Abstract
Trans-sialidases catalyze the transfer of a sialic acid from one sialoside to an acceptor to form a new sialoside. α2,3-Trans-sialidase activity was initially discovered in the parasitic protozoan Trypanosoma cruzi, and more recently was found in a multifunctional Pasteurella multocida sialyltransferase PmST1. α2,8-Trans-sialidase activity was also described for a multifunctional Campylobacter jejuni sialyltransferase CstII. We report here the discovery of the α2,6-trans-sialidase activity of a previously reported recombinant truncated bacterial α2,6-sialyltransferase from Photobacterium damsela (Δ15Pd2,6ST). This is the first time that the α2,6-trans-sialidase activity has ever been identified. Kinetic studies indicate that Δ15Pd2,6ST-catalyzed trans-sialidase reaction follows a ping-pong bi-bi reaction mechanism. Cytidine 5′-monophosphate, the product of sialyltransferase reactions, is not required by the trans-sialidase activity of the enzyme but enhances the trans-sialidase activity modestly as a non-essential activator. Using chemically synthesized Neu5AcαpNP and LacβMU, α2,6-linked sialoside Neu5Acα2,6LacβMU has been obtained in one-step in high yield using the trans-sialidase activity of Δ15Pd2,6ST. In addition to the α2,6-trans-sialidase activity, Δ15Pd2,6ST also has α2,6-sialidase activity. The multifunctionality is thus a common feature of many bacterial sialyltransferases.
Keywords: enzyme, sialidase, sialoside, sialyltransferase, trans-sialidase
Introduction
Sialic acids, a family of α-keto acids with a nine-carbon backbone, often occupy terminal carbohydrate positions in many glycoproteins and glycolipids which play important roles in a number of biological and pathological processes, such as cell–cell communication, signal transduction, bacterial and viral infection, and cancer metastasis (Shen et al. 1999; Schauer 2000; Harduin-Lepers et al. 2001; Angata and Varki 2002; Yu and Chen 2007). N-Acetylneuraminic acid (Neu5Ac) is the most common form of sialic acid. Four major sialyl linkages, Neu5Acα2,3Gal, Neu5Acα2,6Gal(NAc), Neu5Acα2,8Neu5Ac, and Neu5Acα2,9Neu5Ac, have been found in oligosaccharides, polysaccharides, and glycoconjugates. Sialylation in nature is usually catalyzed by a family of enzymes named sialyltransferases. They catalyze the transfer of the sialic acid residue from cytidine 5′-monophosphate sialic acid (CMP-Sia) to an acceptor containing a Gal, GalNAc, or another sialic acid at the non-reducing terminus (Tsuji et al. 1996). In nature, sialylation can also be achieved by trans-sialidases, which catalyze the transfer of a sialic acid from one sialoside to an acceptor to form a new sialoside.
Trans-sialidase was initially discovered in parasitic protozoan Trypanosoma cruzi, the causative agent of Chagas disease, which is an endemic parasitic infection in Latin America and is frequently associated with ventricular tachyarrhythmia and sudden death (Schenkman et al. 1991; Frasch 2000; Bern et al. 2008). The Trypanosoma cruzi trans-sialidase is highly specific toward Neu5Acα2,3Gal linkage and acts as a sialidase to produce free sialic acid in the absence of an acceptor (Cross and Takle 1993; Harrison et al. 2001). The parasite uses this surface enzyme to “steal” sialic acid from host to decorate its own surface and the sialylation is proposed to be necessary for parasite's successful invasion of the host or protecting the parasite from host's immune recognition and defense (Colli 1993). The α2,3-trans-sialidase activity was also found in a multifunctional Pasteurella multocida sialyltransferase PmST1 (Yu et al. 2005). An intramolecular trans-sialidase (neuraminidase B) from Streptococcus pneumonia which is specific toward α2,3-sialyl linkage was recently crystallized with its product 2,3-anhydro-Neu5Ac (Gut et al. 2008). Recently, α2,8-trans-sialidase activity was also described for a multifunctional Campylobacter jejuni sialyltransferase CstII (Cheng et al. 2008).
Photobacterium damsela α2,6-sialyltransferase (Pd2,6ST) was the first bacterial α2,6-sialyltransferase that has been cloned (Yamamoto et al. 1998; Teo et al. 2005; Yu et al. 2006; Sun et al. 2008). Pd2,6ST shares amino acid sequence homology with PmST1. Both belong to glycosyltransferase family 80 (GT80) in the Carbohydrate-Active enZyme database (CAZy, http://www.cazy.org/) (Campbell et al. 1997; Coutinho et al. 2003). Purified Pd2,6ST and its recombinant truncated form (Δ15Pd2,6ST) encoding for amino acid residues 16–497 of the full-length protein (1–675 amino acid residues) have shown to have flexible substrate specificity toward acceptor and/or donor (Kajihara et al. 1996; Teo et al. 2005; Yu et al. 2006; Chokhawala et al. 2008). The truncated form Δ15Pd2,6ST is an N-His6-tagged fusion protein lacking an N-terminal hydrophobic region predicated to be a signal peptide (Bendtsen et al. 2004) and a C-terminal region sharing similarity to phosphate transport system regulatory protein PhoU (Yamamoto et al. 1998; Yu et al. 2006). Herein, we report that Δ15Pd2,6ST has additional α2,6-trans-sialidase and α2,6-sialidase activities. Δ15Pd2,6ST is thus the first α2,6-trans-sialidase that has ever been discovered. It is also the first reported sialidase that is specific for the α2,6-sialyl linkage. Other than α2,6-linked sialyloligosaccharides, para-nitrophenyl α-sialoside such as Neu5AcαpNP can also be used as an alternative sialyl donor by the trans-sialidase activity of Δ15Pd2,6ST for the synthesis of α2,6-linked sialosides in the absence of CMP. Results from kinetic studies indicate that the α2,6-trans-sialidase activity of Δ15Pd2,6ST follows a ping-pong bi-bi reaction mechanism.
Results
pH Profiles of the α2,6-sialidase and the α2,6-trans-sialidase activities of Δ15Pd2,6ST
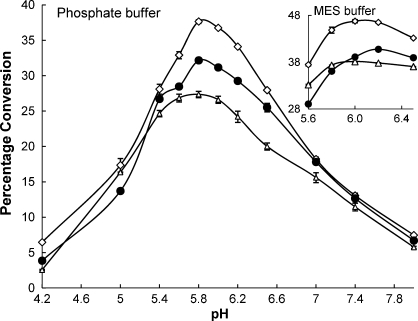
As seen in Figure 1, in phosphate buffers, the pH profiles for the α2,6-sialidase and the α2,6-trans-sialidase activities of Δ15Pd2,6ST were very similar. Both activities were active in a narrow pH range and reached an optimum at pH 5.8. MES buffer (Figure 1 insert), however, was a preferred buffer for both α2,6-sialidase and α2,6-trans-sialidase activities of Δ15Pd2,6ST. In addition, the optimal pH value of the α2,6-sialidase activity of Δ15Pd2,6ST shifted from 5.8 in the phosphate buffer to 6.2 in the MES buffer.
Fig. 1.
The pH profiles of the α2,6-sialidase (filled circles) and the α2,6-trans-sialidase activities of Δ15Pd2,6ST obtained by quantitative HPLC analysis. Both Neu5Acα2,6LacβProN3 (unfilled triangles) and Neu5AcαpNP (unfilled diamonds) were used for the α2,6-trans-sialidase activity assays. 100% conversion was defined as the formation of 1 mM LacβMU for the α2,6-sialidase activity assay or 1 mM Neu5Acα2,6LacβMU for the α2,6-trans-sialidase activity assay.
