Abstract
Background
A strong cumulative effect of five genetic variants and family history on prostate cancer risk was recently reported in a Swedish population (CAPS). We carried out this study to confirm the finding in two U.S. study populations and perform a combined analysis to obtain a more stable estimate of the Odds Ratio (OR) for prostate cancer.
Methods
We evaluated three SNPs at 8q24 and one SNP each at 17q12 and 17q24.3 in two study populations in the U.S. The first was a hospital-based case-control study population at Johns Hopkins Hospital (JHH), including 1,563 prostate cancer patients and 576 control subjects. The second was the National Cancer Institute Cancer Genetic Markers of Susceptibility (CGEMS) Initiative, including 1,172 prostate cancer patients and 1,157 control subjects.
Results
We confirmed a cumulative effect of five risk variants on prostate cancer risk. Based on a total of 5,628 cases and 3,514 controls from JHH, CGEMS, and CAPS, men who carry any combination of 1, 2, 3, and 4 or more of these five risk variants have an estimated OR (95% CI) of 1.41 (1.20-1.67), 1.88 (1.59-2.22), 2.36 (1.95-2.85), and 3.80 (2.77-5.22) for prostate cancer, respectively, compared to men who do not have any of these five risk variants. When family history was included, the cumulative effect was stronger.
Discussion
These results provide an important confirmation for the cumulative effect of five genetic risk variants on prostate cancer risk. The more stable OR estimates of the cumulative effect of these six risk factors are a major step toward individual risk characterization for this disease.
Keywords: association, interaction, 17q12, 17q24.3, 8q24
A strong cumulative effect of five genetic variants on prostate cancer risk was recently reported by our group, based on analyses of Cancer of the Prostate in Sweden (CAPS), a Swedish population-based case-control study [1]. In that study, five prostate cancer risk associated variants [2-7] previously identified through genome-wide association studies were evaluated in 2,893 prostate cancer patients and 1,781 control subjects. While each of these variants was found to be independently and moderately associated with prostate cancer risk, combinations of risk alleles had a stronger cumulative effect on prostate cancer. Compared to men who did not have any of these five risk variants and family history, men who carried any combination of 2, 3, 4, and ≥ 5 of these risk factors had an odds ratio of 2.07, 2.71, 4.76, and 9.46, respectively (P-trend = 3.93 × 10−28).
The strength of the cumulative effect of these common risk variants, if confirmed, would provide an important step in identifying men who have increased risk for prostate cancer. In this study, we performed independent confirmation studies in two additional study populations and subsequently performed a combined analysis to obtain more stable risk estimates.
The first study population consisted of samples from the Johns Hopkins Hospital (JHH). Case patients were 1,563 men of self-reported European American (EA) ancestry who underwent treatment for prostate cancer at the hospital between 1999 and 2006. Each patient’s tumor was graded using the Gleason scoring system [8] and staged using the TNM (tumor–node–metastasis) system [9]. During the same time period, men who were undergoing screening for prostate cancer at the hospital were asked to participate as control subjects. A total of 576 men met our inclusion criteria as control subjects for this study: EA, age above 55 years, normal digital rectal examination, and a serum prostate-specific antigen (PSA) level < 4.0 ng/mL. All five of the risk variants reported in the study were directly genotyped using the same Sequenom iPLEX method [1], including rs4430796 at 17q12, rs1859962 at 17q24.3, rs16901979 at Region 2 of 8q24, rs6983267 at Region 3 of 8q24, and rs1447295 at Region 1 of 8q24.
The second study population was from the National Cancer Institute Cancer Genetic Markers of Susceptibility Initiative for prostate cancer (CGEMS-prostate) [5]. The CGEMS-prostate study included 1,172 prostate cancer case patients and 1,157 control subjects of EA who were selected from the Prostate, Lung, Colon and Ovarian (PLCO) Cancer Screening Trial using an incidence density sampling strategy. Individual genotype data were downloaded from http://cgems.cancer.gov/data/. Four (rs4430796, rs1859962, rs6983267 and rs1447295) of the five SNPs were directly genotyped as part of the 550,000 SNPs in the CGEMS-prostate genome-wide association study [5]. One SNP (rs16901979) was imputed from the adjacent genotyped SNPs at 8q24 using the computer program IMPUTE [10-11]. The average confidence score for the imputed SNP was 1.00, suggesting a reliable imputation for the SNP.
