate reader (Bio-Rad). As previously described, we established conditions under which MCF-7 CM reduced the binding of VEGF165 to heparin-BSA (Uchimura et al. 2006a,b). In brief, heparin-BSA coated wells were treated with MCF-7 CM in the presence of different amounts of PI-88. After the enzyme reaction, 25 μL of 25 nM human VEGF165 was incubated on the wells for 30 min. Heparin-bound human VEGF 165 was detected with 1 μg/mL of primary goat anti-VEGF antibody, 1.2 μg/mL of biotinylated swine anti-goat antibody, and 2 μg/mL of alkali phosphatase-conjugated streptavidin. The concentration of PI-88 that inhibited the Sulf enzymatic activity by 50% (IC50) was determined using the formula: IC50 = 10^(Log [A][B] × (50 − C)/(D − C) + Log [B]), where [A] and [B] are the higher and the lower concentrations nearest to the middle of the curve, respectively, C and D are the Sulf activities at the concentration [A] and [B], respectively. Measurement of the LogIC50 value and depiction of the best-fit curve were performed using Prism software (GraphPad Software, La Jolla, CA).
Immunoblotting
The Ni-NTA agarose-bound FLAG/His-tagged Sulf proteins prepared from 0.8 mL of HEK293 transfectant CM was separated by electrophoresis on a reducing SDS–7.5% polyacrylamide gel (WAKO, Osaka, Japan) and blotted onto PVDF membrane (Millipore). The membrane was blocked with 5% skim milk/PBS-T for 1 h and then incubated overnight with an anti-FLAG tag antibody (0.2 μg/mL) in 5% skim milk/PBS-T at 4°C. The membrane was washed and incubated with horseradish peroxidase-conjugated goat anti-mouse IgG1 (0.016 μg/mL) for 1 h. Bound antibodies were visualized with SuperSignal West Pico Chemiluminescent reagent (Pierce). The H2.3 rabbit anti-HSulf-2 antibody was used to immunblot Sulf-2 in MCF-7 CM as described previously (Morimoto-Tomita et al. 2005).
Ex vivo degradation of the RB4CD12 epitope in cryostat-cut sections
Fresh brains from 12-week-old C57BL/six mice were embedded in the O.C.T. compound (Sakura Finetek, Torrance, CA) and frozen in liquid nitrogen. Cryostat-cut sections (10 μm thick) were prepared onto MAS-coated glass slides (Matsunami, Osaka, Japan), fixed in ice-cold acetone for 15 min, and then air dried for 30 min. Sections were incubated with blocking solution (3% BSA in PBS) for 15 min at RT and then pre-treated with purified HSulfs or MCF-7 CM in PBS at 37°C overnight. Sections were washed twice with PBS and then incubated with a mixture of RB4CD12 (1:100 dilution) and a rabbit anti-laminin antibody (1:100 dilution, Sigma) for 1 h at RT. Then, primary antibodies were detected with Cy3-conjugated monoclonal anti-VSV-G (4 μg/mL) and Cy2-conjugated polyclonal goat anti-rabbit IgG (3 μg/mL). Sections were mounted in FluorSaver™ Reagent (EMD Chemicals, Gibbstown, NJ). Digital images were captured by laser scanning confocal microscopy (model LSM 510, Zeiss) at the same setting for all images. The fluorescence intensities of Cy3-RB4CD12 and Cy2-laminin in stained vessels of digital images were determined semiquantitatively by Image-Pro Plus software (Media Cybernetics, Bethesda, MD).
Statistical analysis
All data are presented as means ± SD unless noted otherwise. The values were analyzed by one-way ANOVA with Tukey's (Figures 1, Figures 2, 4, and 5) or Dunnett's (6 and 7). P-values less than 0.05 were considered to be statistically significant.
Supplementary Data
Supplementary data for this article is available online at http://glycob.oxfordjournals.org/.
Funding
The Japanese Health and Labour Sciences Research Grant (Comprehensive Research on Aging and Health H19-001 to K.U.); the National Institutes of Health (R21 CA122025 to S.D.R., P01 AI053194 to S.D.R., R01 HL075602 to Werb-Rosen); Tobacco-Related Disease Research Program Grant (17RT-0117 to S.D.R.); and in parts by the Mochida Memorial Foundation to (K.U.); the Sumitomo Foundation (to K.U); and the Life Science Foundation of Japan (to K.U.).