Apparent kinetics
As shown in Table I, for the α2,6-sialidase activity of Δ15Pd2,6ST, the Km and kcat of Neu5Acα2,6LacβMU were 7.6 mM and 8.2 min−1, respectively. For the α2,6-trans-sialidase activity of Δ15Pd2,6ST, the Km and kcat of Neu5AcαpNP (53 mM and 53 min−1, respectively) were around 6- and 13-fold, respectively, of those for Neu5Acα2,6LacβProN3 (9.0 mM and 4.0 min−1). Despite a higher Km value, the kcat/Km value of Neu5AcαpNP (1.0 min−1 mM−1) for the α2,6-trans-sialidase activity of Δ15Pd2,6ST was about 2.3-fold of that of Neu5Acα2,6LacβProN3 (0.44 min−1 mM−1).
Table I.
Apparent kinetic parameters for the α2,6-sialidase and α2,6-trans-sialidase activities of Δ15Pd2,6STa
| Activities | α2,6-Sialidase | α2,6-Trans-sialidase | |
|---|---|---|---|
| Substrates | Neu5Acα2,6LacβMU | Neu5Acα2,6LacβProN3 | Neu5AcαpNP |
| Km (mM) | 7.6 ± 0.5 | 9.0 ± 0.6 | 53 ± 9 |
| kcat (min−1) | 8.2 ± 0.3 | 4.0 ± 0.2 | 53 ± 4 |
| kcat/Km (min−1 mM−1) | 1.1 | 0.44 | 1.0 |
aNeu5Acα2,6LacβMU was used for the α2,6-sialidase activity assays. LacβMU was used as an acceptor, and either Neu5Acα2,6LacβProN3 or Neu5AcαpNP was used as a donor for the α2,6-trans-sialidase activity assays.
Kinetic mechanism of the α2,6-trans-sialidase activity of Δ15Pd2,6ST
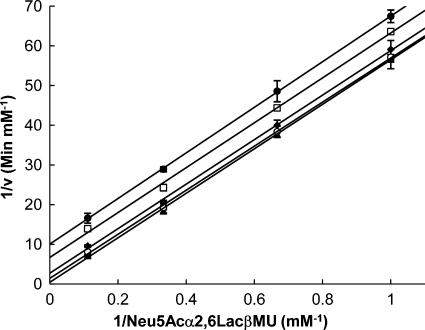
Kinetic mechanism studies for the α2,6-trans-sialidase activity were performed using a series of varied concentrations of Neu5Acα2,6LacβMU and different fixed concentrations of LacβProN3. As shown in Figure 2, Lineweaver–Burk plots yielded a series of parallel lines, which fitted the behavior of a classical ping-pong bi-bi mechanism. The same ping-pong mechanism was described for a non-homologous Trypanosoma cruzi trans-sialidase (Amaya et al. 2004; Damager et al. 2008).
Fig. 2.
Lineweaver–Burk curves for elucidating the kinetic mechanism of the α2,6-trans-sialisase activity of Δ15Pd2,6ST. Double-reciprocal plots of initial velocity were determined for Neu5Acα2,6LacβMU whose concentrations were varied between 1 and 9 mM with the following fixed concentrations of LacβProN3: (•) 0.5 mM; (□) 1 mM; (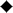 ) 2.5 mM; (○) 5 mM; and (▴) 10 mM. The solid lines corresponded to data fitted to Eq. (1).
) 2.5 mM; (○) 5 mM; and (▴) 10 mM. The solid lines corresponded to data fitted to Eq. (1).
Effects of CMP on the α2,6-sialidase and the α2,6-trans-sialidase activities of Δ15Pd2,6ST
CMP was not required for the α2,6-sialidase activity of Δ15Pd2,6ST. This trans-sialidase activity is different from the reported reverse glycosyltransferase activity for some glycosyltransferases which requires the presence of CMP (Zhang et al. 2006; Lairson et al. 2007; Chandrasekaran et al. 2008). To further exclude the possibility of reverse-glycosyltransferase activity for the trans-sialidase activity observed, Δ15Pd2,6ST was dialyzed extensively against dialysis buffer containing activated charcoal to remove any CMP that may be bound to the enzyme during culturing. Trans-sialidase activity assays carried out in the absence of CMP indicated that similar activities were shown for reactions using enzyme preparations before or after extensive dialysis with the charcoal-containing dialysis buffer. These data confirmed that the activity observed was indeed the trans-sialidase activity.
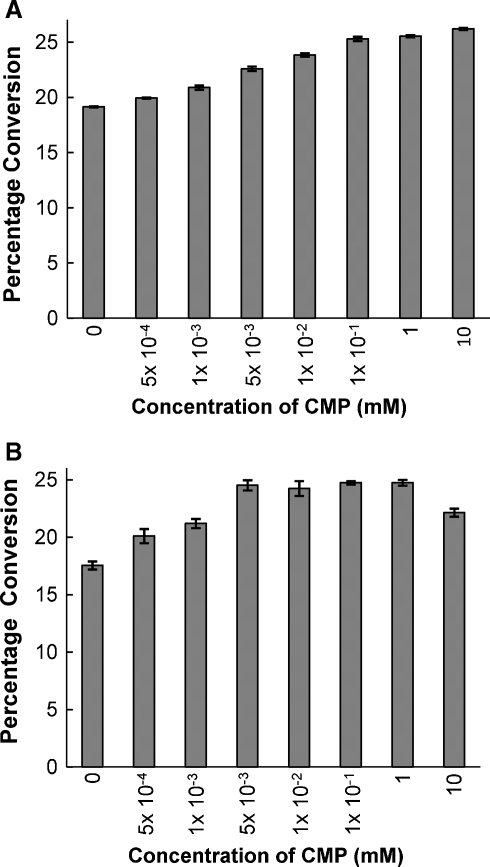
Interestingly, although CMP, a product and an inhibitor of sialyltransferases (Cambron and Leskawa 1993), was not required, CMP was able to enhance the efficiency of the α2,6-sialidase activity of Δ15Pd2,6ST in a dose-dependent manner (Figure 3A). CMP also enhanced the efficiency of the α2,6-trans-sialidase activity of Δ15Pd2,6ST at low concentrations (equal or less than 1 mM) and the activation effect of CMP was weakened when a high concentration (10 mM) of CMP was used (Figure 3B). CMP is thus a non-essential activator of both the α2,6-sialidase and the α2,6-trans-sialidase activities of Δ15Pd2,6ST. In the presence of CMP, the α2,6-trans-sialidase reactions did not produce CMP-Neu5Ac as assayed by capillary electrophoresis studies, further confirming that the observed trans-sialidase activity was not reverse-sialylation activity.
Fig. 3.
The effects of CMP on the α2,6-sialidase (A) and the α2,6-trans-sialidase (B) activities of Δ15Pd2,6ST. Neu5Acα2,6LacβMU was used for the α2,6-sialidase activity assays. Neu5AcαpNP and LacβMU were used for the α2,6-trans-sialidase activity assays.
Unlike CMP which functions as a non-essential activator that improves the efficiency of both the α2,6-sialidase and the α2,6-trans-sialidase activities of Δ15Pd2,6ST, cytidine has no significant effect, while CDP and CTP at a concentration of 1 mM showed obvious inhibition for both sialidase and trans-sialidase activities of Δ15Pd2,6ST (data not shown).
