Abstract
Motivation: DNA cytosine methylation is an important epigenetic regulator, critical for mammalian development and the control of gene expression. Numerous techniques using either restriction enzyme or affinity-based approaches have been developed to interrogate cytosine methylation status genome-wide, however these assays must be validated by a more quantitative approach, such as MALDI-TOF mass spectrometry of bisulphite-converted DNA (commercialized as Sequenom's EpiTYPER assay using the MassArray system). Here, we present an R package (‘MassArray’) that assists in assay design and uses the standard Sequenom output file as the input to a pipeline of analyses not available as part of the commercial software. The tools in this package include bisulphite conversion efficiency calculation, sequence polymorphism flagging and visualization tools that combine multiple experimental replicates and create tracks for genome browser viewing.
Contact: jgreally@aecom.yu.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Cytosine methylation is an epigenetic modification of the DNA, targeted in mammals to CG dinucleotides (Meissner et al., 2005). The distribution of methylcytosine in the genome can be highly variable among different cell types, enabling tissue-specific control of chromatin structure and transcriptional activity (Reik, 2007). Just as it is important for normal development and function, DNA methylation can also be dysregulated in and potentially contributory to a number of disease states, including cancer (Jones and Baylin, 2007).
A number of techniques exist to study DNA methylation. These include restriction enzyme based approaches such as HELP (Khulan et al., 2006), MethylScreen (Holemon et al., 2007) and D-REAM (Yagi et al., 2008). Additionally, there are a number of methyl-cytosine affinity-based techniques, including MeDIP (Weber et al., 2005) and MIRA (Rauch and Pfeifer, 2005). Each of these techniques has its advantages and disadvantages, however all of these approaches lack the degree of quantitative resolution available using bisulphite sequencing approaches, which rely on the preferential conversion of unmethylated cytosine to uracil (measured as thymine following PCR) with preservation of methylcytosine. As the vast majority of mammalian cytosine methylation occurs in the context of CG dinucleotides (Meissner, et al., 2005), unmethylated cytosines outside this context can be used to measure the efficiency of bisulphite treatment, acting as ‘conversion controls’.
There are numerous assays that allow the relative quantification of cytosine to thymine ratios following bisulphite treatment, including cloning and sequencing of DNA PCR-amplified from a locus of interest, or massively parallel sequencing of these amplicons (Korshunova et al., 2008). The same ratios can be measured using SNP quantification platforms such as those based on pyrosequencing (Biotage) (Dupont et al., 2004) or MALDI-TOF mass spectrometry (Ehrich et al., 2005). Whereas pyrosequencing tests relatively shorter sequences, the MALDI-TOF approach can test methylation in amplicons of hundreds of basepairs in length. The latter assay has been commercialized using Sequenom's MassArray platform.
The assay involves an initial base-specific cleavage reaction followed by mass spectrometric analysis. Prior testing of this high-throughput MassCLEAVE assay showed it to allow quantification of DNA methylation with accuracies of ±5% for each informative CG dinucleotide (Coolen et al., 2007). The proprietary software for MassCLEAVE analysis (EpiTYPER) currently provides both graphical and tabular readout of CG methylation calls. Additionally, the software is customizable for the advanced user and enables detailed exploration of raw spectral data. Missing from the current commercial software are several key elements, including a means of testing whether complete conversion of unmethylated cytosines has occurred, and whether the raw data suggest the presence of nucleotide differences in the tested sample compared with the reference genomic sequence.
In this report, we describe how we have built on a prior set of tools (Coolen et al., 2007) to develop a software analytical pipeline that provides a conversion control, flags where sequence polymorphisms may be present, and provides tools for assay design and data representations. The software is written in R and will be made available through BioConductor for open access. Our goal is to support an otherwise useful DNA methylation assay with a suite of analytical tools that significantly improve upon those currently available.