We evaluated the association of prostate cancer risk with each of these five risk variants in the JHH and CGEMS-prostate study populations using single SNP analysis and adjusted for age in each 5-year interval. Using the best genetic model observed in the CAPS [1], we found the risk genotypes of each SNP were more common in cases than in controls in each of these two study populations. When these SNPs were included in a multivariate analysis where all five risk variants, family history (not included in the JHH because of incomplete data), and age were included, three SNPs (rs4430796, rs6983267, and rs1447295) in the JHH and 4 SNPs (rs4430796, rs1859962, rs6983267, and rs1447295) in the CGEMS-prostate were independently and significantly (P < 0.05) associated with prostate cancer risk (Table 1).
Table 1. Adjusted OR of five risk variants and family history for prostate cancer risk in three independent studies.
| Study (case/control) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Variables/SNPs | Chromosomal Region |
Genotypea |
JHH (1,563/576)b |
CGEMS (1,172/1,157)c |
CAPS (2,893/1,781)d |
||||
| Ref. | Risk | OR (95% CI) | Pe | OR (95% CI) | Pe | OR (95% CI) | Pe | ||
| Age | 1.01 (0.93-1.09) | 0.82 | 1.05 (0.96-1.15) | 0.26 | 1.01 (1.00-1.02) | 0.02 | |||
| Family historyf | No | Yes | - | - | 2.01 (1.46-2.78) | 1.21E-05 | 2.22 (1.83-2.68) | 1.15E-17 | |
| rs4430796 | 17q12 | CC/TC | TT | 1.34 (1.07-1.67) | 9.12E-03 | 1.33 (1.10-1.62) | 3.31E-03 | 1.38 (1.21-1.57) | 1.62E-06 |
| rs1859962 | 17q24.3 | GT/TT | GG | 1.08 (0.86-1.36) | 4.93E-01 | 1.31 (1.08-1.60) | 5.84E-03 | 1.28 (1.11-1.47) | 5.49E-04 |
| rs16901979 | 8q24 (Region 2) | CC | AA/CA | 1.11 (0.78-1.58) | 5.49E-01 | 1.06 (0.76-1.47) | 7.29E-01 | 1.53 (1.22-1.92) | 1.83E-04 |
| rs6983267 | 8q24 (Region 3) | TT | GT/GG | 1.38 (1.09-1.76) | 8.62E-03 | 1.47 (1.19-1.81) | 3.05E-04 | 1.37 (1.18-1.59) | 3.44E-05 |
| rs1447295 | 8q24 (Region 1) | GG | CA/AA | 1.80 (1.39-2.34) | 5.05E-06 | 1.48 (1.20-1.83) | 1.99E-04 | 1.22 (1.06-1.40) | 5.31E-03 |
Ref. (refereence) and Risk genotyped were defined based on the CAPS (Zheng 2008).
(JHH) Johns Hopkins Hospital, European Americans
(CGEMS) Cancer Genetic Markers of Susceptibility Initiative , European Americans
(CAPS) CAncer of the Prostate in Sweden, a population-based case-control study in Sweden
Based on likelihood ratio test. Family history and five SNPs are included in the multivariate logistic regression model, adjusting for age in a five-year interval (for all three populations), and geographic region (for CAPS only)
Family history data are incomplete in the JHH study
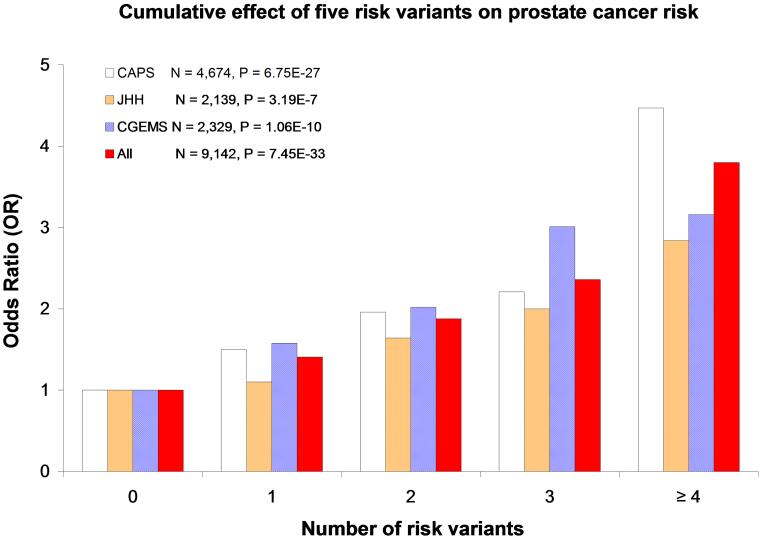
Similar to the results of the CAPS study [1], we observed a stronger cumulative effect of these five risk variants on prostate cancer risk in both of these confirmation study populations (Table 2). Compared to men who do not have any of these five risk variants, men who carry any combination of 1, 2, 3, and 4 or more of these risk genotypes have gradually increased OR for prostate cancer (Figure 1). The trend test was statistically significant in JHH (P = 3.19 × 10−7), in the CGEMS-prostate (P = 1.06 × 10−10), and in the combined CAPS, JHH, and CGEMS-prostate (P = 7.45 × 10−33).
