Abstract
Equivalence testing is growing in use in scientific research outside of its traditional role in the drug approval process. Largely due to its ease of use and recommendation from the United States Food and Drug Administration guidance, the most common statistical method for testing equivalence is the two one-sided tests procedure (TOST). Like classical point-null hypothesis testing, TOST is subject to multiplicity concerns as more comparisons are made. In this manuscript, a condition that bounds the family-wise error rate using TOST is given. This condition then leads to a simple solution for controlling the family-wise error rate. Specifically, we demonstrate that if all pair-wise comparisons of k independent groups are being evaluated for equivalence, then simply scaling the nominal Type I error rate down by (k − 1) is sufficient to maintain the family-wise error rate at the desired value or less. The resulting rule is much less conservative than the equally simple Bonferroni correction. An example of equivalence testing in a non drug-development setting is given.
Keywords: bioequivalence, family-wise error rate, multiple comparisons, t-tests, type I error rate, TOST
1 Introduction
Broadly speaking, scientific research is often thought of as a field that is interested in inductively demonstrating differences between experimental groups while presuming equality under a null (status quo) hypothesis. However, often scientists are not interested in establishing differences, but in proving similarities. It is our experience that questions of similarity or equivalence are as fundamentally important to scientific research as those of differences.
An important example is that of demonstrating the bioequivalence of two formulations, such as an established brand name drug and a new generic equivalent. Bioequivalence refers to establishing a lack of differences in absorption, as measured by blood concentration, of two such formulations. Hence, the natural null hypothesis is that the two formulations have different absorption rates on a scale that is biologically relevant. Typically, the metrics being compared are natural logarithms of areas under plasma/concentration curves obtained by repeated blood samples of subjects having received both drugs in a random order with a suitable washout period.
We refer to this form of evaluation in the drug approval setting as bioequivalence and reserve the term equivalence for more generic settings. Establishing equivalence generally follows two steps; i) first, a setting-specific meaningful difference in population parameters between two groups is selected and ii) statistical inference is used to establish whether empirical estimates of the parameters fall within the bounds of the meaningful limits.
Early related work on equivalence testing using symmetric intervals can be found in Westlake (1976). Anderson and Hauck (1983) and Hauck and Anderson (1984) give a more powerful method for a two-way crossover design. Since these early influential articles, new procedures have been developed based on likelihood intervals (Choi et al., 2007), Bayesian credible intervals (Selwyn and Hall, 1984; Selwyn et al., 1981; Fluehler et al., 1983) and alternative frequentist tests and intervals (Berger and Hsu, 1996; Hsu et al., 1994; Brown et al., 1995), to name a few.
Certainly the most widely used procedure for statistically evaluating equivalence is the two one-sided tests procedure (TOST), which is advocated by the United States FDA for establishing bioequivalence. TOST is a form of equivalence testing proposed by Schuirmann (1987). Part of TOST’s popularity is that it is theoretically and operationally similar to classical normal-theory hypothesis testing of the equality of population means. Despite their close relationship and the ubiquity of (alternative) research hypotheses of similarity, TOST is largely unused in the non-drug development scientific community at large, where classical point-null hypothesis testing of population means is firmly entrenched. A possible reason for the large disparity in usage is not one of utility, but of exposure. Perhaps the greatest evidence supporting this explanation is the frequent misapplication of post-hoc power calculations to data that should be analyzed using equivalence testing (Hoenig and Heisey, 2001; Goodman and Berlin, 1994).
Recently, equivalence testing has made inroads in scientific applications unrelated to drug development (Barnett et al., 2007, 2006). In fact, research papers advocating the use of equivalence testing in a diverse collection of fields have begun to appear (Barker et al., 2002; Tempelman, 2004). We conjecture that as awareness of equivalence testing increases, so will the number of scientists incorporating TOST into their regular statistical toolbox. Hence, it is necessary to develop methods for adapting TOST to the diverse situations scientific data can present.
One example addressed here is that of multiplicity. As in classical hypothesis testing, as more means are compared, the family-wise error rate, the probability of at least one incorrectly rejected null in a family of tests, α F, rises above that of the set nominal type I error rate, α N. If enough means are compared, the family-wise error rate becomes unacceptably high and must be controlled. Because the foundation for equivalence testing is the same as that of classical hypothesis testing, we look to existing solutions for addressing multiplicity.
In this article, we propose an easy multiplicity correction for all pair-wise comparisons in equivalence testing. To develop this correction we show that the nominal error rate is only achieved for the comparison of nearby means. For the more distant means, the error rate is much lower, a fact that can inform the multiplicity correction. This work is motivated by recent examples in cell engineering, where the goal is to establish the equivalence of several labeling agents.