2 METHODS
2.1 Sample preparation
The embryonic stem (ES) cell line V6.5 (C57BL/6x129) was maintained in ES cell proliferation medium and 1000 U/ml of ESGRO [leukemia inhibitory factor (LIF)] on a feeder cell layer. ES cells were then cultured without LIF or feeder cells, collecting embyroid bodies on days 0 and 1. Male Sprague Dawley rats (Charles River Laboratories, Wilmington, MA) were housed and fed identically and sacrificed using humane techniques at ∼7 weeks of age. Genomic DNA was prepared from pancreatic tissue, performing bisulphite conversion using the EZ DNA Methylation Direct Kit (Zymo Research).
2.2 PCR tagging and in vitro transcription
The target regions were amplified by PCR using the primers and cycling conditions described in Supplementary Table 1. Primers were selected with MethPrimer (http://www.urogene.org/methprimer/) using parameters as follows: 200–400 bp amplicon size, temperature 56–60○C, 24–30 bp length, and ≥1 CG in product. 50 μl PCR reactions were carried out using the Roche FastStart High Fidelity Kit. Products were excised from 2% agarose gels, purified by Qiagen Gel Extraction Kit, and eluted with 1X Roche FastStart High Fidelity Reaction Buffer (+MgCl2). PCR products (5 μl) were aliquotted onto 384-well microtiter plates and were treated with 2 μl Shrimp Alkaline Phosphatase (SAP) mix for 20 min at 37○C to dephosphorylate unincorporated dNTPs. Microtiter plates were processed by MassARRAY Matrix Liquid Handler. A 2 μl volume of each SAP-treated sample was then heat-inactivated at 85○C for 5 min and subsequently incubated for 3 h at 37○C with 5 μl of Transcleave mix (3.15 μl RNAse-free water, 0.89 μl 5x T7 Polymerase Buffer, 0.24 μl T or C Cleavage Mix, 0.22 μl 100 mm DTT, 0.44 μl T7 RNA/DNA Polymerase, 0.06 μl RNAse A) for concurrent in vitro transcription and base-specific cleavage.
2.3 MALDI-TOF mass spectrometry
Prior to transfer onto the spectroCHIP array, a 384-well format MALDI-TOF matrix, samples are de-ionized with addition of 6 mg of Sequenom Resin and 20 μl of Millipore de-ionized water. 10–15 nl of de-ionized sample are spotted onto the spectroCHIParray using the Samsung Nanodispenser, calibrated to current temperature and humidity conditions. The spectroCHIP array is analyzed with the Sequenom MALDI-TOF MS Compact Unit following 4-point calibration with oligonuculeotides of different mass provided in the Sequenom kit.
2.4 In silico fragmentation analysis
We implemented an in silico fragmentation analysis for optimal assay design, analogous to one previously demonstrated (Coolen et al., 2007). In the ampliconPrediction() function, in silico RNase A digestion is performed on a target sequence for both the T- and C-cleavage reactions, on both the plus and minus strands. In the T reaction, the RNase A enzyme cleaves 3′ of every rUTP, while in the C reaction, RNase A cleavage occurs 3′ of every rCTP. The theoretical molecular weight is then calculated for each predicted fragment, and is used to determine whether or not the corresponding MALDI-TOF peak occurs within the useable mass window (default is 1500–7000 Da). Additionally, where two fragments share the same predicted mass, molecular weight overlaps are identified and flagged, corresponding to ‘silent peaks’ in the EpiTYPER software. Those overlaps where at least one of the coinciding fragments containing a CG are additionally flagged.
The in silico fragmentation profiles are further analyzed for potential conversion controls, exploitable in ∼91% of assays (Supplementary Fig. 1). In the convControl() function, conversion controls are defined as fragments meeting the following criteria: (i) sequence containing no CGs; (ii) sequence containing at least one non-CG cytosine and (iii) at least one TG; (iv) molecular weight within the useable mass window; (v) no molecular weight overlap with other predicted fragments; (vi) no molecular weight overlap of sequence containing one unconverted cytosine with other predicted fragments. Those fragments meeting the above criteria are flagged appropriately and are treated as if they contained a CG.
