Abstract
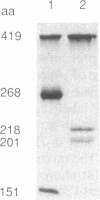
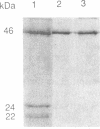
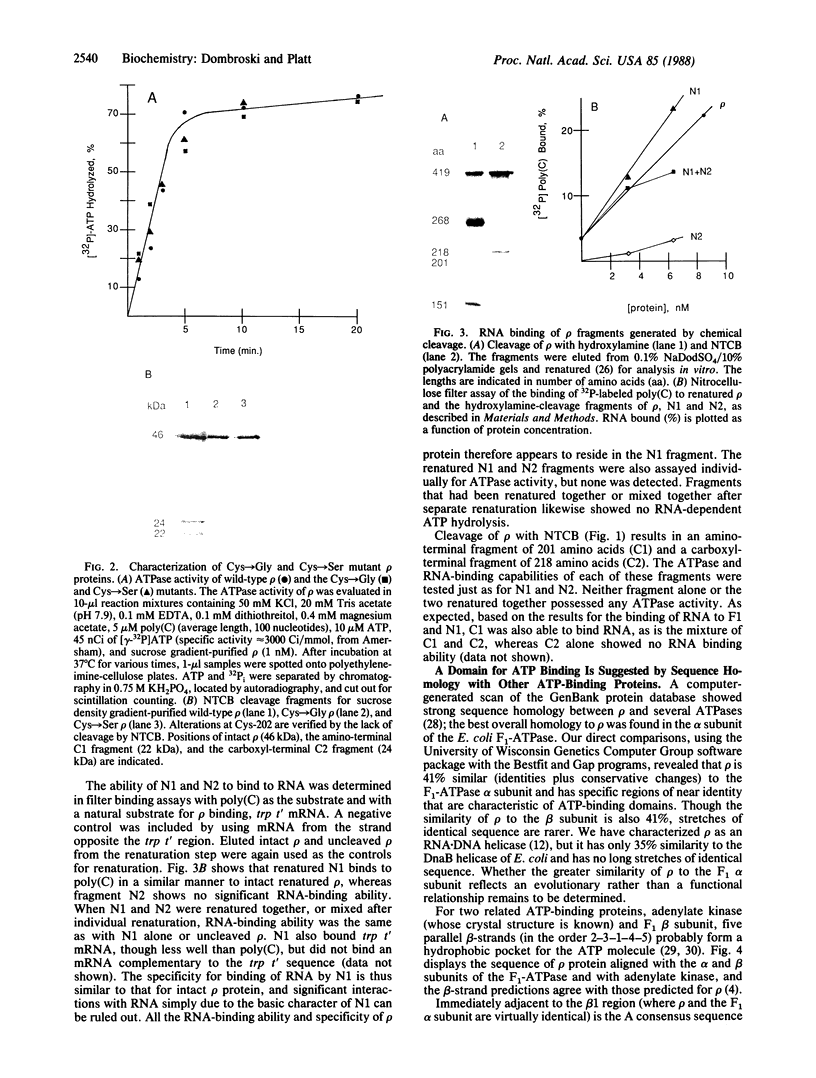
The domain structure of rho protein, a transcription termination factor of Escherichia coli, was analyzed by oligonucleotide site-directed mutagenesis and chemical modification methods. The single cysteine at position 202, previously thought to be essential for rho function, was changed to serine or to glycine with no detectable effects on the protein's hexameric structure, RNA-binding ability, or ATPase, helicase, and transcription termination activities. A 151-residue amino-terminal fragment (N1), generated by hydroxylamine cleavage, and its complementary carboxyl-terminal fragment of 268 amino acids (N2) were extracted from NaDod-SO4/polyacrylamide gels and renatured. The N1 fragment binds poly(C) and mRNA corresponding to the rho-dependent terminator sequence trp t', but not RNA unrecognized by rho; hence, this small renaturable domain retains not only the binding ability but also the specificity of the native protein. Uncleaved rho renatures to regain its RNA-dependent ATPase activity, but neither N1 nor N2 exhibits any detectable ATP hydrolysis. Similarly, the two fragments, isolated separately but renatured together, are unable to hydrolyze ATP. Sequence homology to the alpha subunit of the E. coli F1 membrane ATPase, and to consensus elements of other nucleotide-binding proteins, strongly suggests a structural domain for ATP binding that begins after amino acid 164. The implications of discrete domains for RNA and nucleotide binding are discussed in the context of requirements for specific interactions between RNA-binding and ATP-hydrolysis sites during transcription termination.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bear D. G., Andrews C. L., Singer J. D., Morgan W. D., Grant R. A., von Hippel P. H., Platt T. Escherichia coli transcription termination factor rho has a two-domain structure in its activated form. Proc Natl Acad Sci U S A. 1985 Apr;82(7):1911–1915. doi: 10.1073/pnas.82.7.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggin M. D., Gibson T. J., Hong G. F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornstein P., Balian G. Cleavage at Asn-Gly bonds with hydroxylamine. Methods Enzymol. 1977;47:132–145. doi: 10.1016/0076-6879(77)47016-2. [DOI] [PubMed] [Google Scholar]
- Brennan C. A., Dombroski A. J., Platt T. Transcription termination factor rho is an RNA-DNA helicase. Cell. 1987 Mar 27;48(6):945–952. doi: 10.1016/0092-8674(87)90703-3. [DOI] [PubMed] [Google Scholar]
- Changchien L. M., Craven G. R. The use of hydroxylamine cleavage to produce a fragment of ribosomal protein S4 which retains the capacity to specifically bind 16S ribosomal RNA. Nucleic Acids Res. 1986 Mar 11;14(5):1957–1966. doi: 10.1093/nar/14.5.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doggett P. E., Blattner F. R. Personal access to sequence databases on personal computers. Nucleic Acids Res. 1986 Jan 10;14(1):611–619. doi: 10.1093/nar/14.1.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan T. M., Parsonage D., Senior A. E. Structure of the nucleotide-binding domain in the beta-subunit of Escherichia coli F1-ATPase. FEBS Lett. 1986 Nov 10;208(1):1–6. doi: 10.1016/0014-5793(86)81519-8. [DOI] [PubMed] [Google Scholar]