The manuscript is organized as follows. In Section 2 we review equivalence testing, while in Section 3 we introduce the multiplicity problem and our proposed solution. In Section 4 we present numerical results supporting our correction. We give an example from cell engineering in Section 5, followed by a discussion in Section 6. Appendix A gives proofs of the results used in the paper.
2 Equivalence testing
We describe the TOST procedure for comparing two independent group means from normally distributed data, presuming a common variance and equal sample sizes. This setting for equivalence testing has been described in detail elsewhere (Schuirmann, 1987; Wellek, 2003). Briefly, equivalence testing seeks to test if the difference between the two population means, Δμ, is within some previously defined tolerance interval [θl, θu]. To do this, two sets of disjoint hypotheses are formed. Closely following the description and notation in Schuirmann’s original manuscript, we have:
| (1) |
From each pair of hypotheses, test statistics are formed and compared to critical values from the t-distribution. Specifically, H01 and H02 are rejected if
| (2) |
and
| (3) |
respectively. Here ΔX̄ is the observed difference in means between the two groups, s is the pooled standard deviation, n is the (assumed common) sample size per group, df is the degrees of freedom and ta,b is the b quantile from the t-distribution with a degrees of freedom. The TOST procedure states that if both 2 and 3 are true, then the means are declared equivalent. Equivalently, test (2) rejects if the lower confidence bound is above θl and test (3) rejects if the upper confidence bound is below θu. Hence, the TOST procedure is identical to forming the corresponding (1 − 2αN) confidence interval and declaring the two groups equivalent if the interval lies entirely within the tolerance limits. We emphasize the need for a (1 − 2αN) interval rather than a standard (1− αN) interval (for further discussion see Choi et al., 2007; Berger and Hsu, 1996). This approach is currently recommended by the United States FDA and is motivated by uniformly most powerful tests (see Chapter 3 Section 7 of Lehman, 1986).
For simplicity of the discussion, we assume that the tolerance limit is symmetrically centered around zero; that is−θl = θu = θ. Then hypotheses (1) can be restated as
and equation (2) and equation (3) are restated as
| (4) |
Under our distributional assumptions, in a standard point null hypothesis, the desired type I error rate is obtained exactly. In equivalence testing using TOST, the null hypothesis includes a range of possible parameter values. The desired type I error rate is obtained only with two criteria: i) the true difference in means must be equal to the tolerance limit assumed under the null hypothesis, |Δμ| = θ; ii) the standardized tolerance width, , must be large (with the type I error rate obtained exactly only in the limit Schuirmann, 1987). Here σ is the common group-specific standard deviation and the standardized tolerance width is the tolerance width, 2θ, divided by the standard error of the difference in means, . We prove points i and ii in Appendix A and illustrate the convergence in Section 4. It is crucial to note that the relationship between the type I error rate and the standardized width (criterion ii) only holds when criterion i is true. Otherwise, the procedure is conservative and becomes increasingly so as Δμ gets larger than θ.
Let γℓ be the actual type I error rate for the equivalence test performed for a comparison with a true mean difference equal to ℓ ≥ 1 times the tolerance limit, Δμ = ℓθ. This parameter is derived and discussed in Appendix A. As discussed in criteria i and ii above, γ1 is closest to αN and limits to αN from below as the standardized tolerance width tends to infinity (see Section 4 and Appendix A). Furthermore, the γℓ decrease as ℓ increases, since the chance of incorrectly declaring equivalence decreases as the true distance between the means increases. The rapid decrease of γℓ as ℓ increases will form the basis for our proposed method of multiplicity control.
3 Family-wise error rates for all pair-wise comparisons
An important consideration in multiple comparisons is the family-wise error rate. A useful tool for controlling the family-wise error rate is the Bonferroni inequality, which states that the family-wise error rate for any group of tests is less than or equal to the sum of the individual type I error rates. Below we use the Bonferroni inequality to develop a new method for family-wise error rate control for equivalence testing using TOST and illustrate that a naive application of the standard Bonferroni correction is unnecessarily conservative. Though relevant research in multiple comparisons (see Giani and Strassburger, 2000; Bofinger, 1985; Giani and Strassburger, 1994; Hsu, 1996; Tseng, 2002; Giani and Finner, 1991; Wellek, 2003) may produce more optimal solutions, our interest lies in simple rules that are easily motivated and implemented.
Consider the setting where all pair-wise equivalence comparisons are being made for k groups using TOST; hence there are k(k − 1)/2 tests being considered. Let each group have a population mean μi for i = 1,…,k. Without loss of generality, we presume that the means are ordered from least to greatest. If all tests satisfy the null hypothesis, then the adjacent means must be at least θ apart. Because, as discussed above, the type I error rate for a single comparison increases as the difference in means decreases, the individual error rates are therefore maximized when the means are exactly θ apart (Schuirmann, 1987). The scenario that maximizes the Bonferroni inequality bound is therefore obtained when adjacent means are as close together as possible without violating the null hypothesis. That is, μi − μi−1 = θ for i = 2,…, k. We note that this scenario maximizes the Bonferonni inequality bound on the family-wise error rate because: decreasing the length between any two adjacent means renders them equivalent (a violation of the assumption that all null hypotheses are true) while expanding the distances decreases the individual type I error rates (hence increasing their sum).