2.5 Quantification of methylation status
Matched peak data was exported one assay at a time as a grid from EpiTYPER v.1.0.5 with parameters set to show all ‘matched’ and ‘missing’ peaks. Each assay was then loaded into R with the importEpityperData() function, which can process both the previous (v.1.0) andcurrent (v.1.0.5) EpiTYPER formats. The DNA methylation levels of fragments were then calculated using the weighted formula previously described (Coolen et al., 2007):
where SNR is the signal-to-noise ratio of either the unmethylated peak (NOME) or one of the methylated peaks (1, 2,…, n) associated with a fragment containing n CG sites.
2.6 Single nucleotide polymorphism detection
New peak data, representing signals of molecular weight that are unexpected given the input sequence, is flagged for further analysis. Those peaks that are outside of the assayable range are ignored. The putative base composition of remaining new peaks is identified by the EpiTYPER software and is used to compare with potential deviations from the input sequence. In order to do this, single nucleotide polymorphisms are generated by an exhaustive string substitution approach (identifySNPs()), where every existent base pair in the sequence is substituted with one of the three remaining bases or a gap (representing a single base pair deletion). The local fragmentation pattern is then generated for the appropriate cleavage reaction and base compositional matches to the putative composition of the new peak are flagged. Once these new peaks are mapped to the appropriate fragments, the expected peaks corresponding to these fragments are analyzed (evaluateSNPs()) to determine if they are missing or if there is a diminished signal-to-noise ratio (SNR). This is measured in one of two ways: (i) the expected peak for a given fragment is defined as a ‘missing’ peak by the EpiTYPER software or (ii) the average SNR contribution of each fragment times the number of component fragments for the expected peak is significantly less than the peak's observed SNR. Lastly, the SNR of the new peak itself is compared to the average SNR for the sample and attributed more weight if it exceeds this average.
2.7 Multiple locus integration
Two ‘MassArrayData’ objects, each containing information for a single assay, can be collapsed together in a position-specific manner (combine()). Both objects are first transformed to the forward (plus-strand) orientation. They are then combined positionally with any data averaged across duplicate samples.
3 RESULTS
3.1 In silico assay design and target amplicon analysis
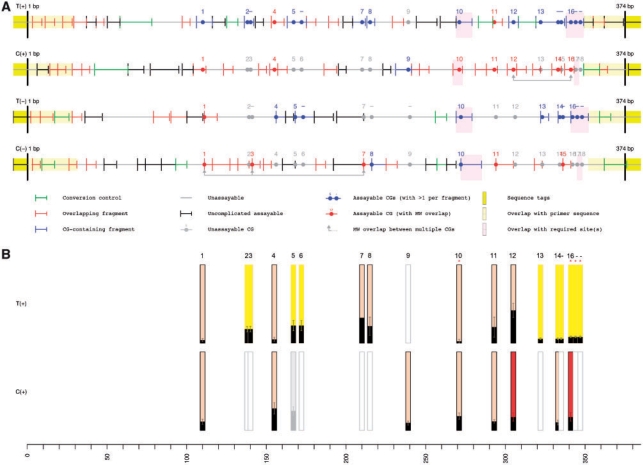
Quantitative methylation analysis can be applied to many CG sites throughout the genome. However, depending upon the target sequence, the MassCLEAVE assay may generate different assayable fragmentation profiles with either the T- or C-cleavage reactions on either the plus or minus strands (Fig. 1A). In CG-rich regions of DNA, such as CG clusters (Glass et al., 2007) and CpG islands, the T-cleavage reaction is often more informative than the C reaction, whereas the opposite may be true for AT-rich, CG-depleted regions. For the majority of assays, however, the differential performance of the T- and C-cleavage reactions on either the plus or minus strands can be selectively employed, individually or in concert, to provide finer assessment of any given CG site.
Fig. 1.