Supplementary Material
Acknowledgments
We thank Durwin Tsay and Mark Singer for their technical assistance, and Zena Werb for helpful suggestions and discussion. We are grateful to Brian Creese, Vito Ferro, and Keith Dredge of Progen Pharmaceutics Ltd for kindly providing PI-88. T.H. is a Research Fellow of the Japan Foundation for Aging and Health.
Conflict of interest statement
None declared.
Abbreviations
- CHO
Chinese hamster ovary
- CM
conditioned medium
- CS
chondroitin sulfate
- ECM
extracellular matrix
- ELISA
enzyme-linked immunosorbent assay
- FACS
fluorescence-activated cell sorting
- HEK
human embryonic kidney
- HS
heparan sulfate
- HSPG
heparan sulfate proteoglycan
- mAb
monoclonal antibody
- MFI
mean fluorescence intensity
- PBS-T
0.1% Tween 20 in PBS
- scFv
single-chain variable fragment
- VEGF
vascular endothelial growth factor
References
- Ai X, Do AT, Kusche-Gullberg M, Lindahl U, Lu K, Emerson CP., Jr Substrate specificity and domain functions of extracellular heparan sulfate 6-O-endosulfatases, QSulf1 and QSulf2. J Biol Chem. 2006;281:4969–4976. doi: 10.1074/jbc.M511902200. [DOI] [PubMed] [Google Scholar]
- Ai X, Do AT, Lozynska O, Kusche-Gullberg M, Lindahl U, Emerson CP., Jr. QSulf1 remodels the 6-O sulfation states of cell surface heparan sulfate proteoglycans to promote Wnt signaling. J Cell Biol. 2003;162:341–351. doi: 10.1083/jcb.200212083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai X, Kitazawa T, Do AT, Kusche-Gullberg M, Labosky PA, Emerson CP., Jr. SULF1 and SULF2 regulate heparan sulfate-mediated GDNF signaling for esophageal innervation. Development. 2007;134:3327–3338. doi: 10.1242/dev.007674. [DOI] [PubMed] [Google Scholar]
- Ambasta RK, Ai X, Emerson CP., Jr. Quail Sulf1 function requires asparagine-linked glycosylation. J Biol Chem. 2007;282:34492–34499. doi: 10.1074/jbc.M706744200. [DOI] [PubMed] [Google Scholar]
- Bernfield M, Gotte M, Park PW, Reizes O, Fitzgerald ML, Lincecum J, Zako M. Functions of cell surface heparan sulfate proteoglycans. Annu Rev Biochem. 1999;68:729–777. doi: 10.1146/annurev.biochem.68.1.729. [DOI] [PubMed] [Google Scholar]
- Bishop JR, Schuksz M, Esko JD. Heparan sulphate proteoglycans fine-tune mammalian physiology. Nature. 2007;446:1030–1037. doi: 10.1038/nature05817. [DOI] [PubMed] [Google Scholar]
- Cosma MP, Pepe S, Annunziata I, Newbold RF, Grompe M, Parenti G, Ballabio A. The multiple sulfatase deficiency gene encodes an essential and limiting factor for the activity of sulfatases. Cell. 2003;113:445–456. doi: 10.1016/s0092-8674(03)00348-9. [DOI] [PubMed] [Google Scholar]
- Dai Y, Yang Y, MacLeod V, Yue X, Rapraeger AC, Shriver Z, Venkataraman G, Sasisekharan R, Sanderson RD. HSulf-1 and HSulf-2 are potent inhibitors of myeloma tumor growth in vivo. J Biol Chem. 2005;280:40066–40073. doi: 10.1074/jbc.M508136200. [DOI] [PubMed] [Google Scholar]
- David G, Bai XM, Van Der Schueren B, Cassiman JJ, Van Den Berghe H. Developmental changes in heparan sulfate expression: In situ detection with mAbs. J Cell Biol. 1992;119:961–975. doi: 10.1083/jcb.119.4.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennissen MA, Jenniskens GJ, Pieffers M, Versteeg EM, Petitou M, Veerkamp JH, van Kuppevelt TH. Large, tissue-regulated domain diversity of heparan sulfates demonstrated by phage display antibodies. J Biol Chem. 2002;277:10982–10986. doi: 10.1074/jbc.M104852200. [DOI] [PubMed] [Google Scholar]
- Dhoot GK, Gustafsson MK, Ai X, Sun W, Standiford DM, Emerson CP., Jr. Regulation of Wnt signaling and embryo patterning by an extracellular sulfatase. Science. 2001;293:1663–1666. doi: 10.1126/science.293.5535.1663. [DOI] [PubMed] [Google Scholar]