Observe that in this most conservative scenario, (k − 1) comparisons occur with a true difference Δμ = 1θ, (k − 2) comparisons with a true difference Δμ = 2θ and in general (k − ℓ) comparisons are made with a true difference Δμ = ℓθ for ℓ = 1,…k − 1. Using these facts and the Bonferroni inequality, αF can be no greater than the sum of the γℓ times the number of comparisons with true difference in the means equal to ℓ. That is,
| (5) |
A naive Bonferroni bound is obtained by the fact that γℓ < αN:
| (6) |
A standard naive Bonferroni correction bounds αF by equating a desired family-wise error rate, αD, to the bound from (6), i.e. αD = αNk(k − 1)/2, and subsequently solving for αN. One obtains a rule that sets the nominal error rate to the desired family-wise error rate divided by the number of tests.
However, since bounding αF by adding the individual error rates is already a conservative procedure, and the γℓ decrease exponentially as ℓ increases, using this naive Bonferroni correction is excessively conservative. Specifically, the naive Bonferroni correction accounts for all possible comparisons, even of the most distal means, which must be at least k − 1 times the tolerance limit apart. Hence, in these cases, the associated γℓ is much smaller than αN and the bound in (6) becomes unnecessarily large. We now create a better Bonferroni correction, by equating the desired family-wise error rate and the more accurate bound given in (5).
Notice that a maximum value for each γℓ, say γ̃ℓ, can be obtained numerically (for known values of n, k, θ and αN) by maximizing over σ, the only unknown quantity in the equation for γℓ. We argue numerically (Section 4) and theoretically (Appendix A) that γ̃1 = αN while γ̃ℓ < αN for ℓ > 1. Using these maximal values, creating a more accurate Bonferroni bound is not conceptually difficult. Specifically, one could equate αD and the bound on αF from (5) using the γ̃ℓ:
| (7) |
and solve for αN. Numerically, this equation could be solved by adaptively modifying αN, such as with a bisection algorithm. Unfortunately, this approach lacks the typical computational ease of the naive Bonferroni correction
Evaluations of the family-wise error rates (described below) illustrate that the first term, γ̃1(k − 1), is equal to αN(k − 1) and completely dominates the right hand side of equation (5). Hence, we propose the following approximation to (7)
Thus, solving for αN, we obtain an easy form of multiplicity control that requires one to set αN to αD/(k − 1). We refer to this multiple comparisons procedure as an ℓ-correction, from our notation for γℓ. To reiterate:
the ℓ-correction controls the family-wise error rate when testing equivalence for all pairs of k groups using TOST by setting the nominal error rate to the desired error rate divided by the number of groups minus one.
Thus we contend that the naive Bonferroni procedure unnecessarily divides αD by an extra factor of k/2. Below, we evaluate this rule and demonstrate that it is much less conservative than a naive Bonferroni correction and is nearly equivalent to a correction based on (5).