In silico assay prediction corresponds to validation data. (A) Putative fragmentation patterns are shown for T- and C-cleavage reactions on both the plus and minus strands of an amplicon of the rat genome (chr1:221405426-221405799, rat rn4 Nov. 2004 assembly, UCSC Genome Browser). CG dinucleotides (filled circles) are numbered and color-coded according to their ability to be assayed, where gray indicates that the CG is located on a fragment whose molecular weight is outside the usable mass window, red indicates a molecular weight overlap with another fragment and blue indicates a uniquely assayable site. Linked arrowheads denote molecular weight overlaps between multiple CG-containing fragments. Fragmentation patterns are shown in corresponding colors, with the addition of green fragments indicating usable conversion controls. Yellow highlights represented or primer sequences, while lavender highlights denote user-specified ‘required’ sites. (B) Methylation data for the corresponding T- and C-cleavage reactions on the plus strand are shown, each as an average of data from fourteen replicates. Bar height denotes percent methylation on a scale from 0% (low) to 100% (high), error bars indicate median absolute deviation, red stars indicate user-defined ‘required’ sites. CG dinucleotides located on a fragment with other CGs are indicated as bars with yellow background, while CG sites having a molecular weight overlap with other CG-containing fragments are colored in red. CG sites that are putatively outside the usable mass window are shown in gray outline, with any data recovered from such locishaded in gray.
In silico RNase A digestion of a target sequence, coupled with in silico mass spectrometry analysis, can be used to predict which combination of cleavage reactions and DNA strands will result in the most informative CG methylation data. This in silico analysis (supplied by the ampliconPrediction() function) is used to filter CG sites that fail to be uniquely informative for any of the following reasons: (i) fragment molecular weight is outside of the useable mass window (for Sequenom EpiTYPER software, the default is 1500–7000 Da); (ii) fragment contains multiple CG sites; (iii) fragment molecular weight overlaps with other predicted CG-containing fragments. The ampliconPrediction() function also identifies and flags molecular weight overlaps with other fragments that do not contain CG sites, as these ‘silent signal’ overlaps can reduce the quantitative resolution of methylation status at affected CG sites.
Amplicon prediction was applied to target sequences in the rat genome and the automated graphical output is shown for an example region (chr1:221405426-221405799) in Figure 1A. The performance of this in silico prediction was then evaluated on the plus strand, for both the T- and C-cleavage reactions (Fig. 1B). In the T-cleavage reaction, 17 out of 18 CG dinucleotides yielded methylation data, with only one (CG #9) failing as predicted. In the C-cleavage reaction, nine out of 10 CG dinucleotides failed to yield any data as predicted, with one borderline CG site (CG #5) rescued by the analysis (note that our algorithm makes an attempt to calculate methylation data from all peaks, even if they occur outside of the user-defined usable mass window). However, this data must be considered as less reliable due to limitations on the resolution at the extremes of molecular weight.
3.2 Single nucleotide polymorphism detection
The MassArray assay fundamentally assumes an accurate amplicon sequence. Any DNA sequence polymorphisms can pose a significant analytical problem, whether in the form of a fragment mass shift or a different fragmentation pattern entirely. Furthermore, such polymorphisms can interrupt the ability to interpret methylation status at one or more CG dinucleotides. Coolen et al. demonstrate the identification of a novel SNP by MassArray, its consequences for spectral data interpretation, and its potential use as a marker for allele-specific DNA methylation status.
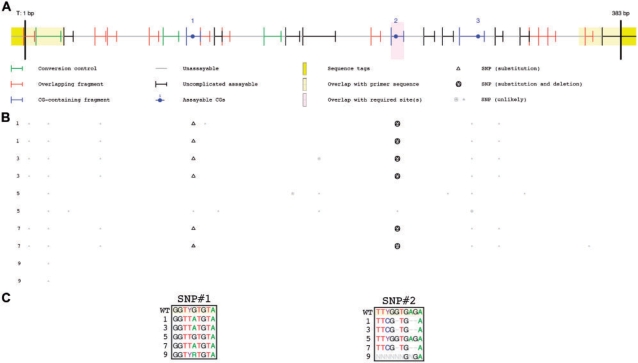
The current version of EpiTYPER software allows neither interrogation of such SNPs nor re-interpretation of a spectrum based on a different sequence without re-running the assay in its entirety. We therefore developed a tool for automated SNP detection using MassArray data (evaluateSNPs() function). We applied this tool to results generated for an amplicon of the rat genome (chr1:27718536-27718918, rn4 Nov. 2004 assembly, UCSC Genome Browser) and show that it correctly identifies two SNPs confirmed by direct sequencing of bisulphite-converted DNA (Fig. 2B and C), further confirmed by genomic DNA sequence (data not shown). The two SNPs each affect quantification of methylation status at a different CG dinucleotide.
