Abstract
Previously we showed that the functional activity of the epithelial chloride channel that is encoded by the cystic fibrosis gene (CFTR) is reciprocally modulated by two components of the vesicle fusion machinery, syntaxin 1A and Munc-18. Here we report that syntaxin 1A inhibits CFTR chloride channels by means of direct and domain-specific protein–protein interactions. Syntaxin 1A stoichiometrically binds to the N-terminal cytoplasmic tail of CFTR, and this binding is blocked by Munc-18. The modulation of CFTR currents by syntaxin 1A is eliminated either by deletion of this tail or by injecting this tail as a blocking peptide into coexpressing Xenopus oocytes. The CFTR binding site on syntaxin 1A maps to the third predicted helical domain (H3) of this membrane protein. Moreover, CFTR Cl− currents are effectively inhibited by a minimal syntaxin 1A construct (i.e., the membrane-anchored H3 domain) that cannot fully substitute for wild-type syntaxin 1A in membrane fusion reactions. We also show that syntaxin 1A binds to and inhibits the activities of disease-associated mutants of CFTR, and that the chloride current activity of recombinant ΔF508 CFTR (i.e., the most common cystic fibrosis mutant) can be potentiated by disrupting its interaction with syntaxin 1A in cultured epithelial cells. Our results provide evidence for a direct physical interaction between CFTR and syntaxin 1A that limits the functional activities of normal and disease-associated forms of this chloride channel.
The cystic fibrosis transmembrane conductance regulator (CFTR) is an epithelial chloride channel that is defective or lacking in patients with cystic fibrosis (1, 2). This polytopic membrane protein is composed of five major cytoplasmic domains (3): the N- and C-terminal tails, two nucleotide-binding domains (NBDs), and a regulatory domain (R domain) with multiple consensus sites for phosphorylation by protein kinase A (PKA) and protein kinase C. Channel activation requires both phosphorylation of the R domain (principally by PKA) and ATP binding and hydrolysis by the NBDs (4, 5). The extent to which CFTR Cl− channels are regulated by other mechanisms (e.g., protein–protein interactions) is largely unknown.
Recently we showed that CFTR-mediated chloride currents are inhibited by syntaxin 1A (6), a member of the syntaxin family of membrane fusion regulators (7, 8) that is expressed highly in neurons and to a lesser extent in intestinal epithelial cells (6). Syntaxin 1A regulates CFTR currents both in Xenopus oocytes that heterologously express these molecules and in epithelial cells that normally produce CFTR and syntaxin 1A (6). This functional interaction is syntaxin 1A-specific, requires the C-terminal membrane anchor of syntaxin 1A, and can be blocked by Munc-18, a soluble syntaxin-binding protein (9, 10). At the neuronal synapse syntaxin 1A is a component of a large molecular complex that controls the docking and/or fusion of synaptic vesicles with the presynaptic plasma membrane (11, 12). Interestingly, the N-type calcium channel is among the multiple syntaxin 1A-binding proteins within this synaptic protein complex (13–15), and several groups have reported that syntaxin 1A negatively modulates the gating of these channels (16–18). These latter observations, coupled with our own studies of the CFTR–syntaxin 1A interaction in gut epithelial cells, indicate that this syntaxin isoform may have the capacity to regulate ion channel function in several tissues.
In the present study we test the hypothesis that syntaxin 1A regulates CFTR currents by means of direct protein–protein interactions. This hypothesis is based on the results of indirect binding experiments which indicated that CFTR and syntaxin 1A can be coimmunoprecipitated from epithelial cells, and that CFTR can be “pulled down” from cell lysates by immobilized recombinant syntaxin 1A (but not by other syntaxin isoforms; ref. 6). We report here that syntaxin 1A directly and stoichiometrically binds to CFTR. We have mapped the interacting domains on each molecule and provide evidence that the binding of syntaxin 1A to CFTR is essential for the regulation of CFTR currents by this molecule. We have also extended our analysis of the syntaxin–CFTR interaction to include several disease-associated mutants of CFTR, and we show that the functional activity of the most common mutant (ΔF508) can be potentiated by disrupting this interaction in an epithelial cell line. Our results indicate that syntaxin 1A directly binds to CFTR and modulates the Cl− currents mediated by wild-type and disease-associated forms of this chloride channel.
MATERIALS AND METHODS
Materials and Constructs.
Bacterial expression vectors were obtained from Pharmacia (pGEX). The syntaxin 1-specific polyclonal antibody was generated by immunizing a rabbit with glutathione S-transferase (GST)-syn1AΔC (syn1AΔC refers to deletion of the C-terminal membrane anchor of syntaxin 1A) and was affinity purified by using immobilized syntaxin 1AΔC cleaved free of GST (6). This antibody was specific for syntaxin 1A in that it did not cross-react with syntaxins 2, 3, or 4 (6). The CFTR C-terminal antibody was obtained from Genzyme (catalog no. 2503-01). Munc-18 was purified as a GST fusion protein, and the GST was then removed by thrombin cleavage (6). CFTR-NBDs were purified as described earlier (19). Munc-18 monoclonal antibody was obtained from Transduction Laboratories (Lexington, KY). All other materials were reagent grade and purchased from common vendors.