4 Numerical evaluations
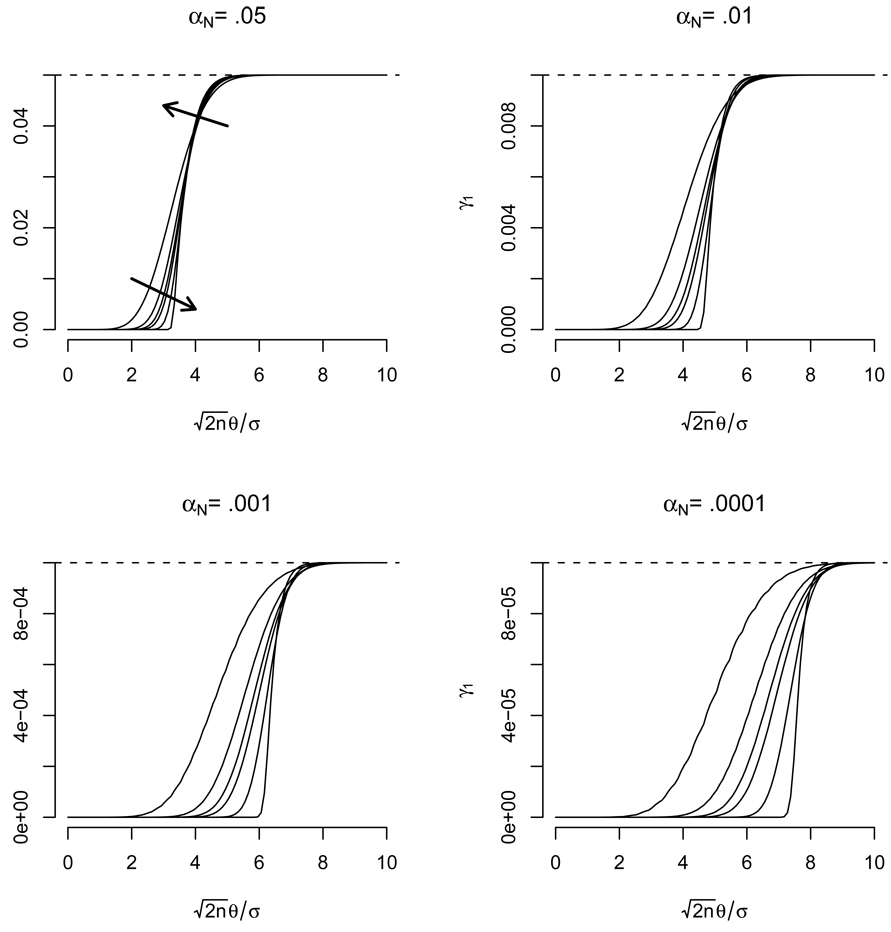
As is argued in Appendix A, we first note that γ1 limits to αN as the standardized tolerance width, ∇, increases. Figure 1 displays the behavior of γ1 as a function of ∇ for various sample sizes and values of αN. This figure illustrates that γ1 tends to the nominal error rate. Because of the square root n in the numerator of ∇, γ1 will typically be near αN. These points are also argued theoretically in Appendix A. Note that in this figure, and the remaining, the smallest possible degrees of freedom under our assumptions (2n − 2) were used.
Figure 1.
The true error rate, γ1, for a TOST test performed when Δμ = 1θ plotted as a function of . Each line represents a different sample size with n = 5, 10, 15, 20, 50, 1000 while each plot considers a different nominal error rate. The arrows point in the direction of increasing n. Dashed reference lines at the nominal error rate are drawn.
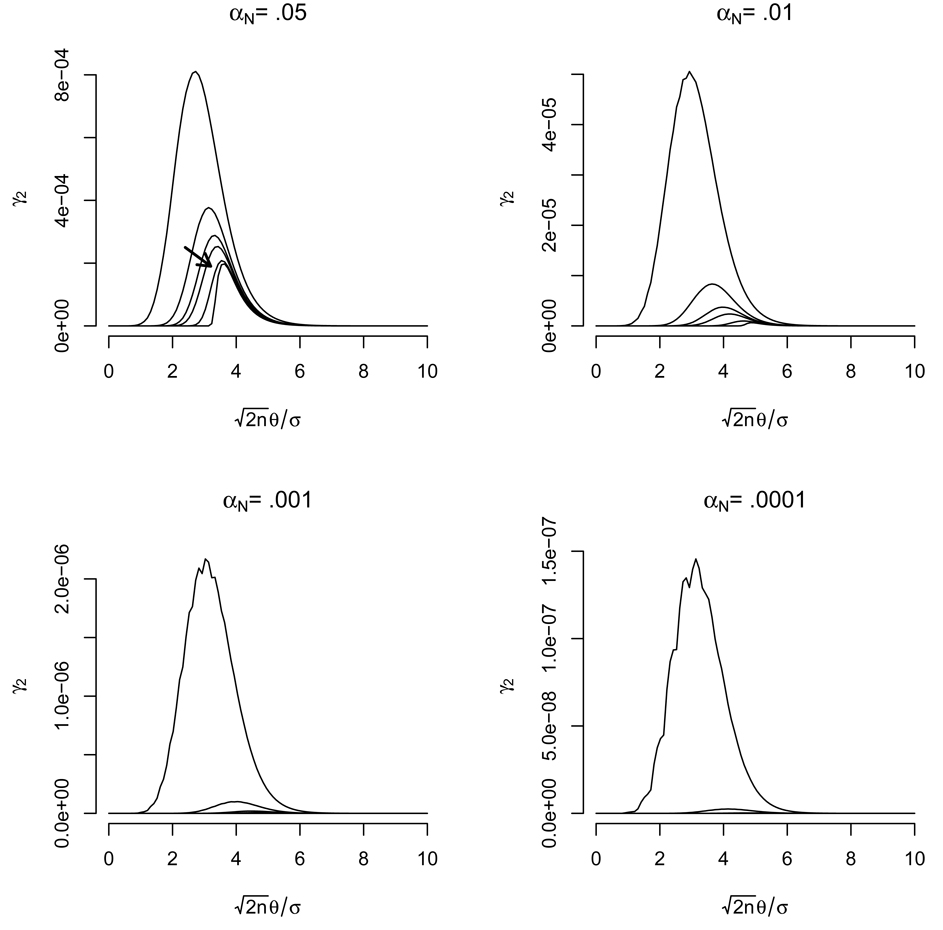
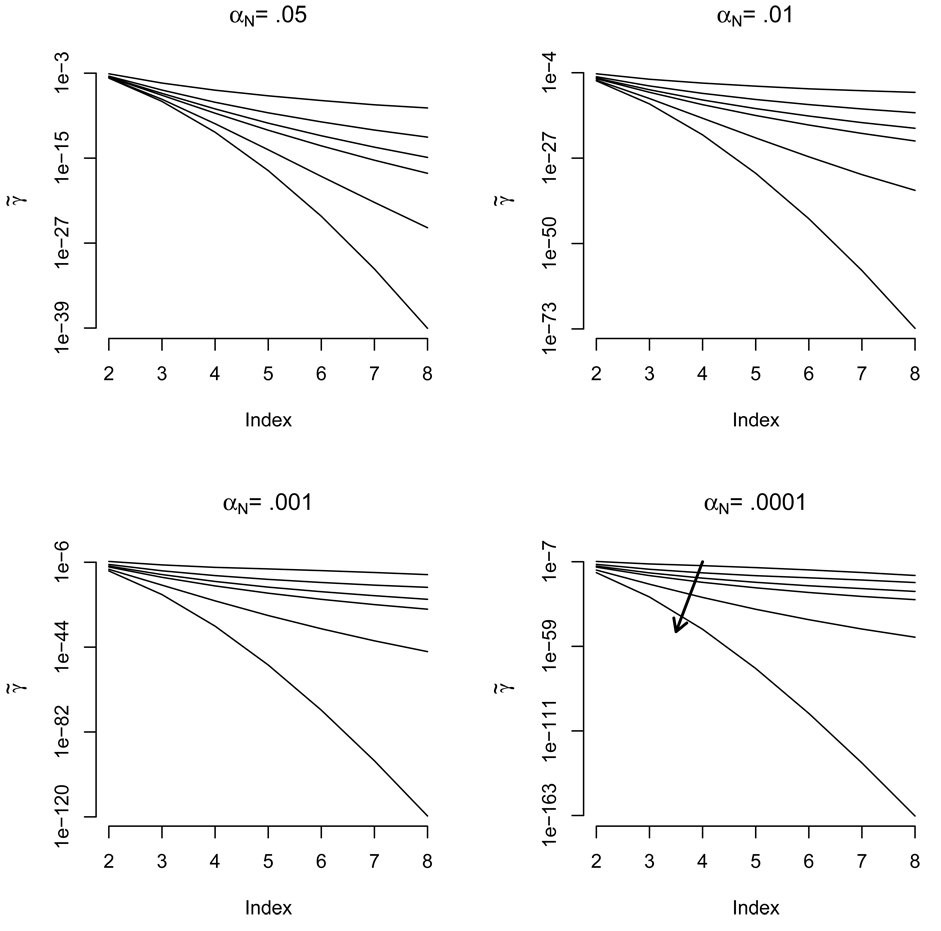
Figure 2 displays the rapid decrease in error rate γ2 for those tests whose mean difference is Δμ = 2θ. Note that the maximum magnitude of these terms, γ̃2, is on the order of 10−4 and becomes much lower as the nominal error rate decreases. Further notice that the error rate peaks, a pattern which persists for all γℓ with ℓ > 1 (see Appendix A). Plots for larger values of ℓ are not shown, as their shape is similar with a rapidly decreasing maximum value. Figure 3 displays this decrease, by plotting the logarithm base 10 of the maximum error rate, γ̃ℓ, as ℓ increases.
Figure 2.
The true error rate, γ2, for a TOST test performed when Δμ = 2θ plotted as a function of . Each line represents a different sample size with n = 5, 10, 15, 20, 50, 1000, while each plot considers a different value of αN. The shape for the ℓ = 2 case is representative of the shapes of all plots for any case ℓ > 1. The arrow points in the direction of increasing n.
Figure 3.
The maximum true error rates, γ̃ℓ, for ℓ = 2,…, for a TOST test performed between two distributions whose difference in means are Δμ= ℓθ. Six different sample sizes n = 5, 10, 15, 20, 50, 1000 are shown within each plot, each plot depicting different nominal error rates, αN. Here ℓ is labeled “index” on the horizontal axes. The arrow in the final plot points in the direction of increasing n. Values on the vertical axis are in log base ten scale.