Fig. 2.
In silico SNP prediction using mass spectral data identifies two polymorphisms, each verified by direct sequencing. (A) The T-cleavage fragmentation profile is shown for an amplicon of the rat genome (chr1:27718536-27718918, rn4 Nov. 2004 assembly, UCSC Genome Browser). CG dinucleotides (filled circles) are numbered and colored in blue. Other fragments are colored according to their ability to be assayed: fragment molecular weight outside the testable mass window (gray), fragment molecular weight overlapping with another fragment (red), fragment containing a potential conversion control (green) or fragment uniquely assayable but containing no CGs (black). (B) Putative SNPs are shown directly below their location within the amplicon fragmentation profile. Each row represents analysis from a single sample (five different biological samples, each with a pair of technical replicates). Small, gray symbols represent potential SNPs that do not have sufficient evidence (presence of a new peak with corresponding absence of an expected peak). Larger black symbols indicate a potential SNP with both new peaks and missing expected peaks. Triangles indicate base pair substitution, while circles indicate single base pair deletion. (C) Direct sequencing data for SNPs from the bisulfite converted amplicon, generated as a combination of both the forward and reverse sequencing reactions. Sample number 5 contains a homozygous wild-type allele, while samples 1, 3 and 7 contain alleles homozygous for two SNPs, the first a G → A substitution at position 109 within the amplicon, the second a multiple base pair polymorphism from position 241–245. Sample number 9 contains a mixture of alleles.
The expected fragment containing SNP#1 shows minimal signal at the expected molecular weights for both its unmethylated (2798.772 Da) and methylated (2814.771 Da) states. Similarly, the two additional fragments introduced by the altered T cleavage at SNP#1 each have elevated signal-to-noise ratios as a result (1272.797 Da, 1561.982 Da). SNP#2 shows a similar pattern, where the expected peaks at 2509.587 Da (unmethylated) and 2525.586 Da (methylated) are both suppressed or absent. Additionally, SNP#2 shows a robust new peak (2236.831 Da) corresponding to the mutated sequence (CACGAAT). A detailed summary of peak intensities for each SNP is presented in Supplementary Table 2.
3.3 Bisulphite conversion control
Accurate measurement of methylation status presupposes that the bisulphite conversion reaction runs to completion. If, however, bisulphite conversion is incomplete, ‘methylation’ measured at any CG dinucleotide will be composed of actual methylation mixed with signal from remnant unconverted, unmethylated cytosines. For many target regions, this issue is mitigated by selective amplification of fully converted templates (primers should contain at least 4 non-CG ‘C’s). Nevertheless, amplicons may still contain some background level of partially unconverted DNA: primer selection criteria occasionally need to be relaxed, and PCR tends to enrich underrepresented sequences (Mathieu-Daude et al., 1996; Suzuki and Giovannoni, 1996).
In order to measure levels of unconverted non-CG cytosines in a given MassArray sample, we designed a tool, evaluateConversion() (a wrapper function for convControl(), see Section 2), to search the in silico fragmentation profile for non-CG cytosines that occur in the absence of CG dinucleotides. Moreover, potential conversion controls are automatically filtered to remove any molecular weight overlaps with other predicted fragments, so that they may be considered in isolation. We applied this tool to data from a number of samples and show detected levels of unconverted cytosines for two examples in Supplementary Figure 2A and B. For the large majority of samples, we find that bisulphite conversion was near-complete (Supplementary Fig. 2A, ranging from 98% to −100%). However, we also show significant retention of unconverted cytosines for rat chr17:48916975-48917295 that approaches 25% (Supplementary Fig. 2B). This incomplete conversion is a relatively rare experimental outcome in our experience; nevertheless it demonstrates the necessity of a conversion control measure as part of each experiment to ensure accuracy of the data.
