Abstract
Motivation: Methods to improve tiling array expression signals are needed to accurately detect genome features. Royce et al. provide statistical normalizations of tile signal based on probe sequence content that promises improved accuracy, and should be independently verified.
Results: Assessment of the sequence content normalization methods identified a problem: confounding of probe sequence content with gene structure (intron/exon) sequence content. Normalization obscured tile signal changes at gene structure boundaries. This and other evidence suggests that simple sequence normalization does not improve detection of genes from tile expression data.
Availability: http://wfleabase.org/genome-summaries/tile-expression/tileseqnorms/
Contact: gilbertd@indiana.edu
1 INTRODUCTION
The paper of Royce et al. (2007) addresses important aspects of artifact in tiling array signal detection: ubiquitous hybridization that varies with probe sequence content. They back this up with uncomplicated, usable R statistics for the methods presented.
Gene structures show changes in sequence GC content: introns and intergene regions generally have lower GC content than exons (Kalari et al., 2006; Mount et al., 1992). The sequence normalization methods do not address well this structure relation, and whether normalization affects accurate discrimination of structures. The authors compare human RefSeq genes versus non-RefSeq regions (control) paired for GC content. This test may not be sufficient to disentangle non-specific signal due to greater hybridization to GC-rich probes, from true signal of transcribed regions.
2 METHODS
We used R source code from the supplement at http://tiling.gersteinlab.org/sequence_effects/ for this article: sequence_normalization_functions.R, both robust least squares (RLS) with iteration, and quantilenorm, the latter seems the better one. These methods have the value of being clear and uncomplicated statistical approaches to adjusting tile signals for effects of probe sequence.
Sample cases of Daphnia pulex scaffolds 1 and 17 and Drosophila melanogaster chromosomes 2L and 4, containing 4300 and 12 900 exons, respectively, were used. Nimblegen tiling array data for Daphnia transcripts (J.K.Colbourne et al., manuscript in preparation) on these scaffolds includes 180 000 tiles of 50 bp, overlapping every 25 bp. Affymetrix tiling data for Drosophila transcripts (Manak et al. 2006; modENCODE transcriptome data, unpublished data) includes 607 000 non-overlapping tiles of 36 bp. Tile signals above median threshold before and after normalizations that overlap exons were counted. This measures sensitivity (exons with tile expression/all exons) and specificity [1 - (high signal tiles outside exons/all high-signal tiles)]. Because one use of tile expression is to detect gene structures, changes at exon/intron bounds were measured, as the difference in successive signals. Intergene regions were not used due their lower certainty of annotation. Maps of gene structures, tile signal and GC content were viewed, which gave a first clue that sequence normalization was affecting clarity of gene structure detection.
A third comparison was with overlapped tiles from Nimblegen arrays for both species. Pairs of overlapped probes (50 bp long, overlapped 25 bp) on were located, 589 900 for Daphnia, and 946 400 for Drosophila. These overlapped tiles were pair-wise compared for GC content and signal to indicate if a correlation exists for sequence effects within the same exon and intron structures.
3 RESULTS
We were able to use and reproduce a GC content effect of probe sequence for both Daphnia (Nimblegen) and Drosophila (Affymetrix) tile expression data. Signals normalized this way do not differ grossly from the raw signals. However, fuzziness at detecting gene sequence structure (exon/intron boundaries) appears to be one result of sequence content normalization. Sequence normalization (quantilenorm) reduced sensitivity and specificity for exon detection by 1% for the Daphnia data, and by 2% for Drosophila data.
3.1 Normalization reduces GC content correlation
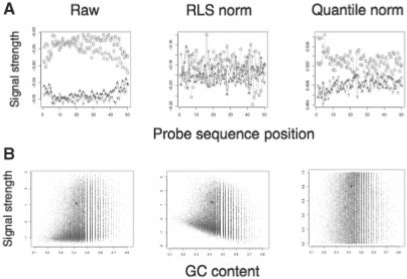
The quantile normalization and RLS methods reproduce generally the GC content effect of probe sequence reported by Royce and colleagues, for both species experiments. The plots in Figure 1 look compelling: raw signal gives a higher signal for GC-rich probes. After normalization by sequence, that effect goes away. Average exon signal and GC values are above those of introns, and this remains after normalizations, although correlation of GC and signals is reduced.
Fig. 1.
(A) Raw and two normalized signals, by base per probe sequence position and (B) as a dot plot of signal strength versus GC content. Plots (A) are as in Royce et al. (Fig. 1) of average signal per base over probe sequence position. Bases G + C in Raw are at the top, A + T at bottom.
3.2 Normalization reduces gene structure signals
For detecting gene structures, the overlap of high-scoring tiles with known exons provides a measure of accuracy for normalization results. Both species data showed a drop off in sensitivity and specificity with normalized signal.
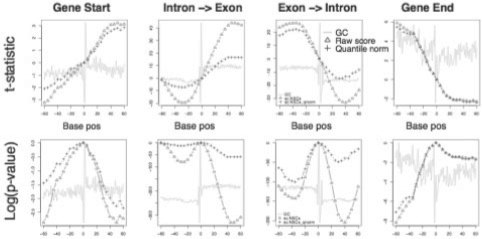
Use of raw signals improves the detection of gene structures as seen with signal changes at exon/intron boundaries. One effect of normalization is to obscure gene structure boundaries, which are often related to sequence changes. Figure 2 plots the statistical power of raw and quantile norm signals to distinguish exon and gene boundaries. The raw signal has a greater statistical discrimination of boundaries. These effects are correlated with GC content, also displayed. With a per-base comparison of GC and score, the major effect is for higher score-GC correlation in intron regions. Quantile normalization reduces this correlation, so that GC-poor introns have a relatively higher tile score.