- Dierks T, Schmidt B, Borissenko LV, Peng J, Preusser A, Mariappan M, von Figura K. Multiple sulfatase deficiency is caused by mutations in the gene encoding the human C(alpha)-formylglycine generating enzyme. Cell. 2003;113:435–444. doi: 10.1016/s0092-8674(03)00347-7. [DOI] [PubMed] [Google Scholar]
- Esko JD, Lindahl U. Molecular diversity of heparan sulfate. J Clin Invest. 2001;108:169–173. doi: 10.1172/JCI13530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esko JD, Selleck SB. Order out of chaos: Assembly of ligand binding sites in heparan sulfate. Annu Rev Biochem. 2002;71:435–471. doi: 10.1146/annurev.biochem.71.110601.135458. [DOI] [PubMed] [Google Scholar]
- Esko JD, Stewart TE, Taylor WH. Animal cell mutants defective in glycosaminoglycan biosynthesis. Proc Natl Acad Sci USA. 1985;82:3197–3201. doi: 10.1073/pnas.82.10.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman SD, Moore WM, Guiral EC, Holme AD, Turnbull JE, Pownall ME. Extracellular regulation of developmental cell signaling by XtSulf1. Dev Biol. 2008;320:436–445. doi: 10.1016/j.ydbio.2008.05.554. [DOI] [PubMed] [Google Scholar]
- Frese MA, Milz F, Dick M, Lamanna WC, Dierks T. Characterization of the human sulfatase sulf1 and its high-affinity heparin/heparan sulfate interaction domain. J Biol Chem. 2009;284:28033–28044. doi: 10.1074/jbc.M109.035808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher JT. Heparan sulfate: Growth control with a restricted sequence menu. J Clin Invest. 2001;108:357–361. doi: 10.1172/JCI13713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habuchi H, Habuchi O, Kimata K. Sulfation pattern in glycosaminoglycan: Does it have a code? Glycoconj J. 2004;21:47–52. doi: 10.1023/B:GLYC.0000043747.87325.5e. [DOI] [PubMed] [Google Scholar]
- Habuchi H, Nagai N, Sugaya N, Atsumi F, Stevens RL, Kimata K. Mice deficient in heparan sulfate 6-O-sulfotransferase-1 exhibit defective heparan sulfate biosynthesis, abnormal placentation, and late embryonic lethality. J Biol Chem. 2007;282:15578–15588. doi: 10.1074/jbc.M607434200. [DOI] [PubMed] [Google Scholar]
- Holst CR, Bou-Reslan H, Gore BB, Wong K, Grant D, Chalasani S, Carano RA, Frantz GD, Tessier-Lavigne M, Bolon B, et al. Secreted sulfatases Sulf1 and Sulf2 have overlapping yet essential roles in mouse neonatal survival. PLoS ONE. 2007;2:e575. doi: 10.1371/journal.pone.0000575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacobuzio-Donahue CA, Maitra A, Olsen M, Lowe AW, van Heek NT, Rosty C, Walter K, Sato N, Parker A, Ashfaq R, et al. Exploration of global gene expression patterns in pancreatic adenocarcinoma using cDNA microarrays. Am J Pathol. 2003;162:1151–1162. doi: 10.1016/S0002-9440(10)63911-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilan N, Elkin M, Vlodavsky I. Regulation, function and clinical significance of heparanase in cancer metastasis and angiogenesis. Int J Biochem Cell Biol. 2006;38:2018–2039. doi: 10.1016/j.biocel.2006.06.004. [DOI] [PubMed] [Google Scholar]
- Jenniskens GJ, Hafmans T, Veerkamp JH, van Kuppevelt TH. Spatiotemporal distribution of heparan sulfate epitopes during myogenesis and synaptogenesis: A study in developing mouse intercostal muscle. Dev Dyn. 2002;225:70–79. doi: 10.1002/dvdy.10138. [DOI] [PubMed] [Google Scholar]
- Jenniskens GJ, Oosterhof A, Brandwijk R, Veerkamp JH, van Kuppevelt TH. Heparan sulfate heterogeneity in skeletal muscle basal lamina: Demonstration by phage display-derived antibodies. J Neurosci. 2000;20:4099–4111. doi: 10.1523/JNEUROSCI.20-11-04099.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce JA, Freeman C, Meyer-Morse N, Parish CR, Hanahan D. A functional heparan sulfate mimetic implicates both heparanase and heparan sulfate in tumor angiogenesis and invasion in a mouse model of multistage cancer. Oncogene. 2005;24:4037–4051. doi: 10.1038/sj.onc.1208602. [DOI] [PubMed] [Google Scholar]