Table 2. Cummulative effect of five risk variants on prostate cancer risk in three independent studies.
| # of risk factorsa |
# (%) of subjects |
||||
|---|---|---|---|---|---|
| Cases | Controls | Odds Ratio (95% CI) | Pb | P-trendc | |
| JHHd | |||||
| 0 | 104 (6.81) | 53 (9.46) | 1 | ||
| 1 | 545 (35.67) | 254 (45.36) | 1.10 (0.76-1.57) | 6.20E-01 | |
| 2 | 579 (37.89) | 180 (32.14) | 1.64 (1.13-2.38) | 8.00E-03 | |
| 3 | 250 (16.36) | 64 (11.43) | 2.00 (1.30-3.07) | 2.00E-03 | |
| ≥ 4 | 50 (3.27) | 9 (1.61) | 2.84 (1.30-6.21) | 9.00E-03 | 3.19E-07 |
| CGEMSe | |||||
| 0 | 73 (6.21) | 125(11.35) | 1.00 | - | |
| 1 | 437 (37.16) | 475 (43.14) | 1.59(1.16-2.19) | 3.75E-03 | |
| 2 | 431 (36.65) | 366 (33.24) | 2.04(1.48-2.82) | 1.04E-05 | |
| 3 | 202 (17.18) | 117 (10.63) | 2.95(2.04-4.28) | 5.05E-09 | |
| ≥ 4 | 33 (2.81) | 18 (1.63) | 3.09(1.62-5.90) | 4.58E-04 | 1.06E-10 |
| CAPSf | |||||
| 0 | 162 (5.64) | 173 (10.12) | 1.00 | - | |
| 1 | 885 (30.80) | 628 (36.75) | 1.49 (1.18-1.90) | 8.27E-04 | |
| 2 | 1123 (39.09) | 617 (36.10) | 1.92 (1.52-2.44) | 2.41E-08 | |
| 3 | 548 (19.07) | 254 (14.86) | 2.27 (1.74-2.93) | 1.06E-09 | |
| ≥ 4 | 155 (5.40) | 37 (2.17) | 4.47 (2.94-6.79) | 3.52E-14 | 8.34348E-18 |
| Combined | |||||
| 0 | 339 (6.08) | 351 (10.42) | 1.00 | ||
| 1 | 1867 (33.47) | 1357 (40.28) | 1.41 (1.20-1.67) | 5.05E-05 | |
| 2 | 2133 (38.24) | 1163 (34.52) | 1.88 (1.59-2.22) | 1.87E-13 | |
| 3 | 1001 (17.95) | 434 (12.88) | 2.36 (1.95-2.85) | 6.15E-19 | |
| ≥ 4 | 238 (4.27) | 64 (1.90) | 3.80 (2.77-5.22) | 3.82E-19 | 7.45E-33 |
We tested the cumulative effects of the five SNPs on prostate cancer by counting the number of prostate cancer risk genotypes (based on the best-fitting genetic model from single SNP analysis in Ref. 1, and coded as 1 if individuals carried risk genotypes and 0 otherwise) for these five SNPs in each subject. The OR for prostate cancer for men carrying any combination of 1, 2, 3, or ≥ 4 risk genotypes was estimated by comparing to men carrying none of the risk genotypes using logistic regression analysis.