Activation effects of CMP by kinetic studies
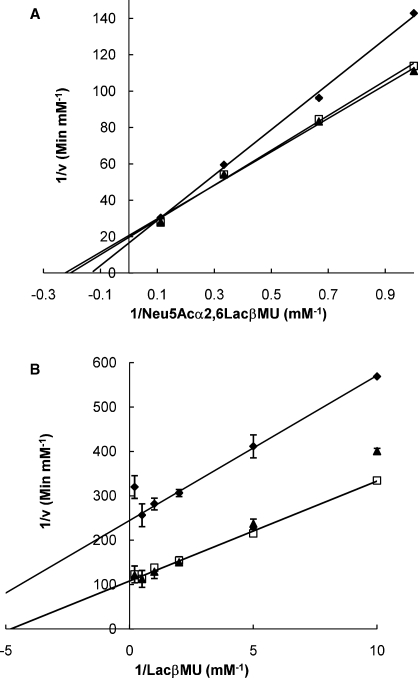
To investigate the kinetic behavior of CMP, a non-essential activator for the α2,6-sialidase and the α2,6-trans-sialidase activities of Δ15Pd2,6ST, initial velocity patterns were obtained as a function of Neu5Acα2,6LacβMU or LacβMU concentrations in the absence or the presence of different concentrations of CMP. As shown in Figure 4A, CMP was able to enhance the α2,6-sialidase activity of Δ15Pd2,6ST by decreasing the apparent Km and increasing Vmax/Km. Fitting the data using the Eq. (2) yielded the following parameters: KS = 7.6 ± 0.5 mM, KA = 0.29 ± 0.02 mM, α = 0.58 ± 0.04, β = 0.82 ± 0.05. For the effect of CMP on the α2,6-trans-sialidase activity of Δ15Pd2,6ST, Lineweaver–Burk plots in Figure 4B indicated that the activation effect of CMP was dependent on the concentrations of CMP and LacβMU presented in the reaction. CMP improved the α2,6-trans-sialidase activity of Δ15Pd2,6ST at low concentrations by increasing the apparent Vmax of LacβMU, though the apparent Km of LacβMU was enhanced at the same time. However, the activation effect of CMP was significantly weakened when a high concentration of CMP was used. Figure 4B also revealed substrate inhibition of LacβMU at high concentrations, which competed with Neu5Acα2,6LacβProN3 for the single substrate binding pocket (Cook and Cleland 2007).
Fig. 4.
Kinetic mechanisms of the CMP effect on the α2,6-sialidase (A) and the α2,6-trans-sialidase (B) of Δ15Pd2,6ST. Double-reciprocal plots were obtained with Neu5Acα2,6LacβMU (A) or LacβMU (B) as the variable substrates at different fixed concentrations of CMP: ( ) 0 mM; (□) 1 mM; (▴) 10 mM. The solid lines corresponded to data fitted to Eq. (2) (data obtained in the presence of 10 mM CMP did not fit).
) 0 mM; (□) 1 mM; (▴) 10 mM. The solid lines corresponded to data fitted to Eq. (2) (data obtained in the presence of 10 mM CMP did not fit).
Δ15Pd2,6ST preferentially cleaves α2,6-linked sialic acid
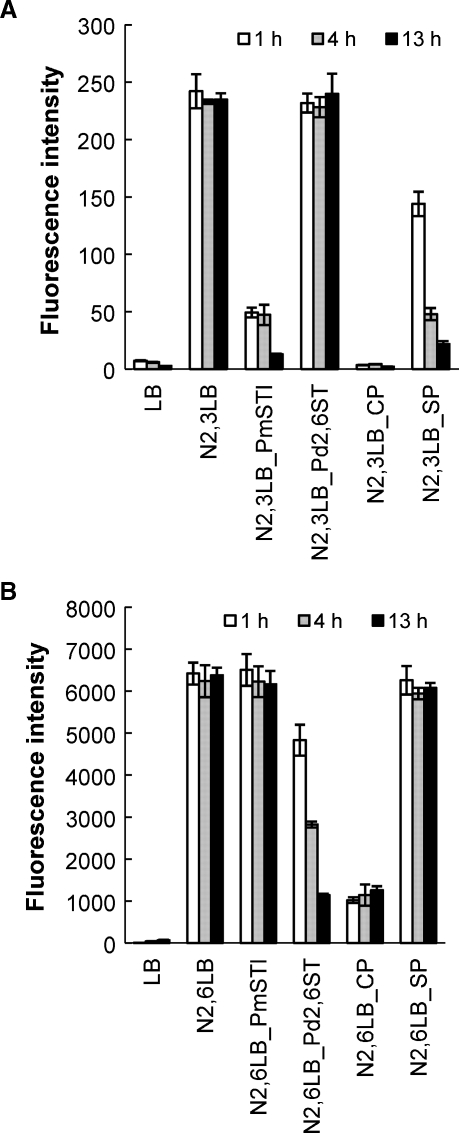
The sialyl linkage preference of the sialidase activity of Δ15Pd2,6ST was assessed using biotinylated Neu5Ac-containing α2,3- and α2,6-linked sialyl lactosides. Fluorescein-labeled Maackia amurensis Lectin (MAA) and fluorescein-labeled Sambucus nigra Lectin (SNA) were used to detect α2,3-linked sialyl lactoside and α2,6-linked sialyl lactoside, respectively. Controls were set up using PmST1 as well as commercially available Clostridium perfingens sialidase and Streptococcus pneumoniae sialidase. Consistent with the results from previous reports (Yu et al. 2005; Chokhawala et al. 2007), the sialidase activity of multifunctional PmST1 specifically catalyzed the cleavage of α2,3-linked sialosides; C. perfingens sialidase hydrolyzed both α2,3- and α2,6-linked sialosides, while S. pneumoniae sialidase was an α2,3-specific sialidase (Chokhawala et al. 2007). As shown in Figure 5, the sialidase activity of the Δ15Pd2,6ST was specific for α2,6-linked sialoside and left α2,3-linked sialoside intact.
Fig. 5.
Δ15Pd2,6ST preferentially cleaves α2,6-linked sialic acid. Δ15Pd2,6ST was incubated with biotinylated Neu5Ac-containing α2,3- (A) and (B) α2,6-linked sialyllactosides for 1 h, 4 h, and 13 h, respectively. PmST1, α2,3/α2,6-sialidase from Clostridium perfingens (CP), and α2,3-sialidase from Streptococcus pneumoniae (SP) were used as controls. Abbreviation: N2,3LB, biotinylated α2,3Neu5Ac-linked lactoside; N2,6LB, biotinylated α2,6Neu5Ac-linked lactoside; LB, biotinylated lactoside. Fluorescein-labeled Maackia amurensis Lectin (MAA) (weaker signals were obtained) and fluorescein-labeled Sambucus nigra Lectin (SNA) (stronger signals were obtained) were used to detect α2,3-linked sialyl lactoside and α2,6-linked sialyl lactoside, respectively.
Application of the α2,6-trans-sialidase activity of Δ15Pd2,6ST in the synthesis of sialosides
In order to confirm the product formed by the α2,6-trans-sialidase reaction by 1H NMR and 13C NMR as well as the practical application of the α2,6-trans-sialidase activity of Δ15Pd2,6ST in the synthesis of sialosides, preparative-scale synthesis of Neu5Acα2,6LacβMU was carried out using Δ15Pd2,6ST at pH 6.0 in a MES buffer (100 mM) containing LacβMU and Neu5AcαpNP. As the kcat/Km values and optimal pH values for the trans-sialidase and the sialidase activities of Δ15Pd2,6ST are close, it was important to control the reaction time to minimize the sialidase-catalyzed hydrolysis of the product formed from the trans-sialidase reaction. To do this, the reaction was monitored by performing thin-layer chromatography (TLC) for every 30 min and the reaction was stopped immediately when the product formation reached a plateau. By doing this, an 89% yield was achieved. The NMR and high-resolution mass spectrometry data of the product obtained were in agreement with those reported previously (Yu et al. 2005, 2006).