- Khachigian LM, Parish CR. Phosphomannopentaose sulfate (PI-88): Heparan sulfate mimetic with clinical potential in multiple vascular pathologies. Cardiovasc Drug Rev. 2004;22:1–6. doi: 10.1111/j.1527-3466.2004.tb00127.x. [DOI] [PubMed] [Google Scholar]
- Kim CW, Lee HM, Lee TH, Kang C, Kleinman HK, Gho YS. Extracellular membrane vesicles from tumor cells promote angiogenesis via sphingomyelin. Cancer Res. 2002;62:6312–6317. [PubMed] [Google Scholar]
- Kreuger J, Spillmann D, Li JP, Lindahl U. Interactions between heparan sulfate and proteins: The concept of specificity. J Cell Biol. 2006;174:323–327. doi: 10.1083/jcb.200604035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai JP, Chien J, Strome SE, Staub J, Montoya DP, Greene EL, Smith DI, Roberts LR, Shridhar V. HSulf-1 modulates HGF-mediated tumor cell invasion and signaling in head and neck squamous carcinoma. Oncogene. 2004;23:1439–1447. doi: 10.1038/sj.onc.1207258. [DOI] [PubMed] [Google Scholar]
- Lai JP, Sandhu DS, Yu C, Han T, Moser CD, Jackson KK, Guerrero RB, Aderca I, Isomoto H, Garrity-Park MM, et al. Sulfatase 2 up-regulates glypican 3, promotes fibroblast growth factor signaling, and decreases survival in hepatocellular carcinoma. Hepatology. 2008;47:1211–1222. doi: 10.1002/hep.22202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamanna WC, Baldwin RJ, Padva M, Kalus I, Ten Dam G, van Kuppevelt TH, Gallagher JT, von Figura K, Dierks T, Merry CL. Heparan sulfate 6-O-endosulfatases: Discrete in vivo activities and functional co-operativity. Biochem J. 2006;400:63–73. doi: 10.1042/BJ20060848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamanna WC, Frese MA, Balleininger M, Dierks T. Sulf loss influences N-, 2-O-, and 6-O-sulfation of multiple heparan sulfate proteoglycans and modulates fibroblast growth factor signaling. J Biol Chem. 2008;283:27724–27735. doi: 10.1074/jbc.M802130200. [DOI] [PubMed] [Google Scholar]
- Li J, Kleeff J, Abiatari I, Kayed H, Giese NA, Felix K, Giese T, Buchler MW, Friess H. Enhanced levels of Hsulf-1 interfere with heparin-binding growth factor signaling in pancreatic cancer. Mol Cancer. 2005;4:14. doi: 10.1186/1476-4598-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X. Functions of heparan sulfate proteoglycans in cell signaling during development. Development. 2004;131:6009–6021. doi: 10.1242/dev.01522. [DOI] [PubMed] [Google Scholar]
- Lum DH, Tan J, Rosen SD, Werb Z. Gene trap disruption of the mouse heparan sulfate 6-O-endosulfatase gene, Sulf2. Mol Cell Biol. 2007;27:678–688. doi: 10.1128/MCB.01279-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto-Tomita M, Uchimura K, Bistrup A, Lum DH, Egeblad M, Boudreau N, Werb Z, Rosen SD. Sulf-2, a proangiogenic heparan sulfate endosulfatase, is upregulated in breast cancer. Neoplasia. 2005;7:1001–1010. doi: 10.1593/neo.05496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto-Tomita M, Uchimura K, Werb Z, Hemmerich S, Rosen SD. Cloning and characterization of two extracellular heparin-degrading endosulfatases in mice and humans. J Biol Chem. 2002;277:49175–49185. doi: 10.1074/jbc.M205131200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita K, Chien J, Mullany SA, Staub J, Qian X, Lingle WL, Shridhar V. Loss of HSulf-1 expression enhances autocrine signaling mediated by amphiregulin in breast cancer. J Biol Chem. 2007;282:14413–14420. doi: 10.1074/jbc.M611395200. [DOI] [PubMed] [Google Scholar]