3.4 Multiple locus integration
While any individual target sequence may be treated in isolation, often such amplicons (typically ranging from 200–400 bp in size) are only a fraction of a larger target region, potentially spanning many thousands of base pairs. Moreover, multiple experimental runs may each have different combinations of samples, further complicating analysis. In order to address both the integration of identical samples across multiple assays, as well as the integration of multiple amplicons across a larger target region, we developed a tool (combine()) for condensing MassArray data objects in a position and sample-specific fashion.
We applied this integrative tool to a collection of individual MassArray experiments, representing both amplicon replicates and separate target sequences. Individual sample data are shown for three adjacent amplicons in Supplementary Figure 3A and the positionally combined data for a set of eight samples, including both T and C reactions, are shown in Supplementary Figure 3B. The combine() function is also able to integrate assays targeted to both DNA strands and is designed to handle varying degrees of overlap, even to the extent of combining data across multiple replicates of the same assay.
3.5 Data visualization
The standard EpiTYPER graphical output (an ‘epigram’) comes in the form of color-coded circles plotted in a linear context as a function of position within the target sequence (Supplementary Fig. 4). While visually appealing, we find that this graphical representation lacks the ability to assess methylation status quantitatively. As an alternative to the epigram, we have implemented another visualization tool that takes methylation data and displays it in the form of color-filled bars, where the height indicates percent methylation (Fig. 1B, Supplementary Fig. 3B). In this graphical depiction, we have also included the ability to display ‘error bars’ which correspond to the median absolute deviation as a measure of variability among a collection of measurements. This graphical functionality is implemented as an extension of the generic plot() function.
4 DISCUSSION
High-throughput, quantitative DNA methylation analysis is crucial for the study of the normal physiology of the epigenome and its dysregulation in disease. The need for such assays is increasing as a means of validating the many emerging genome-wide cytosine methylation techniques that use microarray or massively parallel sequencing technologies. While the MassCLEAVE assay, as commercialized by Sequenom, has great potential for meeting this need, especially when compared with other methodologies such as clonal bisulphite direct sequencing, in this report we focused on several areas where the MassArray technique continues to have analytical shortcomings.
First, we describe the implementation of an assay design tool which allows the user to visualize each of the four different possible reactions for a given sequence (either T or C cleavage on either the plus or minus strand). The graphical and tabular output allows for a highly detailed picture of the nature of results expected for a given assay, enabling the user to maximize the number of CG dinucleotide sites that can be independently assayed (Fig. 1; Supplementary Fig. 5). Sequenom provides the EpiDesigner as a tool for obtaining primer pairs to target a given region with multiple overlapping amplicons. EpiDesigner measures the number of CG sites assayable given a combination of cleavage reaction and DNA strands, however it includes no information about the ability to garner independent methylation status for each CG site. Moreover, EpiDesigner weighs all CG dinucleotides equally, whether or not there are specific CG sites that the user is particularly interested in interrogating (e.g. restriction enzyme cut sites). We therefore enabled the ability to label important (‘required’) sequences, and also provide a detailed graphical output for the relative independent ability to assay each CG dinucleotide in a given amplicon.
Measurement of methylation status by MassArray at a single CG dinucleotide is, in its simplest form (i.e. an isolated site whose containing fragment is of a unique molecular weight), simply a ratio of peak heights. When a given fragment contains multiple CG dinucleotides or when a CG-containing fragment has a molecular weight overlap with another CG-containing fragment, measurement of the constituent CG dinucleotides must be taken in aggregate. Coolen et al. described a modification of MassCLEAVE analysis where the accuracy of measurement can be improved for multiple CG-containing fragments, and further described the potential analysis of allele-specific methylation in these fragments. Additionally, EpiTYPER implements a Monte Carlo-type estimation of methylation states in cases where one or more CG-containing fragments overlap (without overlap due to non-CG fragments) (Deciu, 2008). This approach is of particular value as a measurement of confidence for average methylation values at each overlapping CG site. Despite these improvements, however, no current analytical approaches ascertain independent methylation values for CG dinucleotides that share a single fragment or are obscured by molecular weight overlaps with other CG-containing fragments.
