Abstract
Within 2 months of its discovery last spring, a novel influenza A (H1N1) virus, currently referred to as 2009 H1N1, caused the first influenza pandemic in decades. The virus has caused disproportionate disease among young people with early reports of virulence similar to that of seasonal influenza. This clinical review provides an update encompassing the virology, epidemiology, clinical manifestations, diagnosis, treatment, and prevention of the 2009 H1N1 virus. Because information about this virus, its prevention, and treatment are rapidly evolving, readers are advised to seek additional information. We performed a literature search of PubMed using the following keywords: H1N1, influenza, vaccine, pregnancy, children, treatment, epidemiology, and review. Studies were selected for inclusion in this review on the basis of their relevance. Recent studies and articles were preferred.
CDC = Centers for Disease Control and Prevention; FDA = Food and Drug Administration; GBS = Guillain-Barré Syndrome; HA = hemagglutinin; ILI = influenza-like illness; NA = neuraminidase; PB = basic polymerase; RIDT = rapid influenza diagnostic test; RT-PCR = real-time reverse transcriptase polymerase chain reaction; WHO = World Health Organization
On April 21, 2009, the Centers for Disease Control and Prevention (CDC) confirmed 2 cases (originally identified by the Department of Defense) of a febrile respiratory illness in children from southern California caused by infection with a novel influenza A (H1N1) virus.1 The 2 viral isolates were found to be genetically similar, to be resistant to amantadine and rimantadine, and to contain a novel genetic combination of segments from previous swine influenza viruses that have circulated in the United States since 1999, genes from swine viruses of the Eurasian lineage, and genes from avian influenza viruses. Neither of these children had exposure to swine or to each other, indicating that this virus was capable of human-to-human transmission. Several days later, the CDC reported that H1N1 viruses of the same strain had been confirmed among samples from patients in Mexico, where there was a cluster of 47 cases of rapidly progressive severe pneumonia that resulted in 12 known deaths.2,3
In response to these cases, investigations and enhanced surveillance were implemented by the Mexican Ministry of Health with the assistance of the World Health Organization (WHO). By June 11, 2009, nearly 30,000 cases of 2009 H1N1 virus had been confirmed across 74 countries, compelling the WHO to signal the phase 6 alert level, officially declaring the start of the 2009 influenza pandemic (Table 1).4
TABLE 1.
World Health Organization (WHO) Pandemic Phases
This clinical review provides an update encompassing the virology, epidemiology, clinical manifestations, diagnosis, treatment, and prevention of the 2009 H1N1 virus. To identify relevant literature, we searched PubMed using the following keywords: H1N1, influenza, vaccine, pregnancy, children, treatment, epidemiology, and review. Studies were selected for inclusion in this review on the basis of their relevance and currency.
BACKGROUND
The known historical roots of the influenza A H1N1 virus can be traced to 1918, when a virus currently thought to be of avian origin overcame the complex species barriers required to infect humans.5 Thus began an influenza pandemic that would result in an estimated 50 to 100 million deaths, more than any other influenza pandemic in history. During the second and more severe wave of human disease in 1918, herds of swine were reported to be infected with a respiratory illness of similar scope and severity. Shope,6 a veterinarian, determined that a virus was the causative agent of this swine illness and hypothesized that the human pandemic influenza strain of 1918 must be closely related. His work with mice and other subsequent studies have supported his hypothesis.7,8 Thereafter, the swine and human influenza viruses rapidly diverged antigenically, and H1N1 continued to infect humans in seasonal epidemics.9 In 1957, the H1N1 virus was replaced by a new strain, designated H2N2, that combined genetic material from its H1N1 predecessor and an avian influenza virus.10 However, influenza A/H1N1 reemerged in a 1976 outbreak among 230 Army soldiers at Fort Dix, NJ, resulting in 1 death.11,12 The virus did not extend to the civilian population. One year later, another A/H1N1 strain emerged in China, Hong Kong, and the former Soviet Union.11 This reemergence was thought to result from an unintended laboratory release and caused mild symptoms in predominantly young people.11,13,14 Since 1977, the H1N1 influenza virus has persistently contributed to seasonal epidemics alongside the often more dominant H3N2 subtype.14
In 1998, a novel triple reassortant virus was identified in the North American swine population; it contained 5 gene segments from the classical North American A/H1N1 swine virus that Shope had described with polymerase gene segments from birds and humans.15,16 The first reported human infection with this triple reassortant virus was reported in late 2005 in a 17-year-old male adolescent from Wisconsin who had butchered a pig.16 The patient recovered, as did the 10 subsequent but unrelated cases reported to the CDC before February 2009. Nearly all the patients had exposure to pigs. Although no fatalities occurred, some of the patients had unusually severe lower respiratory tract infection and diarrhea.16
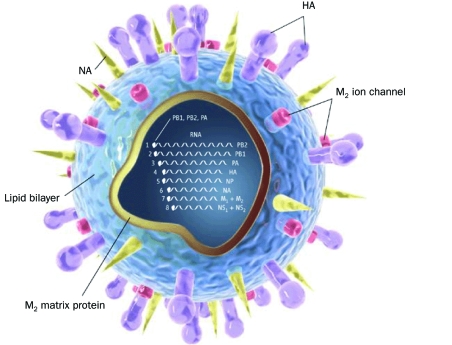
The 2009 H1N1 virus is derived from a reassortment of 6 gene segments from the triple reassortant swine-origin virus and 2 gene segments from the Eurasian influenza A (H1N1) swine virus lineage5,17,18 (Figure 1).
FIGURE 1.
Structural diagram of the H1N1 virus. H = hemagglutinin; M = matrix; N = neuraminidase; NP = nucleoprotein; NS = nonstructural; PA = acidic polymerase; PB = basic polymerase From Science,19 with permission.
VIROLOGY
Like influenza B and C, influenza A belongs to the Orthomyxoviridae family. It contains a genome made up of 8 segments of negative-sense RNA that encodes 11 proteins (Figure 2).20 Standard influenza nomenclature includes the virus type (A, B, or C), geographical origin, strain number, year of isolation, and virus subtype. Thus, the influenza A/H1N1 virus isolated in California in 2009 is identified as influenza A/California/04/2009 (H1N1). Influenza A subtype classification is based on the antigenicity of the 2 major cell surface glycoproteins: hemagglutinin (HA) and neuraminidase (NA). To date, 16 HA (H1-H16) and 9 NA (N1-N9) subtypes have been identified.
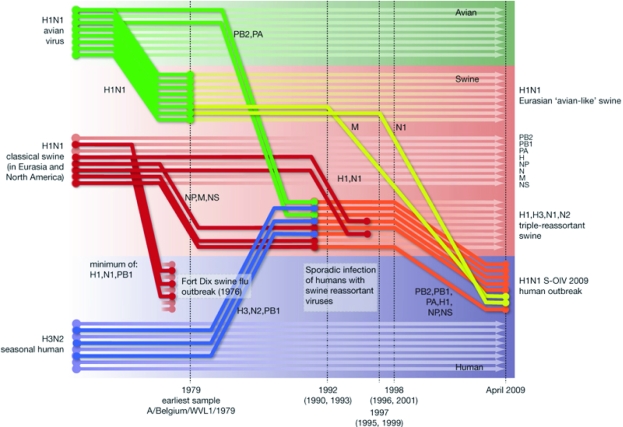
FIGURE 2.
Origins of each of the 8 genomic segments of the 2009 H1N1 virus (designated H1N1 S-OIV 2009 in this figure). Host species are represented by colored boxes: avian (green), swine (red), and human (grey). Interspecies-transmission pathways of influenza genes are represented by colored lines. Note that the N1 and M genomic segments (in yellow) are derived from an H1N1 Eurasian swine virus that is a genomic descendent of the infamous H1N1 avian virus that crossed the species barrier to cause the 1918 influenza pandemic. The remaining 6 genomic segments are derived from the triple-reassortant swine-origin influenza virus that contains genomic material from the classic H1N1 swine influenza virus, the seasonal H3N2 virus, and the H1N1 avian virus. H = hemagglutinin; M = matrix; N = neuraminidase; NP = nucleoprotein; NS = nonstructural; PA = acidic polymerase; PB = basic polymerase.
Reprinted with permission from Macmillan Publishers, Ltd: Nature (http://www.nature.com).20 Copyright © 2009.