Discussion
Multiple activities of bacterial sialyltransferases
Up to date, PmST1 and CstII are the only two bacterial trans-sialidases which have been reported to have α2,3- and α2,8-trans-sialidase activities, respectively. In our study, the recombinant Δ15Pd2,6ST has been found to have α2,6-trans-sialidase and α2,6-sialidase activities in addition to the α2,6-sialyltransferase activity reported previously (Yamamoto et al. 1998; Teo et al. 2005; Yu et al. 2005; Sun et al. 2008). For all these three enzymes, the trans-sialidase and the sialidase activities are weaker than their sialyltransferase activities. For example, the α2,6-sialidase activity of Δ15Pd2,6ST (the kcat/Km value of Neu5Acα2,6LacβMU is 1.1 min−1 mM−1) is 158-fold less efficient than its α2,6-sialyltransferase activity (the kcat/Km value of CMP-Neu5Ac is 2.9 s−1 mM−1) (Sun et al. 2008). The α2,6-trans-sialidase activity of Δ15Pd2,6ST (the kcat/Km value of Neu5AcαpNP is 1.0 min−1 mM−1) is 174-fold weaker than its α2,6-sialyltransferase activity. For multifunctional CstII, its GD3 oligosaccharide sialidase (the kcat/Km value of Neu5Acα2,8Neu5Acα2,3LacβMU is 1.5 min−1 mM−1) and GD3 oligosaccharide trans-sialidase (the kcat/Km value of Neu5Acα2,8Neu5Acα2,3LacβProN3 is 6.0 min−1 mM−1) activities are less active than its GD3 oligosaccharide synthase activity (the kcat/Km value of CMP-Neu5Ac is 30 min−1 mM−1) by 20- and 5-fold, respectively (Cheng et al. 2008). PmST1 also possesses a significantly (about 8-fold) weaker α2,3-sialidase activity (the kcat/Km value of Neu5Acα2,3LacβMU is 9.5 s−1 mM−1) in comparison with its α2,3-sialyltransferase activity (the kcat/Km value of CMP-Neu5Ac is 73 s−1 mM−1) (Yu et al. 2005).
The bacterial trans-sialidase activity that has been found so far co-exists with sialyltransferase and sialidase activities. Pd2,6ST and PmST1 share sequence homology and are grouped into glycosyltransferase (GT) family 80 in Carbohydrate-Active enZyme database (CAZy, http://www.cazy.org/) (Campbell et al. 1997; Coutinho et al. 2003). These two enzymes do not share sequence homology with CstII (GT42 family) nor with the Trypanosoma cruzi trans-sialidase which belongs to Glycoside Hydrolase (GH) family 33. GT42 and GT80 only contain bacterial sialyltransferases, while GH33 includes sialidases from bacteria, parasites, and viruses. Enzymes in GT80 and GT42 are usually multifunctional, such as Pd2,6ST, PmST1, and CstII.
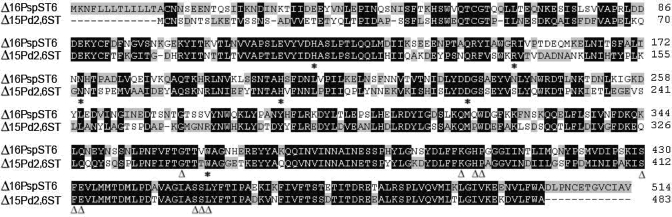
Different from Δ15Pd2,6ST, the sialidase activity has not been detected for Δ16PspST6 (Tsukamoto et al. 2008), which is a truncated α2,6-sialyltransferase from Vibrionaceae Photobacterium sp. JT-ISH-224. Δ16PspST6 is homologous to Δ15Pd2,6ST and belongs to the CAZy GT80 family. The protein x-ray crystal structure of Δ16PspST6 has been recently reported (Kakuta et al. 2008). Δ15Pd2,6ST and Δ16PspST6 share high homology (74% similarity). The alignment of their protein sequences demonstrates that all of the amino acid residues in the CMP- and lactose-binding sites are conserved. Asp232 of Δ16PspST6 is the only amino acid residue within hydrogen-binding distance of the acceptor oxygen at carbon-6 of the Gal residue. It acts as a catalytic base to activate the acceptor for the formation of the sialylated product. A corresponding amino acid residue Asp205 presented in a conserved segment Leu-Tyr-Asp-Asp-Gly-Ser in Δ15Pd2,6ST may function the same as the Asp232 of Δ16PspST6 in the sialyltransferase reaction and may also act as an acid/base catalytic residue in trans-sialidase reaction similar to the function of Asp59 in Trypanosoma cruzi trans-sialidase (Amaya et al. 2004; Damager et al. 2008). Compared to Δ16PspST6, Δ15Pd2,6ST has five separated amino acid deletions in its protein sequence, which may lead to the difference of their structures and catalytic properties to some extent (Figure 6). It will be interesting to test whether Δ16PspST6 also has trans-sialidase activity.
Fig. 6.
Alignment of Δ15Pd2,6ST and Δ16PspST6. CMP- and lactose-binding sites are underlined by open triangles and stars, respectively. Amino acid deletions are presented by dashes.
Optimal pH values for the multiple activities of bacterial sialyltransferases
The sialidase and the trans-sialidase activities of CstII, PmST1, and Δ15Pd2,6ST are optimal under mild acid conditions (pH 5.5–6.5 for CstII and Δ15Pd2,6ST, and pH 5.0–5.5 for PmST1) in comparison with their corresponding sialyltransferase activities which exhibit optimal activities in broad pH ranges (pH 6.5–9.0 for CstII and PmST1, and pH 6.5–10.0 for Δ15Pd2,6ST) (Yu et al. 2005; Cheng et al. 2008; Sun et al. 2008). Other than the multifunctional sialyltransferase CstII in C. jejuni OH4384, the bacterial strain also encodes a complete pathway for de novo Neu5Ac synthesis and a CMP-Neu5Ac synthetase (Cj1143) (Gilbert et al. 2002). Pasteurella multocida Pm70 does not synthesize Neu5Ac de novo, but it can utilize internalized sialic acid by a CMP-Neu5Ac synthetase (NeuA) and sialyltransferases (Scudder et al. 1993). Gathering sialyltransferase, sialidase, and trans-sialidase activities on one enzyme is a possible economic strategy for some pathogenic bacteria to take advantage of both endogenous CMP-Neu5Ac and exogenous sialic acid under different pH conditions. This property could be developed during the long-term evolution. The detailed function of the trans-sialidase activities of these multifunctional sialyltransferases in vivo is currently unclear.
The trans-sialdiase activity of ΔPd2,6ST
Different from the Trypanosoma cruzi trans-sialidase which preferred NeuAcα2,3Lac over NeuAcαpNP as a Neu5Ac donor (25-fold difference) (Scudder et al. 1993), the α2,6-trans-sialidase activity of Δ15Pd2,6ST was around 2.3-fold more active when Neu5AcαpNP, instead of Neu5Acα2,6LacβProN3, was used as the donor for its α2,6-trans-sialidase activity. The trans-sialidase activity and its application in synthesis were confirmed and demonstrated by Δ15Pd2,6ST-catalyzed preparative synthesis of Neu5Acα2,6LacβMU from LacβMU and Neu5AcαpNP in the absence of CMP. This trans-sialidase activity is different from the reported reverse glycosyltransferase activity for some glycosyltransferases which requires the presence of CMP (Zhang et al. 2006; Lairson et al. 2007; Chandrasekaran et al. 2008).