Table 1 displays the maximum bound on the family-wise error rate for various values of k and n for the proposed ℓ-correction and a naive Bonferroni correction for a desired family-wise error rate of αD = .05. The table demonstrates that setting the nominal error rate to the desired family-wise error rate divided by k − 1 accurately controls the bound on the family-wise error rate at .05. The table also demonstrates that the proposed correction, which is slightly anti-conservative at a level that is near the numerical accuracy used in our programs (10−5), provides adequate control of the family-wise error rate. For comparison, we replicate this process for a naive Bonferroni correction, which is shown to be overly conservative.
Table 1.
The maximal bound on αF, , for the ℓ-correction [taking αN = αD/(k − 1)] and naive Bonferroni [taking αN = αD/{k(k − 1)/2}] for various values of k for a desired family-wise error rate (FWER) of αD = .05. The ℓ-correction is shown to closely adhere to the desired bound, while the naive Bonferroni correction is conservative. The sample sizes considered had no effect on the adherence to the bound.
| ℓ-correction | ||||||
|---|---|---|---|---|---|---|
| Maximal bound on the FWER for various values of n | ||||||
| k | 5 | 10 | 15 | 20 | 50 | 1000 |
| 6 | 0.05001957 | 0.05000627 | 0.05000432 | 0.05000361 | 0.05000275 | 0.05000257 |
| 12 | 0.05000192 | 0.05000070 | 0.05000051 | 0.05000045 | 0.05000037 | 0.05000035 |
| 20 | 0.05000032 | 0.05000014 | 0.05000011 | 0.05000010 | 0.05000009 | 0.05000008 |
| Naive Bonferroni | ||||||
| Maximal bound on the FWER for various values of n | ||||||
| k | 5 | 10 | 15 | 20 | 50 | 1000 |
| 6 | 0.01666829 | 0.01666693 | 0.01666680 | 0.01666676 | 0.01666672 | 0.01666671 |
| 12 | 0.00833331 | 0.00833334 | 0.00833334 | 0.00833333 | 0.00833333 | 0.00833333 |
| 20 | 0.00499985 | 0.00500000 | 0.00500000 | 0.00500000 | 0.00500000 | 0.00500000 |
5 Example
We demonstrate the ℓ-correction on an example from the field of cell-engineering. Recently, scientists have been interested in comparing the effects of different labeling agents on what are called microcapsules (Barnett et al., 2007). Briefly, the function of a micro-capsule is to deliver and house healthy xenogenic cells in patients whose own cells do not function properly. An example is injecting porcine pancreatic cells into patients with type II diabetes. In order to monitor microcapsules once inside patients, labels that are either MRI (magnetic resonance imaging), ultrasound, or X-ray visible are added to the microcapsules. However, researchers must assess the labels’ effect on the living cells inside the microcapsule. In one currently unpublished study, a human hepatic cell line (Hep G2 ATCC, Manassas) is encapsulated in contrast-containing polyethylene glycol diacrylate microcapsules and the viability under 6 different labeling conditions is assessed. Included in this study is an unlabeled control, making a total of 7 different conditions. The researchers are concerned with ensuring that the inclusion of a contrast agent did not significantly alter the viability of encapsulated cells and in assessing if cells were equally viable under the different labeling conditions. Hence, interest lies in testing biologically equivalent viability between different labels in order to assess switch-ability. To test viability, cell survival is assessed at different time points after cell encapsulation and equivalence testing performed. Setting θ to 5%, all pair-wise TOST tests are performed, comparing all strata to each other at each time point. The result is a total of 21 comparisons per time point.
In Table 2 the larger of the absolute value of the two confidence endpoints is given for each pair-wise comparison between the seven groups. The ℓ-correction values are given in the points below the diagonal while the naive Bonferroni corrected values are given above. Recall that equivalence is declared if the (1 − 2α) confidence interval is contained within the tolerance interval. Therefore, the TOST test can be performed by comparing these numbers to the tolerance limit, here set to 5%. Any endpoint less than 5 is declared equivalent. Notice that the result of the test reverses after use of the less conservative ℓ-correction in the comparisons: (1, 2), (5, 2), (7, 2), and (6, 4)
Table 2.
The maximum of the absolute value of the (1 − 2α) confidence interval (CI) for Δμ for the example in Section 5. Hence the TOST test, which rejects if the CI is entirely contained within the tolerance limits, can be performed by comparing each number to the tolerance limit. Here the CI limits constructed using the ℓ-correction are given below the diagonal while the limits using the naive Bonferroni are given above the diagonal. For example, the [3, 1] and [1, 3] cells give the comparison of groups one and three for the ℓ-correction and naive Bonferroni, respectively.