The structure and function of the proteins encoded by the 8 genome segments have been defined. The HA protein facilitates binding of the virus to host cell receptors and subsequent endosomal fusion. Polymerase subunits (basic polymerase [PB] 1, PB2, and acidic polymerase) and nucleoprotein implement the replication and transcription of viral RNA. The nuclear export protein and the matrix protein export the viral ribonucleoprotein complexes from the nucleus into the cytoplasm for assembly into new virions at the plasma membrane. Viral release from the cells is facilitated by the NA protein.
Antigenic variation through drift and shift of HA and NA proteins enables the virus to escape host immune responses.21 Drift refers to frequent, minor changes on the HA and/or NA antigens. Antigenic drifts in the HA subtype are associated with seasonal epidemics and often reduce the effectiveness of the previous seasonal vaccines.22 Shift refers to the introduction of an influenza A virus subtype to which the population has no preexisting immunity. Although the precise mechanism is unknown, shifts are widely assumed to be facilitated by the virus's segmented genome and the genetic diversity it achieves by infecting a varied reservoir of animals.21 Antigenic shifts in HA subtypes are associated with pandemics, 3 of which occurred in the past century.11 The most infamous was the “Spanish flu” pandemic of 1918 that resulted in between 50 and 100 million deaths worldwide. In 1957, the introduction of H2N2 resulted in the “Asian flu” pandemic and claimed about 70,000 lives in the United States and about 2 million worldwide. Eleven years later, a novel H3N2 virus caused the “Hong Kong flu” of 1968 and resulted in about 70,000 deaths in the United States and about 1 million worldwide.
The emergence of the 2009 H1N1 virus is an unprecedented event in modern virology. The 2009 H1N1 virus does not fit the classic definition of a new subtype for which most of the population has no previous infection experience. Since 1977, H1N1 viruses have been in continuous circulation, and most persons born before 1956 have previous infection experience with H1N1 strains in the pre-H2N2 era. The 2009 H1N1 virus also does not fit the classic definition of drift because it has no direct evolutionary relationship with recently circulating H1N1 viruses of human origin.23 However, all H1N1 strains share subtype antigens identified by immune-diffusion tests, which is the basis for influenza virus subtype classification. Immune recall exists within each subtype (all H1N1 strains).24 For example, populations born before 1957 (during the period of H1N1 circulation) responded well to 1 dose of swine influenza virus vaccine in 1976, despite low or no preexisting antibodies to the HA, just as persons currently 10 years or older have responded well to a single dose of the 2009 H1N1 vaccine.23
The natural reservoir for all influenza A subtypes is waterfowl, with certain subtypes transmissible among humans, pigs, and 16 other mammals.23,25 Human influenza viruses bind to receptors composed of a sialic acid and galactose linked by an α 2,6 bond (SAα2,6Gal) on epithelial cells within the respiratory tract.26 Avian influenza viruses, however, preferentially bind to receptors composed of sialic acid and galactose linked by an α 2,3 bond (SAα2,3Gal) on epithelial cells within the intestinal tract of waterfowl. The epithelial cells lining the trachea of swine express both receptors, making swine a “mixing vessel” for coinfection with influenza A subtypes and reassortment.27
Once the virus binds to columnar epithelial cells of the respiratory tract, it interferes with host cell protein synthesis and, through unclear mechanisms, induces apoptosis of the host cell.21,28,29 Before cell death, new virions are produced and released to infect adjacent cells.21 Necrotizing bronchitis and intra-alveolar hemorrhage and edema result.30
EPIDEMIOLOGY
Influenza-like illness (ILI) is defined by the CDC as fever (temperature ≥100°F or 37.8°C) and either cough or sore throat in the absence of another known cause.31 A confirmed case of 2009 H1N1 infection is defined by ILI with positive test results for the 2009 H1N1 virus by either real-time reverse transcriptase polymerase chain reaction (RT-PCR) or viral culture. A probable case is ILI with positive influenza A test results but negative results for seasonal H1 and H3 by RT-PCR. A suspected case does not meet either definition but is either a person younger than 65 years hospitalized for ILI or a person of any age with ILI and an epidemiological link to a confirmed or probable case within 7 days of illness onset.
Because of resource limitations, both the CDC and WHO stopped reporting confirmed and probable cases of 2009 H1N1 on July 24, 2009. However, through traditional systems, including surveillance for new viral subtypes, new resistance patterns, geographic spread of the virus, visits to the physician for influenza-related symptoms, hospitalizations for confirmed cases of influenza, and deaths attributable to influenza or pneumonia, the CDC will continue to report weekly statistics on the number of hospitalizations and deaths that can be attributed to the virus. Between August 30, 2009, and November 28, 2009, 31,320 laboratory-confirmed influenza-associated hospitalizations and 1336 laboratory-confirmed influenza-related deaths were reported to the CDC, and more than 99% of the most recently sub-typed influenza viruses were 2009 H1N1 influenza. These data are published weekly in FluView and can be accessed at http://cdc.gov/h1n1flu/reportingqa.htm.
Like seasonal influenza, 2009 H1N1 is thought to be transmissible by 3 routes: contact exposure (when a contaminated hand is exposed to facial membranes), droplet spray exposure (when infectious droplets are projected onto mucous membranes), and airborne exposure (via inhalation of infectious airborne particles). The relative contribution of each of these modes is unknown and likely dependent on such factors as temperature and humidity. The complexities involved with human research to understand influenza transmissibility leave animal studies and mathematical models to inform us. One such model illustrated that the relative importance of each route of transmission differs with the concentration of the virus in saliva.32 Evidence exists for seasonal influenza transmission via fine particles generated during tidal breathing.33 Recent evidence suggests that the 2009 H1N1 virus is transmitted via large particle droplets.34 Because large droplets remain suspended in the air for a short time, transmission via this route requires close contact (<1.83 m [<6 ft]) between the source and the recipient. The possibility of indirect transmission from fomites and contaminated surfaces has prompted the CDC to recommend that all body fluids (eg, stool, respiratory secretions) be treated as potentially infectious. Given the uncertainties surrounding the relative importance of each of these routes of transmission, the Institute of Medicine convened, at the request of the CDC and of the Occupational Safety and Health Administration, to provide recommendations to protect health care workers against 2009 H1N1. These recommendations are discussed in the sections that follow.
The incubation period for the 2009 H1N1 virus has been estimated to be between 1 and 7 days, similar to that of seasonal influenza.35,36 Clinicians should assume that infected persons start shedding virus 1 day before the onset of symptoms and shed at least until symptoms resolve. Recent data have suggested that up to 80% of patients are still shedding virus at 5 days, 40% at 7 days, and 10% at 10 days.37 Children and younger adults may shed for as long as 10 or more days, and immunosuppressed persons may shed virus for weeks.38-40
Most cases have occurred in patients with a median age of 12 to 17 years. However, severe cases also occur in slightly older populations. More recent reports indicate that the median age of cases may increase as infection becomes more widespread in the population.41
To determine whether previous seasonal vaccinations provide cross-protection against the 2009 H1N1 (A/California/04/2009) virus, the CDC performed microneutralization assays on preseasonal and postseasonal stored sera in 100 recipients of the 2005-2006 through 2008-2009 seasonal influenza vaccines.42 Among children, investigators found no prevaccination or postvaccination cross-reactivity; among those aged 18 to 40 years, 6% prevaccination and 7% postvaccination cross-reactivity; among those aged 18 to 64 years, 9% prevaccination and 25% postvaccination cross-reactivity; and among those older than 60 years, 33% prevaccination and 43% postvaccination cross-reactivity. These results suggest that recent seasonal influenza vaccines would not provide adequate protection against the 2009 H1N1 virus, particularly among younger people.
However, virus neutralization test results alone do not fully represent the complex immune-mediated functions involved in protection from or amelioration of influenza disease. Current disease patterns suggest that, despite low levels of cross-reacting neutralizing antibodies, adults and older children who have experienced repeated natural exposure to seasonal H1N1 viruses have an immunologic advantage in protection against disease or in moderation of clinical response to a new H1N1 infection.
CLINICAL MANIFESTATIONS
Analyses have suggested that clinical manifestations of 2009 H1N1 influenza and seasonal influenza are similar.43,44 One review of 44 confirmed 2009 H1N1 cases in a New York City high school revealed that cough (98%), fever (96%), headache (82%), sore throat (82%), rhinorrhea (82%), chills (80%), and muscle aches (80%) were commonly reported.45 Fewer patients had nausea (55%), diarrhea (48%), dyspnea (48%), joint pain (46%), or stomach ache (36%).