P-values are two-sided and were calculated by a likelihood-ratio test, ajusting for age (in 5-year intervals) and study effect for the combined analysis (CAPS, CGEMS, and JHH included as a covariate)
P-values were calculated by the Cochran-Armitage test for trends, adjusting for age and study effect for the combined analysis
(JHH) Johns Hopkins Hospital, European Americans
(CGEMS) Cancer Genetic Markers of Susceptibility Initiative , European Americans
(CAPS) CAncer of the Prostate in Sweden, a population-based case-control study in Sweden
Figure 1.
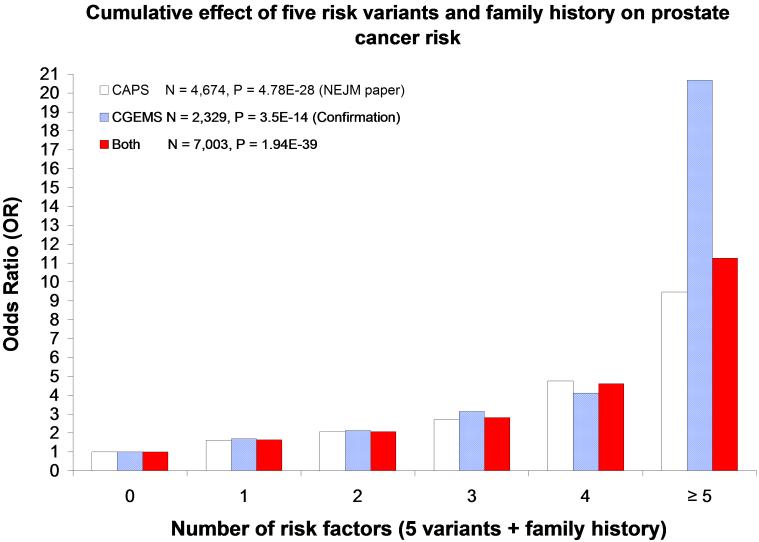
Because family history was independent from the cumulative risk genotype effect, we included it as another risk factor and estimated cumulative effect of these six risk factors on prostate cancer in the CGEMS-prostate population (The JHH population was not included because of incomplete data on family history). Similar to the analysis in CAPS [1], we found a stronger cumulative effect in the CGEMS-prostate study population (Table 3). The estimated ORs for groups of any combination of 1, 2, 3, 4, and 5 or more were similar to that of CAPS. The trend test was statistically significant in the CGEMS-prostate (P = 4.75 × 10−14), and in the combined CAPS and CGEMS-prostate (P = 1.94 × 10−39). The large sample size of the combined analysis provided more stable estimates of OR for prostate cancer. Compared to men who do not have any of these six risk factors, men who carry any combination of 1, 2, 3, 4, and 5 or more of these risk factors have estimated OR (95% CI) of 1.64 (1.34-2.00), 2.07 (1.70-2.51), 2.82 (2.28-3.50), 4.61 (3.40-6.25), and 11.26 (4.74-24.75) for prostate cancer, respectively (Figure 2).
Table 3. Cummulative effect of six risk variants on prostate cancer risk in three independent studies.