As shown here and described elsewhere (Coolen et al., 2007), polymorphisms in the DNA sequence analyzed can be a significant problem for the MassCLEAVE assay, particularly when dealing with outbred rodent strains or clinical samples. The current EpiTYPER software does not allow the interrogation of SNPs, moreover the user is unable to modify the sequence input for a given analysis without completely repeating the experiment. The probability that an amplicon may contain polymorphisms that obscure analysis is variable and depends on the size of the fragment, the density of CG dinucleotides, and the genetic variability of the samples themselves. Our SNP detection tool evaluates their candidacy in an automated fashion. The algorithm makes use of EpiTYPER-identified new peaks and also takes into account missing peak data and average SNR. Currently, we treat each sample and reaction in isolation, but the integration of multiple replicates, as well as data from both the T- and C-cleavage reactions increases the confidence in SNP detection, discriminating genuine SNPs from sequence contamination or experimental artifact. This approach could be combined with that described by Coolen et al. to match candidate SNPs against the public SNP databases to lend additional certainty to this SNP discovery approach. Finally, it is important to note that the current tool is only designed to identify single nucleotide substitutions or deletions. Insertions of any size, duplications, inversions or multiple base pair deletions may not be detected by this current implementation of SNP discovery.
The bisulphite conversion of DNA is a chemical reaction whose equilibrium state skews to the preferential modification of unmethylated cytosine to uracil. However, the reaction may not run to completion under conditions that depart from the ideal, for example, excessive DNA concentration, sample evaporation, reagent deterioration or other thermodynamic factors (Grunau et al., 2001; Hayatsu et al., 2007). While primer design is intended to enrich for completely converted sequences, some proportion of amplicons may contain remnant unconverted (unmethylated) cytosines. Because these remnant cytosines may cause a molecular weight shift in the resultant fragmentation profile, it is essential to measure the extent of bisulphite conversion as the relative signal of unconverted to converted cytosines.
In our hands, the majority of conversion reactions work quite well, with a negligible background (0–2%) of remnant unconverted cytosines detected. However, without directly testing the extent of bisulphite conversion, the methylation readings are inherently unreliable. We show an example of a constitutively hypomethylated locus that appears to be methylated in specific MassArray experiments, due at least in part to incomplete bisulphite conversion, with the control for the same samples showing up to 25% unconverted cytosines (Supplementary Fig. 2).
Our new tools also enable automated analysis of multiple individual amplicons that, when taken together, form a larger target region, potentially spanning thousands of base pairs. Without such automation, comparing data across multiple replicates, assays and amplicons can become especially time consuming, scaling in proportion with the complexity and size of the study. Of additional note, the gel excision protocol used for the assays in this study impedes the high-throughput value of the MassArray approach, but tends to improve overall data quality. We also note, however, that gel excision is not a required step, and that the analyses presented perform as well for samples that have not been gel-purified. We generally recommend the use of non-template PCR products (i.e. water controls) in order to identify instances of potentially contaminating signal. For this purpose, we note that primer-dimer levels can be estimated for a given sample (Supplementary Fig. 6), an additional functionality included with this software.
Statistical analyses for multiple loci are likewise facilitated by this tool, although the t.test() function is not currently supported for MassArray data. We also developed a novel visualization of methylation data, analogous but alternative to the ‘epigrams’ generated by Sequenom's EpiTYPER software. This visualization enables the user to retain the quantitative aspect of methylation data and also to capture the variability of the data (in the form of error bars) for a given CG site. Alternatively, we provide a function to export methylation data in the form of BED tracks for upload and visualization with the UCSC Genome Browser (Kent et al., 2002).
In conclusion, we have implemented a collection of tools for MassArray analysis that provides a number of important improvements. In particular, these new analytical tools allow for the measurement of conversion controls, flag where sequence polymorphisms may be present, and provide for clear assay design and data visualization.