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | |
|---|---|---|---|---|---|---|---|
| 1 | - | 5.49 | 7.39 | 6.34 | 4.44 | 6.36 | 4.95 |
| 2 | 4.52 | - | 7.92 | 7.43 | 5.43 | 7.54 | 5.77 |
| 3 | 6.44 | 6.86 | - | 9.34 | 7.33 | 9.44 | 7.69 |
| 4 | 5.55 | 6.43 | 8.35 | - | 6.49 | 5.01 | 6.93 |
| 5 | 3.68 | 4.45 | 6.38 | 5.65 | - | 6.52 | 4.89 |
| 6 | 5.58 | 6.57 | 8.49 | 4.16 | 5.72 | - | 7.01 |
| 7 | 4.07 | 4.75 | 6.68 | 6.01 | 4.01 | 6.12 | - |
6 Conclusion
The proposed ℓ-correction, simply setting the nominal error rate used in TOST to the desired family-wise error rate divided by the number of groups compared minus one, provides a fast and easy method of multiplicity control for testing the bioequivalence of multiple strata. The basis for this approach comes from a bound on the family-wise error rate by adding the individual error rates and noting that, under a joint null hypothesis for k comparisons, only the k − 1 comparisons with the closest mean differences make any real contribution to this bound. On a practical side, it is important to note that in the case where all strata are compared to a single control, the ℓ-correction and naive Bonferroni correction will be identical. However, in examples where all pair-wise comparisons are made, such as the one considered above, the ℓ-correction will achieve a much tighter bound to the family-wise error rate.
We emphasize that, while a vast improvement over a naive Bonferroni correction, the proposed ℓ-correction is motivated by adding error rates and hence can be very conservative. Its main attractions are its ease of explanation and simple implementation. If these rationales are not of interest to the problem in hand, more optimal procedures should be pursued.
Acknowledgements
We would like to thank Jean-Francois H. Geschwind MD and Bradley P. Barnett for the data set used in the example and Jason Hsu, Jeff Goldsmith, Haley Hedlin and Bruce Swihart for helpful discussions on multiple comparisons. This work was supported by NIH grant K25EB003491.
Appendix
A Derivation of γℓ
We presume that Δμ = ℓθ and, for brevity, we denote the critical value tdf,1−αNsimply by t.
where, recall, df refers to the degrees of freedom and χ and Z represent the square root of a chi-squared (with df degrees of freedom) and an independent Z random variable, respectively. A simple calculation yields that the interior probability is greater than zero only when . Hence this may be written as:
| (8) |
where I(·) is an indicator function. This formula is used for all calculations, with Gauss-Laguerre integration (see Press et al., 1992), implemented in R (Ihaka and Gentleman, 1996), used for the outer expectation.
We now prove several points used in the manuscript. First, we argue that γℓ ≤ αN. To show this, note that the interior of the expectation in (8) satisfies:
Hence, taking expectations we have:
Secondly, we now show that γ̃1 = αN. As the interior of (8) is bounded by a function with a finite expectation in absolute value, we can move limits inside of the expectation. Therefore, plugging ℓ = 1 into (8) and taking the limit as ∇ goes to infinity we have:
Thirdly, it is similarly easy to show that the limit of γℓ is zero as ∇ goes to infinity for ℓ > 1 as follows:
Moreover, γℓ = 0 when ∇ = 0, as the indicator function, , is zero.
Finally, we argue that γℓ converges quickly to 0 as ℓ goes to infinity.
where both aℓ and bℓ converge monotonically and exponentially to 0 with ℓ.
We summarize these points and others apparent from these results as follows:
The only unknown that γℓ depends on is ∇, which in turn only depends on the unknown σ.
γℓ is bounded from below by 0 and above by αN.
γ̃1 is αN and γ̃ℓ < αN for ℓ > 1.
For ℓ > 2, γℓconverges to 0 as ∇ goes to infinity or as ∇ goes to 0.
γℓ converges rapidly to 0 as ℓ gets larger.