Fig. 2.
Tile score statistical power at finding gene/exon boundaries. Student's t-statistic and log10 (probability) for raw (triangle) and quantile norm (cross) scores measure ability to distinguish boundary at base positions away from position 0 (gene or exon boundary ± 60 bp). The t-statistic is score difference from position 0. GC content (line) shows expected spikes at boundaries (x position 0): coding start and end, intron->exon, and exon->intron, with increased GC% in exon regions.
Using partially overlapped tiles of experiments for both species, differences in GC content between overlapped tiles had lower correlation with signal level. The overall correlation of GC and signal strength is 20% in both species. For overlapped tiles this correlation drops to 3% (Drosophila) or 15% (Daphnia). When signal and GC content are measured at exon–intron boundaries, overlapped tiles have a high 60% correlation for Daphnia, and 9% in Drosophila, both about three times higher than outside of boundaries. These species differ in total GC content, and in DNA methylation processing genes associated with variations in GC, so large species differences are not unexpected.
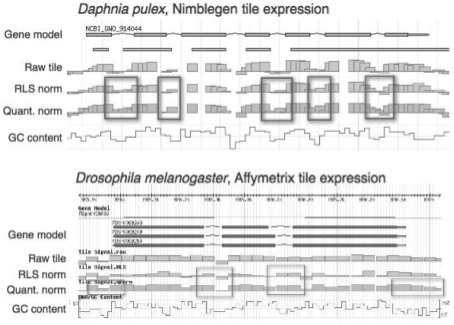
Nornalization problems at detecting gene structures were first evident on gene maps. RLS and quantile normalization down-weighted exons and up-weighted introns so that the normalized signal was strongest for introns of several genes. Figure 3 shows examples of this for two genes. The boxed areas show cases where detection of intron–exon boundaries by tile signal is diminished after normalization compared with the raw tile signals. These areas coincide with changes in GC content. The normalizations have increased the score, and thus noise, of non-expressed introns and intergenic regions.
Fig. 3.
Exon–intron signal loss examples. Genome maps show gene models, the raw and normalized tile signals and GC content, for Daphnia and Drosophila genes. Box (highlighted) areas where normalization has obscured the biological signal.
4 DISCUSSION
Sequence content normalization is a useful concept for improving tile array signal accuracy. Yet, it needs to address gene structure effects if used in transcriptome detection experiments. Royce et al. (2007) describe the technology and motivation for probe sequence normalization. Gene-centric microarray studies can select probes within a gene transcribed region in order to optimize hybridization on arrays. This optimizing selection is not possible with genome tiling, where probes cover the genome in short spans of different sequence content. The results here indicate normalization of tile array scores by sequence content obscures biological signals.
Johnson et al. (2008) find that probe GC content variation is not a significant cause of tile array artifacts. This study used spike-in mixtures with a blind test at several laboratories, with different platforms and measurement algorithms, for ChIP-chip tiling microarrays. One result is that probe GC content does not influence rate of false positives, false negatives or true positives. Simple tandem repeats and segmental duplications are more often associated with false calls.
When tile expression is used to detect gene structures, there is a dilemma because gene structures and ubiquitous hybridization artifacts are confounded with the sequence content. There are cases where sequence normalization improves apparent gene-structure signal in low-GC regions. If there is a way to combine this with gene structure sequence changes, this would be a helpful analysis. One possible use would be to combine sequence normalization with gene structure modeling (e.g. generalized hidden Markov models). Another option may be to estimate transcription fragments without signal normalization, to best detect boundaries, then apply sequence normalization over these fragments to reduce ubiquitous hybridization effects.
ACKNOWLEDGEMENTS
We thank John Colbourne and Peter Cherbas for discussions and use of unpublished tile expression data.
Funding: National Science Foundation and the National Institutes of Health to Don Gilbert.
Conflict of Interest: none declared.
REFERENCES
- Johnson DS, et al. Systematic evaluation of variability in ChIP-chip experiments using predefined DNA targets. Genome Res. 2008;18:393–403. doi: 10.1101/gr.7080508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalari KR, et al. First exons and introns – a survey of GC content and gene structure in the human genome. In Silico Biol. 2006;6:237–242. [PubMed] [Google Scholar]
- Manak JR, et al. Biological function of unannotated transcription during the early development of Drosophila melanogaster. Nat. Genet. 2006;38:1151–1158. doi: 10.1038/ng1875. [DOI] [PubMed] [Google Scholar]
- Mount SM, et al. Splicing signals in Drosophila: intron size, information content, and consensus sequences. Nucleic Acids Res. 1992;20:4255–4262. doi: 10.1093/nar/20.16.4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royce TE, et al. Assessing the need for sequence-based normalization in tiling microarray experiments. Bioinformatics. 2007;23:988–997. doi: 10.1093/bioinformatics/btm052. [DOI] [PubMed] [Google Scholar]