| # of risk factorsa |
# (%) of subjects |
||||
|---|---|---|---|---|---|
| Cases | Controls | Odds Ratio (95% CI) | Pb | P for trendc | |
| CGEMS d | |||||
| 0 | 63 (5.36) | 118 (10.72) | 1.00 | ||
| 1 | 403 (34.27) | 449 (40.78) | 1.69(1.21-2.36) | 1.80E-03 | |
| 2 | 430 (36.56) | 379 (34.42) | 2.13(1.52-2.98) | 6.72E-06 | |
| 3 | 217 (18.45) | 130 (11.81) | 3.15(2.16-4.58) | 9.43E-10 | |
| 4 | 52 (4.42) | 24 (2.18) | 4.11(2.32-7.30) | 5.36E-07 | |
| ? 5 | 11 (0.94) | 1 (0.09) | 20.68(2.61-163.85) | 5.76E-05 | 4.75E-14 |
| CAPS e | |||||
| 0 | 144 (4.97) | 174 (10.10) | 1.00 | - | |
| 1 | 780 (26.93) | 578 (33.57) | 1.64 (1.28-2.10) | 9.88E-05 | |
| 2 | 1053 (36.36) | 621 (36.06) | 2.07 (1.62-2.64) | 5.43E-09 | |
| 3 | 642 (22.17) | 285 (16.55) | 2.72 (2.10-3.55) | 7.94E-14 | |
| 4 | 237 (8.18) | 59 (3.43) | 4.87 (3.38-7.00) | 3.71E-19 | |
| ? 5 | 40 (1.38) | 5 (0.29) | 9.47 (3.62-24.72) | 1.28E-08 | 5.87E-27 |
| Combined | |||||
| 0 | 207 (5.08) | 292 (10.35) | 1.00 | ||
| 1 | 1183 (29.04) | 1027 (36.39) | 1.64 (1.34-2.00) | 8.42E-07 | |
| 2 | 1483 (36.41) | 1000 (35.44) | 2.07 (1.70-2.51) | 2.92E-13 | |
| 3 | 860 (21.11) | 414 (14.67) | 2.82 (2.28-3.50) | 8.37E-22 | |
| 4 | 289 (7.10) | 83(2.94) | 4.61 (3.40-6.25) | 3.24E-25 | |
| ? 5 | 51 (1.25) | 6 (0.21) | 11.26 (4.74-24.75) | 2.57E-12 | 1.94E-39 |
We tested the cumulative effects of six genetic factors (5 SNPs + family history) on prostate cancer risk by counting the number of prostate cancer risk factors (based on the best-fitting genetic model from univariate analysis in Ref 1, and coded as 1 if individuals had the risk factor and 0 otherwise) for each subject. The OR for prostate cancer for men carrying any combination of 1, 2, 3, 4, or ? 5 risk factor was estimated by comparing to men carrying none of the risk factors using logistic regression analysis.
P-values are two-sided and were calculated by a likelihood-ratio test, ajusting for age (in 5-year intervals) and study effect for the combined analysis (CAPS and CGEMS included as a covariate)
P-values were calculated by the Cochran-Armitage test for trends, adjusting for age and study effect for the combined effect
(CGEMS) Cancer Genetic Markers of Susceptibility Initiative , European Americans
(CAPS) CAncer of the Prostate in Sweden, a population-based case-control study in Sweden
Figure 2.
In the case-only analysis, no statistically significant association was found between the five risk variants and Gleason score, age at diagnosis, presence of family history (CGEMS-prostate only), or aggressiveness of prostate cancer, as defined in the study of Duggan and colleagues [12] (data not shown). This was not surprising, considering the original studies which identified these risk variants were focused on overall prostate cancer risk, and not on clinical subsets of this disease.
Results from this study independently confirmed the finding of cumulative effect of the five risk variants and family history on prostate cancer risk reported by Zheng and colleagues [1]. Previously, Yeager et al [5] and Zheng [7] had shown that two independent variants at 8q24 had an additive effect on prostate cancer risk. We now demonstrate that increasing the number of independently associated SNPs increases the observed risk. Thus, it is likely that additional risk-associated SNPs may further improve risk assessment. It is also important to note that the large sample size of the present study provides more stable OR estimates for prostate cancer. Such information may prove useful in predicting individual risk of prostate cancer and identifying men who may benefit from more frequent screening. Additional studies in various races/ethnicities, and preferably prospective studies are urgently needed.
Acknowledgements
The authors thank all the study subjects who participated in the CAPS study and urologists who included their patients in the CAPS study. The Regional Cancer Registries and the CAPS-Steering committee including Drs. Jan Adolfsson, Jan-Erik Johansson and Eberhart Varenhorst. The authors also thank all the study subjects participated in the Johns Hopkins Hospital study and The Prostate, Lung, Colon and Ovarian (PLCO) study. We greatly appreciate that the National Cancer Institute Cancer Genetic Markers of Susceptibility Initiative (CGEMS) made the data available publicly, for this type of research. We also thank for the constructive comments on the manuscript by Drs. Stephen Chanock and Richard Hayes of National Cancer Institute, and by Dr. David Hunter of Harvard School of Public Health.