References
- Anderson S, Hauck W. A new procedure for testing equivalence in comparative bioavailability and other clinical trials. Communications in Statistics-Theory and Methods. 1983;12(23):2663–2692. [Google Scholar]
- Barker L, Luman E, McCauley M, Chu S. Assessing equivalence: An alternative to the use of difference tests for measuring disparities in vaccination coverage. American Journal of Epidemiology. 2002;156(11):1056. doi: 10.1093/aje/kwf149. [DOI] [PubMed] [Google Scholar]
- Barnett B, Arepally A, Karmarkar P, Qian D, Gilson W, Walczak P, Howland V, Lawler L, Lauzon C, Stuber M, et al. Magnetic resonance–guided, real-time targeted delivery and imaging of magnetocapsules immunoprotecting pancreatic islet cells. Nature Medicine. 2007;13:986–991. doi: 10.1038/nm1581. [DOI] [PubMed] [Google Scholar]
- Barnett B, Kraitchman D, Lauzon C, Magee C, Walczak P, Gilson W, Arepally A, Bulte J. Radiopaque alginate microcapsules for X-ray visualization and immunoprotection of cellular therapeutics. Molecular Pharmaceutics. 2006;3(5):531–538. doi: 10.1021/mp060056l. [DOI] [PubMed] [Google Scholar]
- Berger R, Hsu J. Bioequivalence trials, intersection-union tests and equivalence confidence sets. Statistical Science. 1996;11(4):283–319. [Google Scholar]
- Bofinger E. Multiple comparisons and type iii errors. Journal of the American Statistical Association. 1985;80(390):433–437. [Google Scholar]
- Brown L, Casella G, Hwang J. Optimal confidence sets, bioequivalence, and the limacon of Pascal. Journal of the American Statistical Assocication. 1995;90:880–889. [Google Scholar]
- Choi L, Caffo B, Rohde C. Johns Hopkins University, Dept. of Biostatistics Working Papers. 2007. A survey of the likelihood approach to bioequivalence trials; p. 134. [Google Scholar]
- Fluehler H, Grieve A, Mandallaz D, Mau J, Moser H. Bayesian approach to bioequivalence assessment: an example. Journal of Pharmaceutical Science. 1983;72(10):1178–1181. doi: 10.1002/jps.2600721018. [DOI] [PubMed] [Google Scholar]
- Giani G, Finner H. Some general results on least favorable parameter configurations with special reference to equivalence testing and the range statistic. Journal of statistical planning and inference. 1991;28(1):33–47. [Google Scholar]
- Giani G, Strassburger K. Testing and selecting for equivalence with respect to a control. Journal of the American Statistical Assocication. 1994;89:320–329. [Google Scholar]
- Giani G, Strassburger K. Multiple comparison procedures for optimally discriminating between good, equivalent, and bad treatments with respect to a control. Journal of Statistical Planning and Inference. 2000;83(2):413–440. [Google Scholar]
- Goodman S, Berlin J. The use of predicted confidence intervals when planning experiments and the misuse of power when interpreting results. Annals of Internal Medicine. 1994;121(3):200. doi: 10.7326/0003-4819-121-3-199408010-00008. [DOI] [PubMed] [Google Scholar]
- Hauck W, Anderson S. A new statistical procedure for testing equivalence in two-group comparative bioavailability trials. Journal of Pharmacokinetics and Pharmacodynamics. 1984;12(1):83–91. doi: 10.1007/BF01063612. [DOI] [PubMed] [Google Scholar]
- Hoenig J, Heisey D. The abuse of power: the pervasive fallacy of power calculations for data analysis. The American Statistician. 2001;55(1):19–24. [Google Scholar]
- Hsu J. Multiple Comparisons: Theory and Methods. Chapman & Hall/CRC; 1996. [Google Scholar]
- Hsu J, Hwang J, Liu H, Ruberg S. Confidence intervals associated with tests for bioequivalence. Biometrika. 1994;81(1):103–114. [Google Scholar]
- Ihaka R, Gentleman R. R: A language for data analysis and graphics. Journal of Computational and Graphical Statistics. 1996;5(3):299–314. [Google Scholar]
- Lehman E. Testing Statistical Hypotheses. second edition. Springer-Verlag: 1986. [Google Scholar]
- Press W, Teukolsky S, Vetterling W, Flannery B. Numerical Recipes in C. Cambridge, UK: Cambridge University Press; 1992. [Google Scholar]
- Schuirmann D. A comparison of the two one-sided tests procedure and the power approach for assessing the equivalence of average bioavailability. Journal of Pharmacokinetics and Pharmacodynamics. 1987;15(6):657–680. doi: 10.1007/BF01068419. [DOI] [PubMed] [Google Scholar]
- Selwyn M, Dempster A, Hall N. A Bayesian approach to bioequivalence for the 2 × 2 changeover design. Biometrics. 1981;37(1):11–21. [PubMed] [Google Scholar]
- Selwyn M, Hall N. On bayesian methods for bioequivalence. Biometrics. 1984;40(4):1103–1108. [PubMed] [Google Scholar]
- Tempelman R. Experimental design and statistical methods for classical and bioequivalence hypothesis testing with an application to dairy nutrition studies 1. Journal of Animal Science. 2004;82(90130):162–172. doi: 10.2527/2004.8213_supplE162x. [DOI] [PubMed] [Google Scholar]
- Tseng Y. Optimal confidence sets for testing average bioequivalence. Test. 2002;11(1):127–141. [Google Scholar]
- Wellek S. Testing Statistical Hypotheses of Equivalence. CRC Press; 2003. [Google Scholar]
- Westlake W. Symmetrical confidence intervals for bioequivalence trials. Biometrics. 1976;32(4):741–744. [PubMed] [Google Scholar]