The specificity of the α2,6-trans-sialdiase and the α2,6-sialdiase activities of ΔPd2,6ST
When sialyl oligosaccharides are used for the trans-sialidase activity of Δ15Pd2,6ST, the enzyme is specific for the α2,6-sialyl linkage, as Neu5Acα2,6LacβProN3, but not Neu5Acα2,3LacβProN3, can be used to transfer the Neu5Ac to LacβMU to form Neu5Acα2,6LacβMU. Similarly, the sialidase activity of Δ15Pd2,6ST is also specific toward the α2,6-sialyl linkage. The specific α2,6-sialidase activity of Δ15Pd2,6ST can be used to selectively cleave α2,6-linked sialyl glycans from cell surface and glycoproteins, providing a unique tool for glycan analysis and glycan modifications.
Kinetic mechanism of the α2,6-trans-sialdiase activity of ΔPd2,6ST
Different kinetic behaviors of trans-sialidase reactions were observed for Trypanosoma cruzi trans-sialidase when different methods were used to determine the initial velocity. Since the trans-sialidase reaction is accompanied by significant hydrolysis, the reaction rates are dependent on the concentrations of the acceptor (Scudder et al. 1993; Yang et al. 2000). Converging Lineweaver–Burk plots were yielded when the initial rates of Neu5Ac transfer from sialyllactose to an acceptor were measured (Scudder et al. 1993; Ribeirao et al. 1997). In contrast, parallel Lineweaver–Burk plots were yielded when the initial rates of Neu5Ac transfer and sialyllactose hydrolysis were measured (Amaya et al. 2004; Damager et al. 2008). Ping-pong mechanism implicated by the parallel Lineweaver–Burk plot was then demonstrated to be fully consistent with the three-dimensional structure of Trypanosoma cruzi trans-sialidase, in which the donor lactosyl moiety and the acceptor lactose occupy the same binding site (Buschiazzo et al. 2002; Amaya et al. 2004). The ping-pong mechanism was also supported by the detection of a covalent sialyl-enzyme intermediate with release of p-nitrophenol (Damager et al. 2008). In our kinetic study, the initial rate of LacβMU released from Neu5Acα2,6LacβMU was measured as the total flux through the system. This gave rise to the parallel Lineweaver–Burk plot implying a ping-pong mechanism for the α2,6-trans-sialidase activity of Δ15Pd2,6ST. This is the first study to provide a detailed understanding of the kinetic mechanism of the α2,6-trans-sialidase activity of a bacterial protein. The behavior of a non-essential activator CMP was also investigated in a kinetic manner.
Materials and methods
Materials
Histrap FF™ column was purchased from Qiagen (Valencia, CA). Streptococcus pneumoniae sialidase and Clostridium perfingens sialidase (type VI) were from Prozyme (Hayward, CA) and Sigma (St. Louis, MO), respectively. A BCA protein assay kit and NeutrAvidin-coated 384-well plates were purchased from Pierce Biotechnology (Rockford, IL). Fluorescein-labeled MAA and fluorescein-labeled SNA were purchased from Vector Laboratories (Burlingame, CA). Gel filtration chromatography was performed using a column (100 cm × 2.5 cm) packed with BioGel P-2 Fine resins (Bio-Rad, Hercules, CA). Silica gel 60 Å (200–425 mesh, Sorbent technologies, Atlanta, GA) was used for flash column chromatography.
Overexpression and purification of N-His6-tagged Δ15Pd2,6ST
Procedures for overexpression and purification of Δ15Pd2,6ST were the same as described before (Yu et al. 2005). Extensive (24 h) dialysis of the enzyme against a dialysis buffer containing activated charcoal was carried out to eliminate CMP and other nucleotide which may bind to the enzyme during its expression.
pH Profile by HPLC
Assays were performed in a total volume of 20 μL in a buffer (200 mM) with pH varying from 4.2 to 8.0. Neu5Acα2,6LacβMU (1 mM) was incubated with Δ15Pd2,6ST (47 μg) for α2,6-sialidase assays. LacβMU (1 mM) and Neu5Acα2,6LacβProN3 (1.5 mM) or Neu5AcαpNP (1.5 mM) were incubated with Δ15Pd2,6ST (31 μg) for α2,6-trans-sialidase assays. The buffers used were: MES, pH 4.2–6.5; citrate-phosphate, pH 4.2–5.6; phosphate, 5.8–8.0. Reactions were carried out at 37°C for 30 min before being quenched by adding ice-cold 20% acetonitrile (780 μL) to make 40-fold dilutions. The samples were then kept on ice until aliquots of 10 μL were injected and analyzed by a Shimadzu LC-2010A HPLC system equipped with a membrane on-line degasser, a temperature control unit, and a fluorescence detector. A reverse-phase Premier C18 column (250 × 4.6 mm i.d., 5 μm particle size, Shimadzu) protected with a C18 guard column cartridge was used. The mobile phase was 20% acetonitrile. The fluorescent compounds LacβMU and Neu5Acα2,6LacβMU were detected with excitation wavelength at 325 nm and emission wavelength at 372 nm (Kajihara et al. 2004). All assays were carried out in duplicate.
Apparent kinetic parameters of the α2,6-trans-sialidase activity
The assays were performed in a total volume of 20 μL in MES buffer (100 mM, pH 6.0) containing Neu5Acα2,6LacβProN3 or Neu5AcαpNP, LacβMU, and Δ15Pd2,6ST (16 μg when Neu5Acα2,6LacβProN3 was used as the sialyl donor substrate and 1.2 μg when Neu5AcαpNP was used as the sialyl donor substrate). Reactions were allowed to proceed for 20 min at 37°C. Apparent kinetic parameters for Neu5Acα2,6LacβProN3 were obtained by varying the concentrations of Neu5Acα2,6LacβProN3 (0.5, 1, 2, 4, and 8 mM) and a fixed concentration of LacβMU (1 mM). Apparent kinetic parameters for Neu5AcαpNP were obtained by varying the concentrations of Neu5AcαpNP (30, 50, 100, and 150 mM) and a fixed concentration of LacβMU (2 mM). All assays were carried out in duplicate. Kinetic parameters were determined by fitting the rates and substrate concentrations to the Michaelis–Menten equation using GraFit 5.0.
Kinetic mechanism for the α2,6-trans-sialidase activity
The kinetic mechanism was explored by measuring the initial rates of LacβMU formation at a series of concentrations (1, 1.5, 3, and 9 mM) of Neu5Acα2,6LacβMU in the presence of LacβProN3 (0.5, 1.0, 2.5, or 10 mM). The assays were performed in a total volume of 20 μL in MES buffer (100 mM, pH 6.0) containing Neu5Acα2,6LacβMU, LacβProN3, and Δ15Pd2,6ST (12 μg). Reactions were allowed to proceed for 20 min at 37°C. All assays were carried out in duplicate. The data fit well to Eq. (1), which is the rate equation for a bi-bi ping-pong reaction where a parallel initial velocity pattern is observed (Segel 1975):
| (1) |
where V is the maximal velocity, A and B are the concentrations of substrates, and Ka and Kb are the Michaelis–Menten constants for substrates Neu5Acα2,6LacβMU and LacβProN3, respectively.