Patients requiring hospitalization for 2009 H1N1 infection are much more likely to have underlying medical conditions, especially asthma, chronic obstructive pulmonary disease, immunosuppression, diabetes, obesity, or chronic heart conditions.46 Such patients present with fever, cough, dyspnea, vomiting, and/or abnormal chest radiographs and are typically released after short hospital stays.
Hypoxia and chest radiographs consistent with acute respiratory distress syndrome have been characteristic of patients requiring intensive care.47 A prospective observational study of 168 critically ill patients with 2009 H1N1 in Canada found a mean age of 32.3 years and a median time of 4 days between symptom onset and hospitalization.48 Overall mortality among this group was 17.3% at 90 days and correlated with a higher severity of illness and greater organ dysfunction at presentation. All these patients were severely hypoxic at presentation, with a mean PaO2 to fraction of inspired oxygen ratio of 147. Similarly, an observational study of 58 critically ill patients with 2009 H1N1 in Mexico revealed that nonsurvivors were more likely to present with severe organ dysfunction and hypoxia than survivors.49 Both studies validated the use of either SOFA (Sequential Organ Failure Assessment) or APACHE (Acute Physiology and Chronic Health Evaluation) II scores to identify patients at risk of death. A review of medical records of patients with confirmed 2009 H1N1 infection requiring intensive care in Spain showed that chest radiographic findings were abnormal in all patients, with most (72%) demonstrating patchy alveolar opacities affecting 3 of 4 quadrants.50 In all 3 analyses, obese patients (body mass index [calculated as the weight in kilograms divided by height in meters squared] >30 kg/m2) accounted for approximately one-third of the cases requiring intensive care.
Clinicians have been advised to expect that complications from the 2009 H1N1 virus will be similar to those of the seasonal influenza virus.51 These include exacerbations of underlying chronic illness; complications in both the upper (sinusitis, otitis media, croup) and lower (pneumonia, asthma exacerbation, and bronchiolitis) respiratory tracts; neurologic (encephalopathy, encephalitis, febrile seizures, status epilepticus), cardiac (pericarditis and myocarditis), and musculoskeletal (rhabdomyositis) complications; toxic shock syndrome; and secondary bacterial infections with sepsis.
Postmortem lung specimens of 77 fatal H1N1 cases were evaluated by the CDC to determine the role of bacterial coinfection in fatal outcomes.52 Evidence of bacterial coinfection was discovered in 22 cases (29%) by means of histopathologic, molecular, and immunohistochemical analyses. In these 22 cases, Streptococcus pneumoniae predominated (10 cases); however, both methicillin-resistant and -susceptible Staphylococcus aureus, Streptococcus pyogenes, Streptococcus mitis, and Haemophilus influenzae were also discovered, in decreasing order of frequency. These findings highlight the importance of early detection and treatment of bacterial pneumonia in patients with 2009 H1N1 influenza and underscore the importance of the pneumococcal vaccine for those in whom it is indicated. Classically, bacterial pneumonia complicates influenza infection 4 to 14 days after near resolution of influenza symptoms and presents with fever, dyspnea, productive cough, and abnormal chest radiographic findings.53,54 This complication should be treated in accordance with current treatment guidelines.55
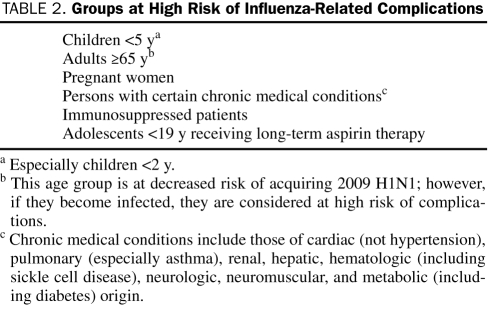
Those at highest risk of developing complications include children younger than 5 years, patients 65 years or older, pregnant women, and patients with chronic underlying medical conditions (particularly asthma, but including other pulmonary, cardiac, hematologic, hepatic, neurologic, and metabolic diseases)56,57 (Table 2). Also at increased risk are immunocompromised patients and residents of long-term care facilities. Clinicians should recall the risk of Reye syndrome, an acute, noninflammatory encephalopathy, in children or adolescents younger than 19 years with influenza who receive aspirin or aspirin-containing products. Obese patients may be at higher risk as well, although the nature of this risk is not yet completely understood.47
TABLE 2.
Groups at High Risk of Influenza-Related Complications
DIAGNOSIS
Persons with suspected or confirmed 2009 H1N1 influenza who require hospitalization should be given priority for 2009 H1N1 testing.58 Others may be eligible for priority testing as determined by state and local health departments. When considering the diagnosis, physicians should keep in mind that atypical presentations occur in infants and in elderly and immunocompromised persons. In addition to the CDC's published guidance on specimen collection, processing, and testing, state and local health department recommendations should be followed.59
A nasopharyngeal or oropharyngeal swab, nasal aspirate, or combined nasopharyngeal and oropharyngeal swab should be used for sampling. Swabs should be made of inert materials, such as synthetic tips and aluminum or plastic shafts. Endotracheal aspirates or bronchoalveolar lavage should be obtained from intubated patients. Samples should be transported in 1 to 3 mL of viral transport media, cooled with ice or refrigerated, and stored at 4°C for no longer than 4 days. Shipped specimens should be transported on wet ice or cold packs and clearly labeled with the information requested by the appropriate state public health laboratory.
Real-time reverse transcriptase polymerase chain reaction is recommended for confirmation of cases of 2009 H1N1 infection. With RT-PCR, 2009 H1N1 will test positive for influenza A and negative for seasonal H1 or H3. Strong reactivity for influenza A (cycle threshold value <30) is indicative of 2009 H1N1. Although not approved by the Food and Drug Administration (FDA) for this indication, RT-PCR assays can be performed by public health laboratories in the United States. However, clinicians should bear in mind the limitations of this testing. In the review of medical records of 32 Spanish patients requiring intensive care described previously, 4 (13%) tested negative for 2009 H1N1 at admission to the intensive care unit.50
Commercially available rapid influenza diagnostic tests (RIDTs) detect influenza viral nucleoprotein antigen and can provide results within 30 minutes. Although some RIDTs can distinguish between influenza A and B viruses, no FDA-approved RIDT can discriminate among viral subtypes (H1 from H3). The sensitivity of RIDTs for detecting 2009 H1N1 has varied from 10% to 70% and is directly related to the amount of virus in the specimen and inversely related to the threshold cycle value of the test.60-63 Thus, negative findings on a test do not “rule out” infection with the 2009 H1N1 virus; when clinical suspicion remains high despite negative test findings, the clinician should empirically treat high-risk patients and institute appropriate infection control measures. Positive results on an RIDT test for influenza type A likely mean that influenza A is present but do not distinguish among 2009 H1N1, seasonal influenza A, and influenza A of animal origin. Thus, for definitive diagnosis of 2009 H1N1 infection, only viral culture or RT-PCR can be used.
TREATMENT
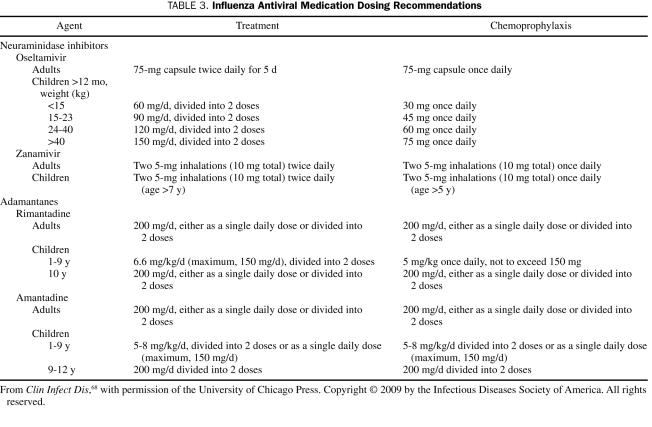
To date, with few exceptions, the 2009 H1N1 virus remains susceptible to NA inhibitors (oseltamivir and zanamivir) and resistant to the adamantanes (amantadine and rimantadine).64-66 Antiviral treatment is recommended for all hospitalized patients with confirmed, probable, or suspected 2009 H1N1 infection and patients at high risk of complications (Table 2). Because low-risk persons with uncomplicated febrile illness are unlikely to derive substantial benefit from antiviral therapy, treatment is not recommended for these patients. Treatment benefit is greatest if antiviral medications are started within 48 hours of illness onset; however, studies have suggested that hospitalized patients benefit from treatment initiation even later.57,67 Treatment dosages and duration are the same as for seasonal influenza (Table 3).68
TABLE 3.