- Finger L. R., Richardson J. P. Stabilization of the hexameric form of Escherichia coli protein rho under ATP hydrolysis conditions. J Mol Biol. 1982 Mar 25;156(1):203–219. doi: 10.1016/0022-2836(82)90467-3. [DOI] [PubMed] [Google Scholar]
- Fry D. C., Kuby S. A., Mildvan A. S. ATP-binding site of adenylate kinase: mechanistic implications of its homology with ras-encoded p21, F1-ATPase, and other nucleotide-binding proteins. Proc Natl Acad Sci U S A. 1986 Feb;83(4):907–911. doi: 10.1073/pnas.83.4.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluppi G. R., Richardson J. P. ATP-induced changes in the binding of RNA synthesis termination protein Rho to RNA. J Mol Biol. 1980 Apr 15;138(3):513–539. doi: 10.1016/s0022-2836(80)80016-7. [DOI] [PubMed] [Google Scholar]
- Garboczi D. N., Shenbagamurthi P., Kirk W., Hullihen J., Pedersen P. L. Mitochondrial ATP synthase. Interaction of a synthetic 50-amino acid, beta-subunit peptide with ATP. J Biol Chem. 1988 Jan 15;263(2):812–816. [PubMed] [Google Scholar]
- Hager D. A., Burgess R. R. Elution of proteins from sodium dodecyl sulfate-polyacrylamide gels, removal of sodium dodecyl sulfate, and renaturation of enzymatic activity: results with sigma subunit of Escherichia coli RNA polymerase, wheat germ DNA topoisomerase, and other enzymes. Anal Biochem. 1980 Nov 15;109(1):76–86. doi: 10.1016/0003-2697(80)90013-5. [DOI] [PubMed] [Google Scholar]
- Higgins C. F., Hiles I. D., Salmond G. P., Gill D. R., Downie J. A., Evans I. J., Holland I. B., Gray L., Buckel S. D., Bell A. W. A family of related ATP-binding subunits coupled to many distinct biological processes in bacteria. Nature. 1986 Oct 2;323(6087):448–450. doi: 10.1038/323448a0. [DOI] [PubMed] [Google Scholar]
- Howard B. H., de Crombrugghe B. ATPase activity required for termination of transcription by the Escherichia coli protein factor rho. J Biol Chem. 1976 Apr 25;251(8):2520–2524. [PubMed] [Google Scholar]
- Kamps M. P., Taylor S. S., Sefton B. M. Direct evidence that oncogenic tyrosine kinases and cyclic AMP-dependent protein kinase have homologous ATP-binding sites. Nature. 1984 Aug 16;310(5978):589–592. doi: 10.1038/310589a0. [DOI] [PubMed] [Google Scholar]
- Kramer B., Kramer W., Fritz H. J. Different base/base mismatches are corrected with different efficiencies by the methyl-directed DNA mismatch-repair system of E. coli. Cell. 1984 Oct;38(3):879–887. doi: 10.1016/0092-8674(84)90283-6. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowery-Goldhammer C., Richardson J. P. An RNA-dependent nucleoside triphosphate phosphohydrolase (ATPase) associated with rho termination factor. Proc Natl Acad Sci U S A. 1974 May;71(5):2003–2007. doi: 10.1073/pnas.71.5.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowery C., Richardson J. P. Characterization of the nucleoside triphosphate phosphohydrolase (ATPase) activity of RNA synthesi termination factor p. I. Enzymatic properties and effects of inhibitors. J Biol Chem. 1977 Feb 25;252(4):1375–1380. [PubMed] [Google Scholar]
- Lowery C., Richardson J. P. Characterization of the nucleoside triphosphate phosphohydrolase (ATPase) activity of RNA synthesis termination factor p. II. Influence of synthetic RNA homopolymers and random copolymers on the reaction. J Biol Chem. 1977 Feb 25;252(4):1381–1385. [PubMed] [Google Scholar]