Effects of CMP
Different concentrations (0.0005, 0.001, 0.005, 0.01, 0.1, 1, and 10 mM) of CMP were used to study its effect on the α2,6-sialidase (1 mM Neu5Acα2,6LacβMU was incubated with 31 μg enzyme in a total volume of 20 μL in 100 mM MES buffer at pH 6.2) and the α2,6-trans-sialidase (1 mM LacβMU and 1.5 mM Neu5AcαpNP were incubated with 16 μg enzyme in a total volume of 20 μL in 100 mM MES buffer at pH 6.0) activities of Δ15Pd2,6ST. Reactions without CMP were used as controls. All reactions were carried out in duplicate at 37°C for 20 min before being quenched by adding ice-cold 20% acetonitrile.
Capillary electrophoresis assays
To detect whether CMP-Neu5Ac was present in the trans-sialidase reactions carried out in the presence of CMP, aliquots of reaction mixtures were analyzed using a P/ACE™ MDQ capillary electrophoresis (CE) system equipped with a UV-Vis detector (Beckman Coulter, Fullerton, CA). CE conditions were as follows: 75 μm i.d. capillary, 25 KV/80 μÅ, 5 s vacuum injections, monitored at 254 nm, and sodium tetraborate (25 mM, pH 9.4) buffer was used as a running buffer. CMP-Neu5Ac standard solution (1 mM) was used as a positive control.
Kinetic mechanisms of CMP activation
To study the CMP activation mechanism of the α2,6-sialidase activity of Δ15Pd2,6ST, initial velocity data were obtained from α2,6-sialidase assays performed in a total volume of 20 μL in MES buffer (100 mM, pH 6.2) containing Neu5Acα2,6LacβMU (at various concentrations: 1.0, 1.5, 3.0, and 9.0 mM) and Δ15Pd2,6ST (8.3 μg) in the absence or the presence of CMP (1.0 and 10 mM). The data obtained in the absence of CMP were used to determine the apparent kinetic parameters for the α2,6-sialidase activity of Δ15Pd2,6ST. To study the CMP activation mechanism of the α2,6-trans-sialidase activity of Δ15Pd2,6ST, initial velocity data were obtained from α2,6-trans-sialidase assays performed in a total volume of 20 μL in MES buffer (100 mM, pH 6.0) containing Neu5Acα2,6LacβProN3 (2.0 mM), LacβMU (0.1, 0.2, 0.5, 1.0, 2.0, and 5.0 mM), and Δ15Pd2,6ST (3.3 μg) in the absence or the presence of CMP (1.0 and 10 mM). All sialidase and trans-sialidase reactions were carried out in duplicate and allowed to proceed for 20 min at 37°C. The data were fit to the following equation:
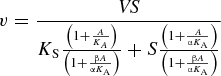 |
(2) |
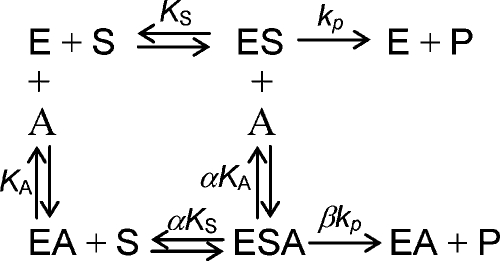
where V is the maximal velocity, S and A are the concentrations of substrate LacβMU and activator CMP, KS and KA are the Michaelis–Menten constants for the substrate and the activator, and α is the factor by which the Michaelis–Menten constants change when activator or substrate occupy the enzyme. The overall equilibrium constant for the formation of enzyme-substrate-activator complex (ESA) must be the same regardless of the paths (E→ES→ESA or E→EA→ESA). β is the factor by which the rate constant changes when activator occupies the enzyme (Figure 7) (Segel 1975).
Fig. 7.
A general scheme for non-essential activation. Abbreviation: E, enzyme; S, substrate; A, activator; P, product; ES, complex of enzyme and substrate; EA, complex of enzyme and activator; ESA, complex of enzyme, substrate, and activator (Segel 1975).
Testing the sialidase substrate specificity of Δ15Pd2,6ST
The assays were performed in a total volume of 22 μL in MES buffer using biotinylated α2,3- or α2,6-linked sialosides (10 μM) as substrates. The conditions used for various sialidases were as follows: PmST1 (12 μg), MES buffer (100 mM, pH 5.5); Δ15Pd2,6ST (8 μg), MES buffer (100 mM, pH 6.2); Clostridium perfingens sialidase (2 mU), MES buffer (100 mM, pH 5.5); Streptococcus pneumoniae sialidase (0.5 mU), MES buffer (100 mM, pH 6.0). All assays were carried out in triplicate. Three sets of assays with same condition were set up in a 384-well plate and performed at 37°C for 1 h, 4 h, and 13 h, respectively. Aliquots (20 μL each) of reaction mixtures were then transferred to NeutrAvidin-coated 384-well plate. After 1 h incubation at room temperature (RT), the plate was washed with three rounds of 1 × PBS buffer containing 0.05% Tween 20 and blocked with 1 × PBS buffer containing 1% BSA for 30 min. Fluorescein-labeled MAA (20 μg mL−1 in 1 × PBS buffer containing 1% BSA) was used to detect the unreacted α2,3-linked sialoside, and fluorescein-labeled SNA (20 μg mL−1 in 1 × PBS buffer containing 1% BSA) was used to detect the unreacted α2,6-linked sialoside in the sialidase reactions as described previously (Chokhawala et al. 2008). Biotek synergy HT microplate reader was used to measure fluorescence with excitation and emission wavelengths of 485 nm and 528 nm, respectively.
Synthesis of sialic acid precursors
Synthesis of biotinylated lactoside and α2,3- and α2,6-linked sialosides was performed as described previously (Yu et al. 2006; Chokhawala et al. 2008). Neu5Acα2,3LacβProN3, Neu5Acα2,6LacβProN3, and LacβMU were synthesized as described before (Yu et al. 2005, 2006).
Chemical synthesis of Neu5AcαpNP
Neu5AcαpNP was chemically synthesized using a previously reported method (Rothermel and Faillard 1990) with modifications. Briefly, the methyl ester of Neu5Ac was prepared in 97% yield using methanol and H+ resin. The glycosyl chloride was prepared by treating the methyl ester with freshly distilled acetyl chloride in acetic acid and directly used for the following transformations. Glycosylation of glycosyl chloride with para-nitrophenol in the presence of a phase-transfer catalyst tetrabutylammonium hydrogensulfate (TBAHS) in aqueous sodium hydroxide solution and dichloromethane gave the protected Neu5AcαpNP in 80% (two steps). Zemplén-deacetylation of the above compound with sodium methoxide in methanol followed by saponification with 0.1 N aqueous NaOH gave the target Neu5AcαpNP in 82% yield.
Preparative synthesis of Neu5Acα2,6LacβMU to confirm the α2,6-trans-sialidase activity of Δ15Pd2,6ST
The reaction was carried out at 37°C in 10 mL of MES buffer (100 mM, pH 6.0) containing LacβMU (25 mg, 50 μmol), Neu5AcαpNP (34 mg, 75 μmol), and Δ15Pd2,6ST (3.3 mg). The reaction was incubated at 37°C for around 2 h when TLC analysis (EtOAc:MeOH:H2O:HOAc = 4:2:1:0.2, v/v) indicated the completion of the reaction. The reaction was quenched by adding an equal volume of ice-cold ethanol, and the mixture was kept on ice for 30 min. The precipitates were removed by centrifugation, and the supernatant was concentrated. The purification was carried using BioGel P-2 gel filtration chromatography and silica gel chromatography to give desired Neu5Acα2,6LacβMU (36 mg, 89% yield). The product and the purity were confirmed by comparing the 1H NMR and high-resolution mass spectrometry data to the data reported previously for Neu5Acα2,6LacβMU (Yu et al. 2005, 2006).