Influenza Antiviral Medication Dosing Recommendations
Because infants are at high risk of influenza-related complications and limited safety data are favorable, the FDA issued an Emergency Use Authorization approving the use of oseltamivir for treatment and prophylaxis in pediatric patients younger than 1 year (Table 4).69
TABLE 4.
Emergency Use Authorization by the Food and Drug Administration for Oseltamivir in Infants <12 Mo
Postexposure chemoprophylaxis with oral oseltamivir or inhaled zanamivir can be considered for high-risk persons, health care workers, public health workers, or first responders who have, within the past 48 hours, been in close contact with a case of confirmed, probable, or suspected 2009 H1N1 during the infectious period.58 Although the infectious period starts 1 day before and can extend at least 7 to 10 days after illness onset, for purposes of postexposure prophylaxis the infectious period is considered to begin 1 day before symptoms and to end 24 hours after resolution of fever. The prophylactic course should extend for 10 days from the most recent contact with the infectious person. The patient should be informed that influenza can be acquired despite prophylaxis and to seek medical attention if influenza-like symptoms develop. Alternatively, clinicians can consider forgoing postexposure prophylaxis in appropriate patients reliable enough to seek immediate care if symptoms develop.
Preexposure prophylaxis with oseltamivir or zanamivir should generally be considered for high-risk persons with frequent exposures (health care workers, public health workers, or first responders) to 2009 H1N1 after consultation with local public health authorities. For this indication, the antiviral agent should be given during the potential exposure and continued for 10 days after the last known exposure.
If an outbreak in a nursing home or long-term care facility were to occur, chemoprophylaxis with oseltamivir or zanamivir should be administered to all healthy residents and continued for a minimum of 7 days after illness onset in the last patient or 2 weeks, whichever comes later. This strategy also applies for use in other closed settings, such as correctional facilities; however, it does not apply to settings where lower-risk populations congregate, such as schools and workplaces.
Special treatment considerations exist for pregnant women because of their high risk of complications from both seasonal and 2009 H1N1 influenza.70-73 They should receive oseltamivir or zanamivir for 5 days, and the drug should ideally be initiated within 48 hours of illness onset. Although both of these agents are pregnancy category C (controlled human studies are unavailable), pregnancy should not be considered a contraindication to their use.74 Pregnant women in contact with a confirmed, probable, or suspected case of infection with the 2009 H1N1 virus should receive 10 days of prophylactic therapy with zanamivir or oseltamivir. Because of its limited systemic absorption, inhaled zanamivir may be the preferred prophylactic agent in these women who do not otherwise have contraindications for its use.
All patients should be counseled regarding potential adverse effects and reactions from these antiviral agents. Oseltamivir can cause nausea and vomiting that can be alleviated if taken with food. Transient neuropsychiatric events, such as self-injury and delirium, have been reported from postmarketing surveillance studies of oseltamivir.75,76 Zanamivir is licensed only for persons without underlying respiratory (especially asthma) or cardiac disease. Cases of respiratory deterioration and allergic reactions (including angioedema) have been reported from postmarketing surveillance of zanamivir.77
Clinicians should be aware that use of peramivir, an intravenous NA inhibitor, has been authorized by the FDA under an Emergency Use Authorization.69 Although still being evaluated in phase 3 trials, limited data regarding its safety and efficacy are available (Fact Sheet for Health Care Providers, available at http://www.cdc.gov/h1n1flu/eua/Final%20HCP%20Fact%20sheet%20Peramivir%20IV_CDC.pdf). Its use is restricted to hospitalized patients who are not responding to either oral or inhaled antiviral therapy or to those for whom oral or inhaled therapy is not expected to be effective. The standard dose is 600 mg intravenously once daily for 5 to 10 days. Although no children have received the drug in clinical trials, its use in children has been permitted under emergency investigational new drug procedures. Peramivir has not been administered to pregnant women or nursing mothers in clinical trials. Adverse effects have included nausea, vomiting, diarrhea, and neutropenia.
VACCINATION/PREVENTION
Vaccination is the most effective means of preventing influenza-associated morbidity and mortality. Because previous seasonal vaccinations do not appear to confer protection against 2009 H1N1, new vaccines have been licensed and are available.78 The manufacturing and licensure process for this vaccine was based on the same standards as the seasonal influenza vaccines. The vaccine is based on the A/California/07/2009 (H1N1) strain and is available in both live-attenuated and inactivated formulations. Given the prior broad H1N1 infection experience in the population, a single dose is adequate for those older than 9 years.79 With a single administration of the 2009 H1N1 vaccine, a robust immune response was seen in 80% to 96% of adults aged 18 to 64 years and in 56% to 80% of adults aged 65 years or older.80 Children younger than 10 years will require 2 administrations of the vaccine separated by at least 21 days. Although clinicians are advised to start providing the 2009-2010 seasonal vaccine as soon as it becomes available, some patients will likely present for vaccination for both seasonal and 2009 H1N1 influenza. Although the 2009 H1N1 vaccine in either form can be given simultaneously with the inactivated 2009-2010 seasonal vaccine, live-attenuated versions of both vaccines should not be used simultaneously. The live-attenuated vaccine is only licensed for persons aged 2 through 49 years who are not pregnant, are not immunocompromised, and have no chronic medical conditions. Children younger than 5 years who have asthma, those in close contact with immunosuppressed persons, and children receiving long-term aspirin therapy should not receive the live vaccine. The inactivated vaccine should not be given to patients with severe allergic reaction to eggs or any component of the vaccine.
In early studies, the immunogenicity and safety of the 2009 H1N1 vaccine have been similar to the seasonal influenza vaccine.78 Concerns regarding the risk of Guillain-Barré syndrome (GBS) after influenza vaccination have been raised.81 These concerns stem from the suspension of the 1976 H1N1 National Influenza Immunization Program because of reports of vaccine-related GBS. Subsequent analysis estimated an attributable risk of developing vaccine-related GBS from the 1976 H1N1 vaccine at just less than 1 per 100,000 persons in the adult population.82 Studies have been unable to show a consistent association between GBS and influenza vaccination, and studies suggest a higher risk of GBS from influenza itself rather than from the vaccine.83,84 Patients should be advised that adverse effects from the 2009 H1N1 vaccine are expected to be similar to those of the seasonal vaccine and notably involve the possibility of self-limited tenderness at the injection site of the inactivated vaccine and nasal congestion, rhinorrhea, or cough with the live-attenuated vaccine.
On July 29, 2009, the CDC's Advisory Committee on Immunization Practices convened to review current epidemiological and clinical data to establish priority groups to receive 2009 H1N1 vaccine.85 Five such groups were established: pregnant women, caregivers for and cohabitants of infants younger than 6 months, health care and emergency services personnel, persons aged 6 months through 24 years, and persons aged 25 through 64 years with medical conditions that increase risk of influenza-related complications (Table 5). Members of these 5 groups constitute approximately half of the US population and are at higher risk of influenza-related complications, have an occupational risk of acquiring and subsequently transmitting influenza, or are contacts of infants younger than 6 months who are too young to be vaccinated.
TABLE 5.
Initial Target Groups for 2009 H1N1 Vaccine
If vaccine production does not meet the demand for these 5 priority groups, the Advisory Committee on Immunization Practices has designated 5 population subsets for highest priority in receiving vaccine: pregnant women, caregivers for and cohabitants of infants younger than 6 months, health care and emergency services personnel in direct contact with patients or infectious material, children aged 6 months to 4 years, and children or adolescents aged 5 to 18 years at higher risk of influenza-related complications. In the event of any local vaccine shortages, members of these highest priority subsets, numbering approximately 42 million in the United States, should be vaccinated first. Once demand among all high-priority groups has been met, vaccination should be extended to other persons aged 25 through 64 years, followed by those older than 65 years.
Unfortunately, supplies of the 2009 H1N1 vaccine have been limited. Several reasons for the shortage can be identified (http://cdc.gov/h1n1flu/vaccination/qa_vac_supply.htm). Not only has the virus necessary for vaccine production grown more slowly than expected, larger than anticipated quantities of the virus were necessary to produce an acceptably potent vaccine. These issues have been addressed and, as of December 1, 2009, nearly 60 million doses of vaccine had been shipped from the manufacturers for distribution (http://cdc.gov/h1n1flu/vaccination/vaccinesupply.htm).