5 IMPLEMENTATION
These analytical tools are implemented in the R Statistical Package (R Development Core Team, 2005). The scripts have been tested on the Mac platform (OS X 10.5) using R version 2.8.0 with base packages installed and enabled. R source code is currently available at http://greallylab.aecom.yu.edu/∼greally/MassArray/ and will be made available through BioConductor (Gentleman, et al., 2004).
Supplementary Material
ACKNOWLEDGEMENTS
The authors acknowledge the contribution of the Genomics Core Facility at the Albert Einstein College of Medicine, and also thank Rebecca Simmons of the Childrens' Hospital of Pennsylvania (CHoP) for generously providing the rat samples.
Funding: National Institutes of Health (NIH) (NHGRI R01 HG004401 and NICHD R01 HD044078 to J.M.G.). NIH MSTP Training (grant GM007288 to R.F.T.).
REFERENCES
- Coolen MW, et al. Genomic profiling of CpG methylation and allelic specificity using quantitative high-throughput mass spectrometry: critical evaluation and improvements. Nucleic Acids Res. 2007;35:e119. doi: 10.1093/nar/gkm662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deciu C. A heuristic equipartion model for estimating individual methylation ratios in the case of isobaric CpG units. Sequenom Technical Report. 2008:1–10. [Google Scholar]
- Dupont JM, et al. De novo quantitative bisulfite sequencing using the pyrosequencing technology. Anal. Biochem. 2004;333:119–127. doi: 10.1016/j.ab.2004.05.007. [DOI] [PubMed] [Google Scholar]
- Ehrich M, et al. Quantitative high-throughput analysis of DNA methylation patterns by base-specific cleavage and mass spectrometry. Proc. Natl Acad. Sci. USA. 2005;102:15785–15790. doi: 10.1073/pnas.0507816102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass JL, et al. CG dinucleotide clustering is a species-specific property of the genome. Nucleic Acids Res. 2007;35:6798–6807. doi: 10.1093/nar/gkm489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunau C, et al. Bisulfite genomic sequencing: systematic investigation of critical experimental parameters. Nucleic Acids Res. 2001;29:E65–E65. doi: 10.1093/nar/29.13.e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayatsu H, et al. Chemistry of bisulfite genomic sequencing; advances and issues. Nucleic Acids Symp. Ser. (Oxf) 2007:47–48. doi: 10.1093/nass/nrm024. [DOI] [PubMed] [Google Scholar]
- Holemon H, et al. MethylScreen: DNA methylation density monitoring using quantitative PCR. Biotechniques. 2007;43:683–693. doi: 10.2144/000112597. [DOI] [PubMed] [Google Scholar]
- Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khulan B, et al. Comparative isoschizomer profiling of cytosine methylation: The HELP assay. Genome Res. 2006;16:1046–1055. doi: 10.1101/gr.5273806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korshunova Y, et al. Massively parallel bisulphite pyrosequencing reveals the molecular complexity of breast cancer-associated cytosine-methylation patterns obtained from tissue and serum DNA. Genome Res. 2008;18:19–29. doi: 10.1101/gr.6883307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu-Daude F, et al. DNA rehybridization during PCR: the ‘Cot effect’ and its consequences. Nucleic Acids Res. 1996;24:2080–2086. doi: 10.1093/nar/24.11.2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A, et al. Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic Acids Res. 2005;33:5868–5877. doi: 10.1093/nar/gki901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2005. [Google Scholar]
- Rauch T, Pfeifer GP. Methylated-CpG island recovery assay: a new technique for the rapid detection of methylated-CpG islands in cancer. Lab. Invest. 2005;85:1172–1180. doi: 10.1038/labinvest.3700311. [DOI] [PubMed] [Google Scholar]
- Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- Suzuki MT, Giovannoni SJ. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl. Environ. Microbiol. 1996;62:625–630. doi: 10.1128/aem.62.2.625-630.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M, et al. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat. Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- Yagi S, et al. DNA methylation profile of tissue-dependent and differentially methylated regions (T-DMRs) in mouse promoter regions demonstrating tissue-specific gene expression. Genome Res. 2008;18:1969–1978. doi: 10.1101/gr.074070.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.