- Nawroth R, van Zante A, Cervantes S, McManus M, Hebrok M, Rosen SD. Extracellular sulfatases, elements of the Wnt signaling pathway, positively regulate growth and tumorigenicity of human pancreatic cancer cells. PLoS ONE. 2007;2:e392. doi: 10.1371/journal.pone.0000392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parish CR. The role of heparan sulphate in inflammation. Nat Rev Immunol. 2006;6:633–643. doi: 10.1038/nri1918. [DOI] [PubMed] [Google Scholar]
- Parish CR, Freeman C, Brown KJ, Francis DJ, Cowden WB. Identification of sulfated oligosaccharide-based inhibitors of tumor growth and metastasis using novel in vitro assays for angiogenesis and heparanase activity. Cancer Res. 1999;59:3433–3441. [PubMed] [Google Scholar]
- Powell AK, Yates EA, Fernig DG, Turnbull JE. Interactions of heparin/heparan sulfate with proteins: Appraisal of structural factors and experimental approaches. Glycobiology. 2004;14:17R–30R. doi: 10.1093/glycob/cwh051. [DOI] [PubMed] [Google Scholar]
- Saad OM, Ebel H, Uchimura K, Rosen SD, Bertozzi CR, Leary JA. Compositional profiling of heparin/heparan sulfate using mass spectrometry: Assay for specificity of a novel extracellular human endosulfatase. Glycobiology. 2005;15:818–826. doi: 10.1093/glycob/cwi064. [DOI] [PubMed] [Google Scholar]
- Tang R, Rosen SD. Functional consequences of the subdomain organization of the sulfs. J Biol Chem. 2009;284:21505–21514. doi: 10.1074/jbc.M109.028472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thery C, Zitvogel L, Amigorena S. Exosomes: Composition, biogenesis and function. Nat Rev Immunol. 2002;2:569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- Uchimura K, Morimoto-Tomita M, Bistrup A, Li J, Lyon M, Gallagher J, Werb Z, Rosen SD. HSulf-2, an extracellular endoglucosamine-6-sulfatase, selectively mobilizes heparin-bound growth factors and chemokines: Effects on VEGF, FGF-1, and SDF-1. BMC Biochem. 2006a;7:2. doi: 10.1186/1471-2091-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchimura K, Morimoto-Tomita M, Rosen SD. Measuring the activities of the Sulfs: Two novel heparin/heparan sulfate endosulfatases. Methods Enzymol. 2006b;416:243–253. doi: 10.1016/S0076-6879(06)16015-2. [DOI] [PubMed] [Google Scholar]
- Van Den Born J, Salmivirta K, Henttinen T, Ostman N, Ishimaru T, Miyaura S, Yoshida K, Salmivirta M. Novel heparan sulfate structures revealed by monoclonal antibodies. J Biol Chem. 2005;280:20516–20523. doi: 10.1074/jbc.M502065200. [DOI] [PubMed] [Google Scholar]
- van Kuppevelt TH, Dennissen MA, van Venrooij WJ, Hoet RM, Veerkamp JH. Generation and application of type-specific anti-heparan sulfate antibodies using phage display technology. Further evidence for heparan sulfate heterogeneity in the kidney. J Biol Chem. 1998;273:12960–12966. doi: 10.1074/jbc.273.21.12960. [DOI] [PubMed] [Google Scholar]
- Viviano BL, Paine-Saunders S, Gasiunas N, Gallagher J, Saunders S. Domain-specific modification of heparan sulfate by Qsulf1 modulates the binding of the bone morphogenetic protein antagonist Noggin. J Biol Chem. 2004;279:5604–5611. doi: 10.1074/jbc.M310691200. [DOI] [PubMed] [Google Scholar]
- Vlodavsky I, Ilan N, Naggi A, Casu B. Heparanase: Structure, biological functions, and inhibition by heparin-derived mimetics of heparan sulfate. Curr Pharm Des. 2007;13:2057–2073. doi: 10.2174/138161207781039742. [DOI] [PubMed] [Google Scholar]
- Zito E, Fraldi A, Pepe S, Annunziata I, Kobinger G, Di Natale P, Ballabio A, Cosma MP. Sulphatase activities are regulated by the interaction of sulphatase-modifying factor 1 with SUMF2. EMBO Rep. 2005;6:655–660. doi: 10.1038/sj.embor.7400454. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.