According to an Internet-based survey of 2067 US adults conducted by the RAND corporation between May 26, 2009, and June 8, 2009, approximately half (49.6%) of the respondents indicated that they intended to receive the 2009 H1N1 vaccine when it became available.86 Intention to receive the vaccine was strongly correlated with receipt of the seasonal influenza vaccine. Similarly, anonymous questionnaires completed by 2255 health care professionals in Hong Kong indicated that only 47.9% were willing to accept 2009 H1N1 influenza vaccine.87 In this study, willingness to receive pandemic influenza vaccination was not affected by changes in the WHO pandemic alert level.
Patients should be educated regarding other preventive measures, including using tissues to cover their mouth and nose when coughing and sneezing, developing good handwashing technique, using alcohol-based hand cleaners, avoiding contact with ill persons if possible, and staying home when ill unless medical attention must be sought.
The CDC has provided recommendations for infection control of 2009 H1N1 in health care settings.88 In order of importance, they are as follows: elimination of potential exposures, use of engineering controls (ie, partitions in triage areas), administrative controls (ie, promoting and providing vaccinations and enforcing policies for ill staff members), and use of personal protective equipment. Standard isolation precautions, including the use of nonsterile gloves for any patient contact and the use of gown and eye protection for activities involving respiratory secretions or other infectious material, should also be observed. Aerosol-generating procedures (Table 6)88 should ideally be performed in an airborne infection isolation room with as few people present as necessary. Isolation precautions should be continued for 7 days after illness onset in confirmed or suspected cases in whom 2009 H1N1 infection has not been excluded. If the patient is transported from the room, he or she should wear a mask if tolerable and should be encouraged to perform hand and cough hygiene practices frequently. Health care personnel should observe standard precautions and wear respiratory protection equivalent to a fitted N95 respirator plus eye protection during aerosol-generating procedures.
TABLE 6.
Aerosol-Generating Procedures
After review of the evidence regarding the efficacy of various types of available respiratory protective equipment, the Institute of Medicine has recommended that health care workers use fit-tested N95 respirators or the equivalent when in close contact with persons who have ILIs or are infected with the 2009 H1N1 virus. Supporting this recommendation are data that fit-tested N95 respirators filter 95% to 99% of infectious particles, whereas variable filtration results have been reported for surgical masks. Despite this, perhaps for economic considerations, some institutions have implemented policies that call for the use of masks for routine patient care activities and reserve N95 masks for aerosol-generating activities only.
At the end of the patient encounter, immediately after removing gloves, health care workers should wash their hands with soap and water or alcohol-based sanitizer. Health care personnel with febrile respiratory illness should notify their supervisor or infection control officer and be instructed not to work or to avoid patient care duties for 7 days from illness onset and until they have been asymptomatic for 24 hours, whichever is longer. Finally, all health care personnel should receive both seasonal and pandemic influenza vaccines.89,90
Readers should be advised that these infection control recommendations are subject to rapid change and should consult their local health departments and infection control officers for further guidance.
FUTURE CONSIDERATIONS
Influenza activity in the southern hemisphere can be informative regarding the impact of 2009 H1N1 in the months ahead. Reports from Peru indicate that 8381 cases were confirmed between May 9, 2009, and September 27, 2009, with 137 confirmed influenza-related deaths.91 This translated to an estimated case fatality rate of 1.71% based on confirmed cases. Because the number of confirmed cases is undoubtedly lower than actual, this estimated case fatality rate is likely higher than actual. By comparison, the crude case fatality rate of the 1918 influenza pandemic is estimated to have been greater than 2.5%.92 The basic reproduction number (R0), used to define the average number of secondary cases generated in a susceptible population from 1 primary case, was estimated to be between 1.2 and 1.7, consistent with other reports.93-95 If the R0 is greater than 1, a pandemic can occur; however, when R0 is less than 1, a pandemic will not occur.96,97 By comparison, the R0 of the 1918 virus was estimated to have exceeded 1.3 and may have approached 3.1.98
On the basis of historical interpretations of the waves of impact during the 1918 pandemic, some have speculated that this current 2009 H1N1 virus has potential to become more virulent.99 The first wave in 1918 was mild. The second was more virulent and within weeks had spread to most of the world's cities. The first wave seemed to provide seroprotection for the second; however, neither the first nor the second wave provided seroprotection for the third.100 Current data do not show increasing influenza virulence over time with this virus; however, different countries, regions, and populations have experienced differing degrees of epidemic severity depending on multiple host and environmental factors. No evidence of the evolution of the 2009 H1N1 virus toward a more transmissible or pathogenic phenotype has been reported. Experience in the southern hemisphere has been similar to observations in the northern hemisphere between April and June 2009.
Despite efficient human transmission, the 2009 H1N1 virus lacks the molecular determinants usually associated with transmission.18 For example, 2009 H1N1 contains a glutamate at position 627 in the PB2 protein, characteristic of avian influenza viruses; human influenza viruses traditionally have a lysine at that position.101-103 Such findings suggest that this virus could enhance our knowledge of molecular determinants for transmissibility.
Although it has maintained susceptibility to NA inhibitors, multiple cases of oseltamivir-resistant 2009 H1N1 have been reported.104 Two immunocompromised patients infected with 2009 H1N1 in the state of Washington with no epidemiological link developed oseltamivir resistance during treatment. Isolates from both patients were initially susceptible to oseltamivir; however, during treatment with the drug, a mutation involving the substitution of a histidine for tyrosine at position 275 (H275Y) of the NA gene occurred, rendering the virus resistant. Zanamivir remains active against isolates with the H275Y mutation. Subsequently, the first epidemiologically linked cases of oseltamivir-resistant 2009 H1N1 infection were reported in 2 adolescent girls in North Carolina who had stayed in the same cabin during a summer camp.65 These isolated viruses contained the H275Y mutation in addition to a second previously unreported 2009 H1N1 mutation (I223V). If oseltamivir resistance is suspected, physicians may choose to treat with zanamivir or a combination of oseltamivir and amantadine or rimantadine.
CONCLUSION
The 2009 H1N1 virus has created the first influenza pandemic in more than 4 decades. Unlike previous pandemics, some of the population has had prior infection experience with this subtype. Thus far, in contrast to seasonal influenza viruses, the 2009 H1N1virus has disproportionately affected younger populations. Its virulence is similar to that of seasonal influenza viruses. The 2009 H1N1 virus is resistant to adamantanes but has remained susceptible to the NA inhibitors with few exceptions. Vaccine trials confirm that only 1 dose is required for persons older than 9 years and 2 doses for children aged 6 months to 9 years. Larger vaccine trials are in progress. Whether this virus will displace or cocirculate with the current seasonal strains is unknown. Clinicians are advised to stay up to date regarding H1N1 by frequently checking Web sites authored by the CDC and state health departments. An effective response to this pandemic and the amelioration of the associated morbidity and mortality must be predicated on a vaccinated, working, and informed health care population.
Supplementary Material
REFERENCES
- 1.Centers for Disease Control and Prevention (CDC) Swine influenza A (H1N1) infection in two children—Southern California, March-April 2009. MMWR Morb Mortal Wkly Rep. 2009;58(15):400-402 [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention (CDC) Update: swine influenza A (H1N1) infections—California and Texas, April 2009. MMWR Morb Mortal Wkly Rep. 2009;58(16):435-437 [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention (CDC) Update: novel influenza A (H1N1) virus infection—Mexico, March-May, 2009. MMWR Morb Mortal Wkly Rep. 2009;58(21):585-589 [PMC free article] [PubMed] [Google Scholar]
- 4.World Health Organization Current WHO phase of pandemic alert. World Health Organization website. http://www.who.int/csr/disease/avian_influenza/phase/en/index.html. http://www.who.int/csr/disease/avian_influenza/phase/en/index.html Accessed November 10, 2009.