Sequence analysis
Sequences were aligned using Clustal X (version 2.0) (Larkin et al. 2007) and aligned sequences were presented using Genedoc (version 3.2).
Funding
National Institutes of Health (Award no R01GM076360) and National Science Foundation CAREER (Award CHE-0548235).
Acknowledgments
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences, the National Institutes of Health, or the National Science Foundation. X.C. is a Beckman Young Investigator, an Alfred P. Sloan Research Fellow, a Camille Dreyfus Teacher-Scholar, and a UC-Davis Chancellor's Fellow.
Conflict of interest statement
None declared.
Abbreviations
- CMP
cytidine 5′-monophosphate
- CMP-Neu5Ac
cytidine 5′-monophosphate-N-acetylneuraminic acid
- GT
glycosyltransferase
- HPLC
high-performance liquid chromatography
- LacβMU
4-methylumbelliferyl-β-d-lactoside
- MAA
Maackia amurensis Lectin
- MES
2-(N-morpholino)ethanesulfonic acid
- Neu5Ac
N-acetylneuraminic acid
- Neu5AcαpNP
para-nitrophenyl α-d-Neu5Ac
- pNP
para-nitrophenyl
- SNA
Sambucus nigra Lectin
- ST
sialyltransferase
- TLC
thin-layer chromotography
References
- Amaya MF, Watts AG, Damager I, Wehenkel A, Nguyen T, Buschiazzo A, Paris G, Frasch AC, Withers SG, Alzari PM. Structural insights into the catalytic mechanism of Trypanosoma cruzi trans-sialidase. Structure. 2004;12:775–784. doi: 10.1016/j.str.2004.02.036. [DOI] [PubMed] [Google Scholar]
- Angata T, Varki A. Chemical diversity in the sialic acids and related alpha-keto acids: An evolutionary persepective. Chem Rev. 2002;102:439–469. doi: 10.1021/cr000407m. [DOI] [PubMed] [Google Scholar]
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- Bern C, Montgomery SP, Katz L, Caglioti S, Stramer SL. Chagas disease and the US blood supply. Curr Opin Infect Dis. 2008;21:476–482. doi: 10.1097/QCO.0b013e32830ef5b6. [DOI] [PubMed] [Google Scholar]
- Buschiazzo A, Amaya MF, Cremona ML, Frasch AC, Alzari PM. The crystal structure and mode of action of trans-sialidase, a key enzyme in Trypanosoma cruzi pathogenesis. Mol Cell. 2002;10:757–768. doi: 10.1016/s1097-2765(02)00680-9. [DOI] [PubMed] [Google Scholar]
- Cambron LD, Leskawa KC. Inhibition of CMP-N-acetylneuraminic acid: lactosylceramide sialyltransferase by nucleotides, nucleotide sugars and nucleotide dialdehydes. Biochem Biophys Res Commun. 1993;193:585–590. doi: 10.1006/bbrc.1993.1664. [DOI] [PubMed] [Google Scholar]
- Campbell JA, Davies GJ, Bulone V, Henrissat B. A classification of nucleotide-diphospho-sugar glycosyltransferases based on amino acid sequence similarities. Biochem J. 1997;326(Pt 3):929–939. doi: 10.1042/bj3260929u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekaran EV, Xue J, Xia J, Locke RD, Matta KL, Neelamegham S. Reversible sialylation: synthesis of cytidine 5′-monophospho-N-acetylneuraminic acid from cytidine 5′-monophosphate with alpha2,3-sialyl O-glycan-, glycolipid-, and macromolecule-based donors yields diverse sialylated products. Biochemistry. 2008;47:320–330. doi: 10.1021/bi701472g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Yu H, Lau K, Huang S, Chokhawala HA, Li Y, Tiwari VK, Chen X. Multifunctionality of Campylobacter jejuni sialyltransferase CstII: Characterization of GD3/GT3 oligosaccharide synthase, GD3 oligosaccharide sialidase, and trans-sialidase activities. Glycobiology. 2008;18:686–697. doi: 10.1093/glycob/cwn047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chokhawala HA, Huang S, Lau K, Yu H, Cheng J, Thon V, Hurtado-Ziola N, Guerrero JA, Varki A, Chen X. Combinatorial chemoenzymatic synthesis and high-throughput screening of sialosides. ACS Chem Biol. 2008;3:567–576. doi: 10.1021/cb800127n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chokhawala HA, Yu H, Chen X. High-throughput substrate specificity studies of sialidases by using chemoenzymatically synthesized sialoside libraries. Chembiochem. 2007;8:194–201. doi: 10.1002/cbic.200600410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colli W. Trans-sialidase: A unique enzyme activity discovered in the protozoan Trypanosoma cruzi. FASEB J. 1993;7:1257–1264. doi: 10.1096/fasebj.7.13.8405811. [DOI] [PubMed] [Google Scholar]
- Cook PF, Cleland WW. Enzyme Kinetics and Mechanism. New York: Garland Science; 2007. [Google Scholar]
- Coutinho PM, Deleury E, Davies GJ, Henrissat B. An evolving hierarchical family classification for glycosyltransferases. J Mol Biol. 2003;328:307–317. doi: 10.1016/s0022-2836(03)00307-3. [DOI] [PubMed] [Google Scholar]
- Cross GA, Takle GB. The surface trans-sialidase family of Trypanosoma cruzi. Annu Rev Microbiol. 1993;47:385–411. doi: 10.1146/annurev.mi.47.100193.002125. [DOI] [PubMed] [Google Scholar]
- Damager I, Buchini S, Amaya MF, Buschiazzo A, Alzari P, Frasch AC, Watts A, Withers SG. Kinetic and mechanistic analysis of Trypanosoma cruzi trans-sialidase reveals a classical ping-pong mechanism with acid/base catalysis. Biochemistry. 2008;47:3507–3512. doi: 10.1021/bi7024832. [DOI] [PubMed] [Google Scholar]
- Frasch AC. Functional diversity in the trans-sialidase and mucin families in Trypanosoma cruzi. Parasitol Today. 2000;16:282–286. doi: 10.1016/s0169-4758(00)01698-7. [DOI] [PubMed] [Google Scholar]
- Gilbert M, Karwaski MF, Bernatchez S, Young NM, Taboada E, Michniewicz J, Cunningham AM, Wakarchuk WW. The genetic bases for the variation in the lipo-oligosaccharide of the mucosal pathogen, Campylobacter jejuni. Biosynthesis of sialylated ganglioside mimics in the core oligosaccharide. J Biol Chem. 2002;277:327–337. doi: 10.1074/jbc.M108452200. [DOI] [PubMed] [Google Scholar]
- Gut H, King SJ, Walsh MA. Structural and functional studies of Streptococcus pneumoniae neuraminidase B: An intramolecular trans-sialidase. FEBS Lett. 2008;582:3348–3352. doi: 10.1016/j.febslet.2008.08.026. [DOI] [PubMed] [Google Scholar]
- Harduin-Lepers A, Vallejo-Ruiz V, Krzewinski-Recchi MA, Samyn-Petit B, Julien S, Delannoy P. The human sialyltransferase family. Biochimie. 2001;83:727–737. doi: 10.1016/s0300-9084(01)01301-3. [DOI] [PubMed] [Google Scholar]