The study is supported by National Cancer Institute CA105055, CA106523, and CA95052 to J.X., CA112517 and CA58236 to W.B.I., CA86323 to AWP, Department of Defense grant PC051264 to J.X, Swedish Cancer Society (Cancerfonden) to HG and Swedish Academy of Sciences (Vetenskapsrådet) to HG. The support of William T Gerrard, Mario A Duhon, John and Jennifer Chalsty, and David Koch to W.B.I is gratefully acknowledged.
References
- 1.Zheng SL, Sun J, Wiklund F, Smith S, Stattin P, Li G, Adami HO, Hsu FC, Zhu Y, Bälter K, Kader AK, Turner AR, Liu W, Bleecker ER, Meyers DA, Duggan D, Carpten JD, Chang BL, Isaacs WB, Xu J, Grönberg H. Cumulative association of five genetic variants with prostate cancer. N Engl J Med. 2008;358(9):910–919. doi: 10.1056/NEJMoa075819. [DOI] [PubMed] [Google Scholar]
- 2.Amundadottir LT, Sulem P, Gudmundsson J, Helgason A, Baker A, Agnarsson BA, Sigurdsson A, Benediktsdottir KR, Cazier JB, Sainz J, Jakobsdottir M, Kostic J, Magnusdottir DN, Ghosh S, Agnarsson K, Birgisdottir B, Le Roux L, Olafsdottir A, Blondal T, Andresdottir M, Gretarsdottir OS, Bergthorsson JT, Gudbjartsson D, Gylfason A, Thorleifsson G, Manolescu A, Kristjansson K, Geirsson G, Isaksson H, Douglas J, Johansson JE, Bälter K, Wiklund F, Montie JE, Yu X, Suarez BK, Ober C, Cooney KA, Gronberg H, Catalona WJ, Einarsson GV, Barkardottir RB, Gulcher JR, Kong A, Thorsteinsdottir U, Stefansson K. A common variant associated with prostate cancer in European and African populations. Nat Genet. 2006;38:652–658. doi: 10.1038/ng1808. [DOI] [PubMed] [Google Scholar]
- 3.Gudmundsson J, Sulem P, Manolescu A, Amundadottir LT, Gudbjartsson D, Helgason A, Rafnar T, Bergthorsson JT, Agnarsson BA, Baker A, Sigurdsson A, Benediktsdottir KR, Jakobsdottir M, Xu J, Blondal T, Kostic J, Sun J, Ghosh S, Stacey SN, Mouy M, Saemundsdottir J, Backman VM, Kristjansson K, Tres A, Partin AW, Albers-Akkers MT, Godino-Ivan Marcos J, Walsh PC, Swinkels DW, Navarrete S, Isaacs SD, Aben KK, Graif T, Cashy J, Ruiz-Echarri M, Wiley KE, Suarez BK, Witjes JA, Frigge M, Ober C, Jonsson E, Einarsson GV, Mayordomo JI, Kiemeney LA, Isaacs WB, Catalona WJ, Barkardottir RB, Gulcher JR, Thorsteinsdottir U, Kong A, Stefansson K. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nat Genet. 2007;39:631–637. doi: 10.1038/ng1999. [DOI] [PubMed] [Google Scholar]
- 4.Haiman CA, Patterson N, Freedman ML, Myers SR, Pike MC, Waliszewska A, Neubauer J, Tandon A, Schirmer C, McDonald GJ, Greenway SC, Stram DO, Le Marchand L, Kolonel LN, Frasco M, Wong D, Pooler LC, Ardlie K, Oakley-Girvan I, Whittemore AS, Cooney KA, John EM, Ingles SA, Altshuler D, Henderson BE, Reich D. Multiple regions within 8q24 independently affect risk for prostate cancer. Nat Genet. 2007;39:638–644. doi: 10.1038/ng2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yeager M, Orr N, Hayes RB, Jacobs KB, Kraft P, Wacholder S, Minichiello MJ, Fearnhead P, Yu K, Chatterjee N, Wang Z, Welch R, Staats BJ, Calle EE, Feigelson HS, Thun MJ, Rodriguez C, Albanes D, Virtamo J, Weinstein S, Schumacher FR, Giovannucci E, Willett WC, Cancel-Tassin G, Cussenot O, Valeri A, Andriole GL, Gelmann EP, Tucker M, Gerhard DS, Fraumeni JF, Jr, Hoover R, Hunter DJ, Chanock SJ, Thomas G. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat Genet. 2007;39:645–649. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 6.Gudmundsson J, Sulem P, Steinthorsdottir V, Bergthorsson JT, Thorleifsson G, Manolescu A, Rafnar T, Gudbjartsson D, Agnarsson BA, Baker A, Sigurdsson A, Benediktsdottir KR, Jakobsdottir M, Blondal T, Stacey SN, Helgason A, Gunnarsdottir S, Olafsdottir A, Kristinsson KT, Birgisdottir B, Ghosh S, Thorlacius S, Magnusdottir D, Stefansdottir G, Kristjansson K, Bagger Y, Wilensky RL, Reilly MP, Morris AD, Kimber CH, Adeyemo A, Chen Y, Zhou J, So WY, Tong PC, Ng MC, Hansen T, Andersen G, Borch-Johnsen K, Jorgensen T, Tres A, Fuertes F, Ruiz-Echarri M, Asin L, Saez B, van Boven E, Klaver S, Swinkels DW, Aben KK, Graif T, Cashy J, Suarez BK, van Vierssen Trip O, Frigge ML, Ober C, Hofker MH, Wijmenga C, Christiansen C, Rader DJ, Palmer CN, Rotimi C, Chan JC, Pedersen O, Sigurdsson G, Benediktsson R, Jonsson E, Einarsson GV, Mayordomo JI, Catalona WJ, Kiemeney LA, Barkardottir RB, Gulcher JR, Thorsteinsdottir U, Kong A, Stefansson K. Two variants on chromosome 17 confer prostate cancer risk, and the one in TCF2 protects against type 2 diabetes. Nat Genet. 2007;39:977–983. doi: 10.1038/ng2062. [DOI] [PubMed] [Google Scholar]
- 7.Zheng SL, Sun J, Cheng Y, Li G, Hsu FC, Zhu Y, Chang BL, Liu W, Kim JW, Turner AR, Gielzak M, Yan G, Isaacs SD, Wiley KE, Sauvageot J, Chen HS, Gurganus R, Mangold LA, Trock BJ, Gronberg H, Duggan D, Carpten JD, Partin AW, Walsh PC, Xu J, Isaacs WB. Additive effects of two unlinked loci at 8q24 are associated with a considerable fraction of prostate cancer among European Americans. JNCI. 2007;99:1525–1533. doi: 10.1093/jnci/djm169. [DOI] [PubMed] [Google Scholar]
- 8.Epstein JI, Allsbrook WC, Jr, Amin MB, Egevad LL, ISUP Grading Committee Am J Surg Pathol. 2005;29:1228–1242. doi: 10.1097/01.pas.0000173646.99337.b1. The 2005 International Society of Urological Pathology (ISUP) Consensus Conference on Gleason Grading of Prostatic Carcinoma. [DOI] [PubMed] [Google Scholar]
- 9.Hoedemaeker RF, Vis AN, Van Der Kwast TH. Staging prostate cancer. Microsc Res Tech. 2000;51:423–429. doi: 10.1002/1097-0029(20001201)51:5<423::AID-JEMT4>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 10.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 11.Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duggan D, Zheng SL, Knowlton M, Benitez D, Dimitrov L, Wiklund F, Robbins C, Isaacs SD, Cheng Y, Li G, Sun J, Chang BL, Marovich L, Wiley KE, Bälter K, Stattin P, Adami HO, Gielzak M, Yan G, Sauvageot J, Liu W, Kim JW, Bleecker ER, Meyers DA, Trock BJ, Partin AW, Walsh PC, Isaacs WB, Grönberg H, Xu J, Carpten JD. Two genome-wide association studies of aggressive prostate cancer implicate putative prostate tumor suppressor gene (DAB2IP) JNCI. 2007;99:1836–1844. doi: 10.1093/jnci/djm250. [DOI] [PubMed] [Google Scholar]