- Mott J. E., Grant R. A., Ho Y. S., Platt T. Maximizing gene expression from plasmid vectors containing the lambda PL promoter: strategies for overproducing transcription termination factor rho. Proc Natl Acad Sci U S A. 1985 Jan;82(1):88–92. doi: 10.1073/pnas.82.1.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda T., Takanami M. Observations on the structure of the termination factor rho and its attachment to DNA. J Mol Biol. 1972 Nov 28;71(3):799–802. doi: 10.1016/s0022-2836(72)80041-x. [DOI] [PubMed] [Google Scholar]
- Pinkham J. L., Platt T. The nucleotide sequence of the rho gene of E. coli K-12. Nucleic Acids Res. 1983 Jun 11;11(11):3531–3545. doi: 10.1093/nar/11.11.3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt T. Transcription termination and the regulation of gene expression. Annu Rev Biochem. 1986;55:339–372. doi: 10.1146/annurev.bi.55.070186.002011. [DOI] [PubMed] [Google Scholar]
- Profy A. T., Schimmel P. A sulfhydryl presumed essential is not required for catalysis by an aminoacyl-tRNA synthetase. J Biol Chem. 1986 Nov 25;261(33):15474–15479. [PubMed] [Google Scholar]
- Richardson J. P. Activation of rho protein ATPase requires simultaneous interaction at two kinds of nucleic acid-binding sites. J Biol Chem. 1982 May 25;257(10):5760–5766. [PubMed] [Google Scholar]
- Richardson J. P., Conaway R. Ribonucleic acid release activity of transcription termination protein rho is dependent on the hydrolysis of nucleoside triphosphates. Biochemistry. 1980 Sep 2;19(18):4293–4299. doi: 10.1021/bi00559a022. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp J. A., Galloway J. L., Platt T. A kinetic mechanism for the poly(C)-dependent ATPase of the Escherichia coli transcription termination protein, rho. J Biol Chem. 1983 Mar 25;258(6):3482–3486. [PubMed] [Google Scholar]
- Shigesada K., Wu C. W. Studies of RNA release reaction catalyzed by E. coli transcription termination factor rho using isolated ternary transcription complexes. Nucleic Acids Res. 1980 Aug 11;8(15):3355–3369. doi: 10.1093/nar/8.15.3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark G. R. Cleavage at cysteine after cyanylation. Methods Enzymol. 1977;47:129–132. doi: 10.1016/0076-6879(77)47015-0. [DOI] [PubMed] [Google Scholar]
- Walker J. E., Saraste M., Runswick M. J., Gay N. J. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1(8):945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierenga R. K., Hol W. G. Predicted nucleotide-binding properties of p21 protein and its cancer-associated variant. Nature. 1983 Apr 28;302(5911):842–844. doi: 10.1038/302842a0. [DOI] [PubMed] [Google Scholar]
- Zoller M. J., Smith M. Oligonucleotide-directed mutagenesis using M13-derived vectors: an efficient and general procedure for the production of point mutations in any fragment of DNA. Nucleic Acids Res. 1982 Oct 25;10(20):6487–6500. doi: 10.1093/nar/10.20.6487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoller M. J., Smith M. Oligonucleotide-directed mutagenesis: a simple method using two oligonucleotide primers and a single-stranded DNA template. DNA. 1984 Dec;3(6):479–488. doi: 10.1089/dna.1.1984.3.479. [DOI] [PubMed] [Google Scholar]
- von Hippel P. H., Bear D. G., Morgan W. D., McSwiggen J. A. Protein-nucleic acid interactions in transcription: a molecular analysis. Annu Rev Biochem. 1984;53:389–446. doi: 10.1146/annurev.bi.53.070184.002133. [DOI] [PubMed] [Google Scholar]