- 5.Zimmer SM, Burke DS. Historical perspective—emergence of influenza A (H1N1) viruses. N Engl J Med 2009;361(3):279-285 [DOI] [PubMed] [Google Scholar]
- 6.Shope RE. The incidence of neutralizing antibodies for swine influenza virus in the sera of human beings of different ages. J Exp Med 1936;63(5):669-684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakajima K, Nobusawa E, Nakajima S. Genetic relatedness between A/Swine/Iowa/15/30(H1N1) and human influenza viruses. Virology 1984;139(1):194-198 [DOI] [PubMed] [Google Scholar]
- 8.Gorman OT, Bean WJ, Kawaoka Y, Donatelli I, Guo YJ, Webster RG. Evolution of influenza A virus nucleoprotein genes: implications for the origins of H1N1 human and classical swine viruses. J Virol 1991;65(7):3704-3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kanegae Y, Sugita S, Shortridge KF, Yoshioka Y, Nerome K. Origin and evolutionary pathways of the H1 hemagglutinin gene of avian, swine and human influenza viruses: cocirculation of two distinct lineages of swine virus. Arch Virol 1994;134(1-2):17-28 [DOI] [PubMed] [Google Scholar]
- 10.Scholtissek C, Rohde W, Von Hoyningen V, Rott R. On the origin of the human influenza virus subtypes H2N2 and H3N2. Virology 1978;87(1):13-20 [DOI] [PubMed] [Google Scholar]
- 11.Kilbourne ED. Influenza pandemics of the 20th century. Emerg Infect Dis 2006;12(1):9-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaydos JC, Top FH, Jr, Hodder RA, Russell PK. Swine influenza a outbreak, Fort Dix, New Jersey, 1976. Emerg Infect Dis 2006;12(1):23-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scholtissek C, von Hoyningen V, Rott R. Genetic relatedness between the new 1977 epidemic strains (H1N1) of influenza and human influenza strains isolated between 1947 and 1957 (H1N1). Virology 1978;89(2):613-617 [DOI] [PubMed] [Google Scholar]
- 14.Rizza SR, Tangalos EG, McClees MD, et al. Nelfinavir monotherapy increases naïve T-cell numbers in HIV-negative healthy young adults. Front Biosci 2008January1;13:1605-1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vincent AL, Lager KM, Ma W, et al. Evaluation of hemagglutinin subtype 1 swine influenza viruses from the United States. Vet Microbiol 2006;118(3-4):212-222 [DOI] [PubMed] [Google Scholar]
- 16.Shinde V, Bridges CB, Uyeki TM, et al. Triple-reassortant swine influenza A (H1) in humans in the United States, 2005-2009 [published correction appears in N Engl J Med. 2009;361(1):102] N Engl J Med 2009;360(25):2616-2625 [DOI] [PubMed] [Google Scholar]
- 17.Novel Swine-Origin Influenza A (H1N1) Virus Investigation Team Emergence of a novel swine-origin influenza A (H1N1) virus in humans [published correction appears in N Engl J Med. 2009;361(1):102] N Engl J Med 2009;360(25):2605-2615 [DOI] [PubMed] [Google Scholar]
- 18.Garten RJ, Davis CT, Russell CA, et al. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science 2009;325(5937):197-201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaiser J. A one-size-fits-all flu vaccine [published corrections appear in Science. 2006;312(5779):1472 and 2006;312(5776):999]?Science 2006;312(5772):380-382 www.sciencemag.org/cgi/reprint/312/5772/380.pdf Accessed November 30, 2009 [DOI] [PubMed] [Google Scholar]
- 20.Smith GJ, Vijaykrishna D, Bahl J, et al. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature 2009;459(7250):1122-1125 [DOI] [PubMed] [Google Scholar]
- 21.Mandell GL, Bennett JE, Dolin R, eds. Mandell, Douglas, and Bennett's Principles and Practice of Infectious Diseases 6th ed.Philadelphia, PA: Elsevier Churchill Livingstone; 2005. [Google Scholar]
- 22.Boni MF. Vaccination and antigenic drift in influenza. Vaccine 2008;26(suppl 3):C8-C14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neumann G, Noda T, Kawaoka Y. Emergence and pandemic potential of swine-origin H1N1 influenza virus. Nature 2009;459(7249):931-939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Noble GR, Kaye HS, Kendal AP, Dowdle WR. Age-related heterologous antibody responses to influenza virus vaccination. J Infect Dis 1977;136(suppl):S686-S692 [DOI] [PubMed] [Google Scholar]
- 25.Gatherer D. The 2009 H1N1 influenza outbreak in its historical context. J Clin Virol 2009;45(3):174-178 [DOI] [PubMed] [Google Scholar]
- 26.Rogers GN, Paulson JC. Receptor determinants of human and animal influenza virus isolates: differences in receptor specificity of the H3 hemagglutinin based on species of origin. Virology 1983;127(2):361-373 [DOI] [PubMed] [Google Scholar]
- 27.Ito T, Couceiro JN, Kelm S, et al. Molecular basis for the generation in pigs of influenza A viruses with pandemic potential. J Virol 1998;72(9):7367-7373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hinshaw VS, Olsen CW, Dybdahl-Sissoko N, Evans D. Apoptosis: a mechanism of cell killing by influenza A and B viruses. J Virol 1994;68(6):3667-3673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takizawa T, Matsukawa S, Higuchi Y, Nakamura S, Nakanishi Y, Fukuda R. Induction of programmed cell death (apoptosis) by influenza virus infection in tissue culture cells. J Gen Virol 1993;74(pt 11):2347-2355 [DOI] [PubMed] [Google Scholar]
- 30.Noble RL, Lillington GA, Kempson RL. Fatal diffuse influenzal pneumonia: premortem diagnosis by lung biopsy. Chest 1973;63(4):644-646 [DOI] [PubMed] [Google Scholar]
- 31.Centers for Disease Control and Prevention (CDC) Interim guidance on case definitions to be used for investigations of novel influenza A (H1N1) cases. CDC website. http://www.dphss.guam.gov/docs/swine_flu/H1N1_Guidance/case%20def%20june%201.pdf. http://www.dphss.guam.gov/docs/swine_flu/H1N1_Guidance/case%20def%20june%201.pdf Accessed November 10, 2009.
- 32.Nicas M, Jones RM. Relative contributions of four exposure pathways to influenza infection risk. Risk Anal 2009;29(9):1292-1303 [DOI] [PubMed] [Google Scholar]
- 33.Fabian P, McDevitt JJ, DeHaan WH, et al. Influenza virus in human exhaled breath: an observational study. PLoS ONE 2008;3(7):e2691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maines TR, Jayaraman A, Belser JA, et al. Transmission and pathogenesis of swine-origin 2009 A(H1N1) influenza viruses in ferrets and mice. Science 2009;325(5939):484-487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Temte JL. Basic rules of influenza: how to combat the H1N1 influenza (swine flu) virus [editorial]. Am Fam Physician 2009;79(11):938-939 [PubMed] [Google Scholar]
- 36.Jeannot AC, Hamoudi M, Bourayou N, et al. First cases of secondary transmission of a novel swine-origin influenza A (H1N1) virus in France [in French] Med Mal Infect. doi: 10.1016/j.medmal.2009.06.014. [published online ahead of print July 23, 2009] doi:10.1016/j.medmal.2009.06.014. [DOI] [PubMed] [Google Scholar]
- 37.Lye DC, Chow A, Tan A, et al. Oseltamivir therapy and viral shedding in pandemic (H1N1) 2009. In: Program and Abstracts of the 49th Interscience Conference on Antimicrobial Agents and Chemotherapy, San Francisco, California, September 12-15, 2009[abstract V-1269c] [Google Scholar]
- 38.Klimov AI, Rocha E, Hayden FG, Shult PA, Roumillat LF, Cox NJ. Prolonged shedding of amantadine-resistant influenzae A viruses by immunodeficient patients: detection by polymerase chain reaction-restriction analysis. J Infect Dis 1995;172(5):1352-1355 [DOI] [PubMed] [Google Scholar]
- 39.Ison MG, Gubareva LV, Atmar RL, Treanor J, Hayden FG. Recovery of drug-resistant influenza virus from immunocompromised patients: a case series. J Infect Dis 2006;193(6):760-764 [DOI] [PubMed] [Google Scholar]
- 40.Weinstock DM, Gubareva LV, Zuccotti G. Prolonged shedding of multidrug-resistant influenza A virus in an immunocompromised patient [letter]. N Engl J Med 2003;348(9):867-868 [DOI] [PubMed] [Google Scholar]
- 41.World Health Organization Preliminary information important for understanding the evolving situation: pandemic (H1N1) 2009 briefing note 4. WHO website. http://www.who.int/csr/disease/swineflu/notes/h1n1_situation_20090724/en/index.html. http://www.who.int/csr/disease/swineflu/notes/h1n1_situation_20090724/en/index.html Accessed November 11, 2009.