- Harrison JA, Kartha KP, Turnbull WB, Scheuerl SL, Naismith JH, Schenkman S, Field RA. Hydrolase and sialyltransferase activities of Trypanosoma cruzi trans-sialidase towards NeuAc-alpha-2,3-gal-Gal-beta-O-pNP. Bioorg Med Chem Lett. 2001;11:141–144. doi: 10.1016/s0960-894x(00)00611-9. [DOI] [PubMed] [Google Scholar]
- Kajihara Y, Kamiyama D, Yamamoto N, Sakakibara T, Izumi M, Hashimoto H. Synthesis of 2-[(2-pyridyl)amino]ethyl beta-d-lactosaminide and evaluation of its acceptor ability for sialyltransferase: A comparison with 4-methylumbelliferyl and dansyl beta-d-lactosaminide. Carbohydr Res. 2004;339:1545–1550. doi: 10.1016/j.carres.2004.03.015. [DOI] [PubMed] [Google Scholar]
- Kajihara Y, Yamamoto T, Nagae H, Nakashizuka M, Sakakibara T, Terada I. A novel a-2,6-sialyltransferase: Transfer of sialic acid to fucosyl and sialyl trisaccharides. J Org Chem. 1996;61:8632–8635. [Google Scholar]
- Kakuta Y, Okino N, Kajiwara H, Ichikawa M, Takakura Y, Ito M, Yamamoto T. Crystal structure of Vibrionaceae Photobacterium sp. JT-ISH-224 alpha2,6-sialyltransferase in a ternary complex with donor product CMP and acceptor substrate lactose: Catalytic mechanism and substrate recognition. Glycobiology. 2008;18:66–73. doi: 10.1093/glycob/cwm119. [DOI] [PubMed] [Google Scholar]
- Lairson LL, Wakarchuk WW, Withers SG. Alternative donor substrates for inverting and retaining glycosyltransferases. Chem Commun. 2007;4:365–367. doi: 10.1039/b614636h. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Ribeirao M, Pereira-Chioccola VL, Eichinger D, Rodrigues MM, Schenkman S. Temperature differences for trans-glycosylation and hydrolysis reaction reveal an acceptor binding site in the catalytic mechanism of Trypanosoma cruzi trans-sialidase. Glycobiology. 1997;7:1237–1246. doi: 10.1093/glycob/7.8.1237. [DOI] [PubMed] [Google Scholar]
- Rothermel J, Faillard H. Phase-transfer-catalyzed synthesis of aryl alpha-ketosides of N-acetylneuraminic acid. A 2-methylfluoran-6-yl glycoside of N-acetylneuraminic acid, 2-methyl-6-(5-acetamido-3,5-dideoxy-alpha-d-glycero-d-galacto-nonulopyranosylonic acid)xanthene-9-spiro-1′-isobenzofuran-3′-one, a new substrate for neuraminidase assay. Carbohydr Res. 1990;196:29-40. doi: 10.1016/0008-6215(90)84104-3. [DOI] [PubMed] [Google Scholar]
- Schauer R. Achievements and challenges of sialic acid research. Glycoconj J. 2000;17:485–499. doi: 10.1023/A:1011062223612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenkman S, Jiang MS, Hart GW, Nussenzweig V. A novel cell surface trans-sialidase of Trypanosoma cruzi generates a stage-specific epitope required for invasion of mammalian cells. Cell. 1991;65:1117–1125. doi: 10.1016/0092-8674(91)90008-m. [DOI] [PubMed] [Google Scholar]
- Scudder P, Doom JP, Chuenkova M, Manger ID, Pereira ME. Enzymatic characterization of beta-d-galactoside alpha 2,3-trans-sialidase from Trypanosoma cruzi. J Biol Chem. 1993;268:9886–9891. [PubMed] [Google Scholar]
- Segel IH. Enzyme Kinetics: Behavior and Analysis of Rapid Equilibrium and Steady-State Enzyme Systems. New York: Wiley; 1975. [Google Scholar]
- Shen GJ, Datta AK, Izumi M, Koeller KM, Wong CH. Expression of alpha2,8/2,9-polysialyltransferase from Escherichia coli K92. Characterization of the enzyme and its reaction products. J Biol Chem. 1999;274:35139–35146. doi: 10.1074/jbc.274.49.35139. [DOI] [PubMed] [Google Scholar]
- Sun M, Li Y, Chokhawala HA, Henning R, Chen X. N-Terminal 112 amino acid residues are not required for the sialyltransferase activity of Photobacterium damsela alpha2,6-sialyltransferase. Biotechnol Lett. 2008;30:671-676. doi: 10.1007/s10529-007-9588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo C, Hwang T, Chen P, Hung C, Gao H, Chang L, Lin C. Synthesis of sialyl TN glycopeptides – Enzymatic sialylation by α2,6-sialyltransferase from Photobacterium damsela. Adv Synth Catal. 2005;347:967–972. [Google Scholar]
- Tsuji S, Datta AK, Paulson JC. Systematic nomenclature for sialyltransferases. Glycobiology. 1996;6:v-vii. doi: 10.1093/glycob/6.7.647. [DOI] [PubMed] [Google Scholar]
- Tsukamoto H, Takakura Y, Mine T, Yamamoto T. Photobacterium sp. JT-ISH-224 produces two sialyltransferases, alpha-/beta-galactoside alpha2,3-sialyltransferase and beta-galactoside alpha2,6-sialyltransferase. J Biochem. 2008;143:187–197. doi: 10.1093/jb/mvm208. [DOI] [PubMed] [Google Scholar]
- Yamamoto T, Nakashizuka M, Terada I. Cloning and expression of a marine bacterial beta-galactoside alpha2,6-sialyltransferase gene from Photobacterium damsela JT0160. J Biochem. 1998;123:94–100. doi: 10.1093/oxfordjournals.jbchem.a021921. [DOI] [PubMed] [Google Scholar]
- Yang J, Schenkman S, Horenstein BA. Primary 13C and beta-secondary 2H KIEs for trans-sialidase. A snapshot of nucleophilic participation during catalysis. Biochemistry. 2000;39:5902–5910. doi: 10.1021/bi000061+. [DOI] [PubMed] [Google Scholar]
- Yu H, Chen X. Carbohydrate post-glycosylational modifications. Org Biomol Chem. 2007;5:865–872. doi: 10.1039/b700034k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Chokhawala H, Karpel R, Yu H, Wu B, Zhang J, Zhang Y, Jia Q, Chen X. A multifunctional Pasteurella multocida sialyltransferase: A powerful tool for the synthesis of sialoside libraries. J Am Chem Soc. 2005;127:17618–17619. doi: 10.1021/ja0561690. [DOI] [PubMed] [Google Scholar]
- Yu H, Huang S, Chokhawala H, Sun M, Zheng H, Chen X. Highly efficient chemoenzymatic synthesis of naturally occurring and non-natural alpha-2,6-linked sialosides: A P. damsela alpha-2,6-sialyltransferase with extremely flexible donor-substrate specificity. Angew Chem Int Ed Engl. 2006;45:3938–3944. doi: 10.1002/anie.200600572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Griffith BR, Fu Q, Albermann C, Fu X, Lee IK, Li L, Thorson JS. Exploiting the reversibility of natural product glycosyltransferase-catalyzed reactions. Science. 2006;313:1291–1294. doi: 10.1126/science.1130028. [DOI] [PubMed] [Google Scholar]