- 42.Centers for Disease Control and Prevention (CDC) Serum cross-reactive antibody response to a novel influenza A (H1N1) virus after vaccination with seasonal influenza vaccine. MMWR Morb Mortal Wkly Rep 2009;58(19):521-524 [PubMed] [Google Scholar]
- 43.Komiya N, Gu Y, Kamiya H, et al. Clinical features of cases of influenza A (H1N1)v in Osaka prefecture, Japan, May 2009. Euro Surveill 2009;14(29). pii: 19272 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19272 Accessed December 9, 2009 [DOI] [PubMed] [Google Scholar]
- 44.Belgian Working Group on Influenza A(H1N1)v Influenza A(H1N1)v virus infections in Belgium, May-June 2009. Euro Surveill 2009;14(28). pii19270 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19270 Accessed December 9, 2009 [DOI] [PubMed] [Google Scholar]
- 45.Centers for Disease Control and Prevention(CDC) Swine-origin influenza A (H1N1) virus infections in a school—New York City, April 2009. MMWR Morb Mortal Wkly Rep 2009;58(17):470-472 [PubMed] [Google Scholar]
- 46.Centers for Disease Control and Prevention (CDC) Hospitalized patients with novel influenza A (H1N1) virus infection—California, April-May, 2009. MMWR Morb Mortal Wkly Rep 2009;58(19):536-541 [PubMed] [Google Scholar]
- 47.Centers for Disease Control and Prevention (CDC) Intensive-care patients with severe novel influenza A (H1N1) virus infection—Michigan, June 2009. MMWR Morb Mortal Wkly Rep 2009;58(27):749-752 [PubMed] [Google Scholar]
- 48.Kumar A, Zarychanski R, Pinto R, et al. Canadian Critical Care Trials Group H1N1 Collaborative Critically ill patients with 2009 influenza A(H1N1) infection in Canada. JAMA 2009;302(17):1872-1879 [DOI] [PubMed] [Google Scholar]
- 49.Domínguez-Cherit G, Lapinsky SE, Macias AE, et al. Critically ill patients with 2009 influenza A(H1N1) in Mexico. JAMA 2009;302(17):1880-1887 [DOI] [PubMed] [Google Scholar]
- 50.Rello J, Rodriguez A, Ibanez P, et al. H1N1 SEMICYUC Working Group Intensive care adult patients with severe respiratory failure caused by Influenza A (H1N1)v in Spain [published online ahead of print September 11, 2009]. Crit Care 2009;13(5):R148 doi:10.1186/cc8044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Centers for Disease Control and Prevention (CDC) Interim guidance for clinicians on identifying and caring for patients with swine-origin influenza A (H1N1) virus infection. CDC Web site. www.cdc.gov/h1n1flu/identifyingpatients.htm. www.cdc.gov/h1n1flu/identifyingpatients.htm Accessed November 11, 2009.
- 52.Centers for Disease Control and Prevention (CDC) Bacterial coinfections in lung tissue specimens from fatal cases of 2009 pandemic influenza A (H1N1)—United States, May-August 2009. MMWR Morb Mortal Wkly Rep 2009;58(38):1071-1074 [PubMed] [Google Scholar]
- 53.Rothberg MB, Haessler SD, Brown RB. Complications of viral influenza. Am J Med 2008;121(4):258-264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Treanor JJ. Influenza virus. In: Mandell GL, Bennett JE, Dolin R, eds. Mandell, Douglas, and Bennett's Principles and Practice of Infectious Diseases 6th ed.Philadelphia, PA: Elsevier Churchill Livingstone; 2005:2060-2085 [Google Scholar]
- 55.Mandell LA, Wunderink RG, Anzueto A, et al. Infectious Diseases Society of America/American Thoracic Society consensus guidelines on the management of community-acquired pneumonia in adults. Clin Infect Dis 2007;44(suppl 2):S27-S72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vaillant L, La Ruche G, Tarantola A, Barboza P, Epidemic Intelligence Team at InVS Epidemiology of fatal cases associated with pandemic H1N1 influenza 2009. Euro Surveill 2009;14(33). pii19309 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19309 Accessed December 9, 2009 [DOI] [PubMed] [Google Scholar]
- 57.Hanshaoworakul W, Simmerman JM, Narueponjirakul U, et al. Severe human influenza infections in Thailand: oseltamivir treatment and risk factors for fatal outcome. PLoS ONE 2009;4(6):e6051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Centers for Disease Control and Prevention (CDC) H1N1 flu: Updated interim recommendations for the use of antiviral medications in the treatment and prevention of influenza for the 2009-2010 season. CDC Web site. www.cdc.gov/h1n1flu/recommendations.htm. www.cdc.gov/h1n1flu/recommendations.htm Accessed November 11, 2009.
- 59.Centers for Disease Control and Prevention (CDC) H1N1 flu: Interim guidance on specimen collection, processing, and testing for patients with suspected novel influenza A (H1N1) virus infection. CDC Web site. www.cdc.gov/h1n1flu/specimencollection.htm. www.cdc.gov/h1n1flu/specimencollection.htm Accessed November 11, 2009.
- 60.Hurt AC, Baas C, Deng YM, Roberts S, Kelso A, Barr IG. Performance of influenza rapid point-of-care tests in the detection of swine lineage A(H1N1) influenza viruses. Influenza Other Respi Viruses 2009;3(4):171-176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chan KH, Lai ST, Poon LL, Guan Y, Yuen KY, Peiris JS. Analytical sensitivity of rapid influenza antigen detection tests for swine-origin influenza virus (H1N1). J Clin Virol 2009;45(3):205-207 [DOI] [PubMed] [Google Scholar]
- 62.Faix DJ, Sherman SS, Waterman SH. Rapid-test sensitivity for novel swine-origin influenza A (H1N1) virus in humans [letter]. N Engl J Med 2009;361(7):728-729 [DOI] [PubMed] [Google Scholar]
- 63.Centers for Disease Control and Prevention (CDC) Evaluation of rapid influenza diagnostic tests for detection of novel influenza A (H1N1) virus—United States, 2009. MMWR Morb Mortal Wkly Rep 2009;58(30):826-829 [PubMed] [Google Scholar]
- 64.Itoh Y, Shinya K, Kiso M, et al. In vitro and in vivo characterization of new swine-origin H1N1 influenza viruses. Nature 2009;460(7258):1021-1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Centers for Disease Control and Prevention (CDC) Oseltamivir-resistant 2009 pandemic influenza A (H1N1) virus infection in two summer campers receiving prophylaxis—North Carolina, 2009. MMWR Morb Mortal Wkly Rep 2009;58(35):969-972 [PubMed] [Google Scholar]
- 66.Leung TWC, Tai ALS, Cheng PKC, Kong MSY, Lim W. Detection of an oseltamivir-resistant pandemic influenza A/H1N1 virus in Hong Kong [letter]. J Clin Virol 2009;46:298-299 [DOI] [PubMed] [Google Scholar]
- 67.McGeer A, Green KA, Plevneshi A, et al. Toronto Invasive Bacterial Diseases Network Antiviral therapy and outcomes of influenza requiring hospitalization in Ontario, Canada. Clin Infect Dis 2007;45(12):1568-1575 [DOI] [PubMed] [Google Scholar]
- 68.Harper SA, Bradley JS, Englund JA, et al. Seasonal influenza in adults and children—diagnosis, treatment, chemoprophylaxis, and institutional outbreak management: clinical practice guidelines of the Infectious Diseases Society of America. Clin Infect Dis 2009;48(8):1003-1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Food and Drug Administration Emergency use authorization (EUA) review oseltammivir phosphate for swine influenza A. FDA Web site. www.fda.gov/downloads/Drugs/DrugSafety/ImformationbyDrugClass/UCM153547.pdf. www.fda.gov/downloads/Drugs/DrugSafety/ImformationbyDrugClass/UCM153547.pdf Accessed November 11, 2009.
- 70.Dodds L, McNeil SA, Fell DB, et al. Impact of influenza exposure on rates of hospital admissions and physician visits because of respiratory illness among pregnant women. CMAJ 2007;176(4):463-468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Neuzil KM, Reed GW, Mitchel EF, Simonsen L, Griffin MR. Impact of influenza on acute cardiopulmonary hospitalizations in pregnant women. Am J Epidemiol 1998;148(11):1094-1102 [DOI] [PubMed] [Google Scholar]
- 72.Rasmussen SA, Jamieson DJ, Bresee JS. Pandemic influenza and pregnant women [editorial]. Emerg Infect Dis 2008;14(1):95-100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Centers for Disease Control and Prevention (CDC) Novel influenza A (H1N1) virus infections in three pregnant women—United States, April-May 2009 [published correction appears in MMWR Morb Mortal Wkly Rep. 2009;58(19):541] MMWR Morb Mortal Wkly Rep 2009;58(18):497-500 [PubMed] [Google Scholar]
- 74.Tanaka T, Nakajima K, Murashima A, Garcia-Bournissen F, Koren G, Ito S. Safety of neuraminidase inhibitors against novel influenza A (H1N1) in pregnant and breastfeeding women. CMAJ 2009;181(1-2):55-58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Toovey S, Rayner C, Prinssen E, et al. Assessment of neuropsychiatric adverse events in influenza patients treated with oseltamivir: a comprehensive review. Drug Saf 2008;31(12):1097-1114 [DOI] [PubMed] [Google Scholar]
- 76.Moscona A. Neuraminidase inhibitors for influenza. N Engl J Med 2005;353(13):1363-1373 [DOI] [PubMed] [Google Scholar]
- 77.Loughlin JE, Alfredson TD, Ajene AN, et al. Risk for respiratory events in a cohort of patients receiving inhaled zanamivir: a retrospective study. Clin Ther 2002;24(11):1786-1799 [DOI] [PubMed] [Google Scholar]
- 78.Centers for Disease Control and Prevention (CDC) Update on influenza A (H1N1) 2009 monovalent vaccines. MMWR Morb Mortal Wkly Rep 2009;58(39):1100-1101 [PubMed] [Google Scholar]
- 79.Greenberg ME, Lai MH, Hartel GF, et al. Response after one dose of a monovalent influenza A (H1N1) 2009 vaccine—preliminary report. N Engl J Med. http://content.nejm.org/cgi/reprint/NEJMoa0907413.pdf?resourcetype-HWCIT. http://content.nejm.org/cgi/reprint/NEJMoa0907413.pdf?resourcetype-HWCIT [published online ahead of print September 10, 2009] doi:10.1056/NEJMoa0907413. Accessed November 12, 2009.
- 80.Fauci AS.Statement by Dr. Anthony Fauci, Director, National Institute of Allergy and Infectious Diseases, NIH, regarding early results from clinical trails of 2009 H1N1 influenza vaccines in healthy adults. US Dept Health and Human Services Web site. www.hhs.gov/news/press/2009pres/09/20090911a.html. www.hhs.gov/news/press/2009pres/09/20090911a.html Accessed November 12, 2009.
- 81.Price LC. Should I have an H1N1 flu vaccination after Guillain-Barré syndrome? BMJ 2009;339:b3577 [DOI] [PubMed] [Google Scholar]
- 82.Schonberger LB, Bregman DJ, Sullivan-Bolyai JZ, et al. Guillain-Barré syndrome following vaccination in the National Influenza Immunization Program, United States, 1976-1977. Am J Epidemiol 1979;110(2):105-123 [DOI] [PubMed] [Google Scholar]
- 83.Stowe J, Andrews N, Wise L, Miller E. Investigation of the temporal association of Guillain-Barré syndrome with influenza vaccine and influenzalike illness using the United Kingdom General Practice Research Database. Am J Epidemiol 2009;169(3):382-388 [DOI] [PubMed] [Google Scholar]
- 84.Tam CC, O'Brien SJ, Petersen I, Islam A, Hayward A, Rodrigues LC. Guillain-Barré syndrome and preceding infection with Campylobacter, influenza and Epstein-Barr virus in the general practice research database. PLoS ONE 2007;2(4):e344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Centers for Disease Control and Prevention (CDC) Use of influenza A (H1N1) 2009 monovalent vaccine: recommendations of the Advisory Committee on Immunization Practices (ACIP), 2009. MMWR Morb Mortal Wkly Rep 2009;58(RR10):1-8 [PubMed] [Google Scholar]
- 86.Maurer J, Harris KM, Parker A, Lurie N. Does receipt of seasonal influenza vaccine predict intention to receive novel H1N1 vaccine: evidence from a nationally representative survey of U.S. adults. Vaccine 2009;27(42):5732-5734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chor JS, Ngai KL, Goggins WB, et al. Willingness of Hong Kong healthcare workers to accept pre-pandemic influenza vaccination at different WHO alert levels: two questionnaire surveys. BMJ 2009;339:b3391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Centers for Disease Control and Prevention (CDC) Interim guidance for infection control for care of patients with confirmed or suspected novel influenza A (H1N1) virus infection in a healthcare setting. CDC Web site. http://www.cdc.gov/h1n1flu/guidelines_infection_control.htm. http://www.cdc.gov/h1n1flu/guidelines_infection_control.htm Accessed on September 13, 2009.
- 89.Poland GA, Tosh P, Jacobson RM. Requiring influenza vaccination for health care workers: seven truths we must accept. Vaccine 2005;23:2251-2255 [DOI] [PubMed] [Google Scholar]
- 90.Poland GA. Health care workers and influenza vaccine: first do no harm, then do the right thing. J Am Pharm Assoc (2003) 2004;44(5):637-638 [DOI] [PubMed] [Google Scholar]
- 91.Gomez J, Munayco C, Arrasco J, et al. Pandemic influenza in a southern hemisphere setting: the experience in Peru from May to September, 2009. Euro Surveill 2009October22;14(42): pii19371 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19371 Accessed December 9, 2009 [DOI] [PubMed] [Google Scholar]
- 92.Taubenberger JK, Morens DM. 1918 Influenza: the mother of all pademics. Emerg Infect Dis 2006;12(1):15-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fraser C, Donnelly CA, Cauchemez S, et al. Pandemic potential of a strain of influenza A (H1N1): early findings. Science 2009;324(5934):1557-1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Boëlle PY, Bernillon P, Desenclos JC. A preliminary estimation of the reproduction ratio for new influenza A(H1N1) from the outbreak in Mexico, March-April 2009. Euro Surveill 2009;14(19). pii19205 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19205 Accessed December 9, 2009 [DOI] [PubMed] [Google Scholar]
- 95.Nishiura H, Wilson N, Baker MG. Estimating the reproduction number of the novel influenza A virus (H1N1) in a Southern Hemisphere setting: preliminary estimate in New Zealand [letter]. N Z Med J 2009;122(1299):73-77 [PubMed] [Google Scholar]
- 96.Anderson RM, May RM, Anderson B. Infectious Diseases of Humans: Dynamics and Control New York, NY: Oxford University Press; 1991:viii-757 [Google Scholar]
- 97.Diekmann O, Heesterbeek JAP. Mathematical Epidemiology of Infectious Diseases: Model Building, Analysis and Interpretation New York, NY: John Wiley and Sons, Ltd; 2000:xi-301 [Google Scholar]
- 98.Sertsou G, Wilson N, Baker M, Nelson P, Roberts MG. Key transmission parameters of an institutional outbreak during the 1918 influenza pandemic estimated by mathematical modelling. Theor Biol Med Model 2006November30;3:38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Barry JM, Massachusetts Institute of Technology Engineering Systems Division White paper on novel H1N1—prepared for the MIT Center for Engineering Systems Fundamentals. Working paper series. http://esd.mit.edu/WPS/2009/esd-wp-2009-07-072709.pdf. http://esd.mit.edu/WPS/2009/esd-wp-2009-07-072709.pdf Revised July 27, 2009. Accessed November 13, 2009.
- 100.Barry JM, Viboud C, Simonsen L. Cross-protection between successive waves of the 1918-1919 influenza pandemic: epidemiological evidence from US Army Camps and from Britain. J Infect Dis 2008;198(10):1427-1434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Conenello GM, Zamarin D, Perrone LA, Tumpey T, Palese P. A single mutation in the PB1-F2 of H5N1 (HK/97) and 1918 influenza A viruses contributes to increased virulence. PLoS Pathog 2007;3(10):1414-1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Steel J, Lowen AC, Mubareka S, Palese P. Transmission of influenza virus in a mammalian host is increased by PB2 amino acids 627K or 627E/701N. PLoS Pathog 2009;5(1):e1000252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zamarin D, Ortigoza MB, Palese P. Influenza A virus PB1-F2 protein contributes to viral pathogenesis in mice. J Virol 2006;80(16):7976-7983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Centers for Disease Control and Prevention (CDC) Oseltamivir-resistant novel influenza A (H1N1) virus infection in two immunosuppressed patients—Seattle, Washington, 2009. MMWR Morb Mortal Wkly Rep 2009;58(32):893-896 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.