The N-terminal deletion mutant of human CFTR (Δ2–79 CFTR) was generated as follows: The full-length CFTR cDNA in pGT1 was digested with XmaI and XbaI and the resulting fragment (containing nucleotides 1–279 of the coding region) was subcloned into pSK-Bluescript (Stratagene). The region corresponding to amino acids 2–79 was deleted from this fragment by oligonucleotide-directed mutagenesis. The resulting fragment was then ligated back into pGT1-CFTR that had been cut with XmaI and XbaI. For expression studies in COS-7 cells and oocytes, the Δ2–79 CFTR construct was subcloned into pCDNA3 (Invitrogen).
Transfection of COS-7 Cells.
COS-7 cells grown in 6-well plates to about 70% confluence were washed with Opti-MEM (GIBCO/BRL) and infected with vaccinia virus at a multiplicity of infection of 5. After 40 min, 5 μg of pTM1 plasmid (4) or pCDNA3 plasmid expressing wild-type or mutant CFTR was mixed with 20 μg of lipid—N-[1-(2,3-dioleoyloxy)propyl]-N,N,N-trimethylammonium chloride/dioleoyl phosphatidylethanolamine (DOTAP/DOPE) at 1:1 wt/wt ratio—in Opti-MEM (200 μl) for 20 min, diluted to 1 ml in tissue culture medium, and then added to the cells. After 24 hr the medium was removed and the cells were lysed as described below.
Binding Assays.
The cDNAs encoding the various syntaxin 1A and CFTR cytosolic domains were amplified by polymerase chain reaction and subcloned into an appropriate pGEX bacterial expression vector. These domains were expressed as GST fusion proteins in Escherichia coli, purified on glutathione-agarose, eluted in 20 mM glutathione, and dialyzed extensively against PBS. For pairwise binding assays, syntaxin 1A cytosolic domain (syntaxin 1AΔC) was cleaved from GST with thrombin, dialyzed in binding buffer (0.2% Triton X-100 in PBS), and mixed with the appropriate GST-CFTR fusion protein in this buffer for 3 hr at 4°C. The GST fusion protein was then precipitated with excess glutathione-agarose, washed extensively in binding buffer, and analyzed for syntaxin 1AΔC binding by immunoblotting using a syntaxin 1-specific polyclonal antibody (6).
CFTR pull-down assays were performed as described previously (6). Briefly, soluble eluted GST-syn1AΔC was added to a Triton X-100 lysate (1% Triton X-100 in PBS plus 1 mM phenylmethanesulfonyl fluoride and 1 μg/ml each of leupeptin, pepstatin, and aprotinin) of HT29-CL19A colonic epithelial cells expressing native CFTR or of COS-7 cells transiently expressing recombinant CFTR. After diluting the samples in PBS to bring the final Triton concentration to 0.2%, the samples were mixed for 3–12 hr at 4°C. The bound proteins were then precipitated with excess glutathione-agarose, washed extensively, and analyzed for CFTR by immunoblotting using a monoclonal antibody to the C terminus (Genzyme). Duplicate lysate samples were assayed for CFTR protein amount by immunoprecipitation followed by immunoblotting using the same CFTR antibody. The efficiency of CFTR immunoprecipitation was greater than 90% for all CFTR constructs examined. The amount of CFTR was quantitated by densitometry and analyzed by using ip lab software (Signal Analytics, Vienna, VA).
Electrophysiology.
Oocyte preparation, injection, and two-electrode voltage-clamp methods are described in detail elsewhere (6, 20). Using the message machine kit (Ambion), we transcribed complementary RNA (cRNA) in vitro from linearized templates. Unless otherwise noted, final injected cRNA amounts were wild-type CFTR, 1 ng; ΔF508 and Δ2–79 CFTR, 25 ng; syntaxin 1A, syntaxin 3, H3-TMD, and Munc-18, 5 ng. Oocytes were assayed 48–72 hr after cRNA injection. Peptides and botulinum neurotoxin C1 were injected in 25-nl volumes 30 min prior to assay. Final concentrations were estimated assuming an oocyte volume of 1 μl. Oocytes were voltage clamped at −50 mV and CFTR chloride currents were activated by using 20 μM forskolin, 100 μM dibutryl-cAMP, and 100 μM 3-isobutyl-1-methylxanthine (IBMX) (Figs. 1E, 3, and 4) or 20 μM forskolin, 200 μM dibutryl-cAMP, and 5 mM IBMX (Figs. 2B and 5C). All currents mediated by wild-type CFTR or the CFTR mutants were cAMP dependent, exhibited an approximately linear current–voltage relation, and were inhibitable by the chloride channel blocker diphenylamine carboxylate (data not shown).
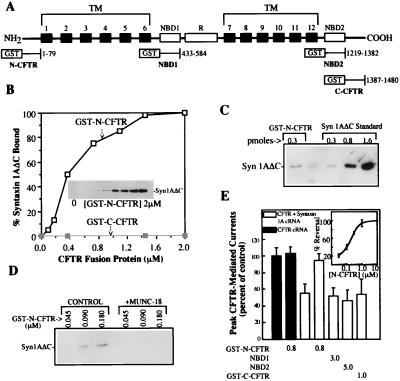
Figure 1.
Syntaxin 1A directly binds to the N-terminal cytoplasmic tail of CFTR. (A) Schematic view of the major CFTR domains and the locations of the GST fusion proteins used in this study. TM, transmembrane. (B) Binding of syntaxin 1A cytosolic domain (0.35 nmol) to increasing concentrations of GST-N-CFTR (Inset and □) in a 0.2-ml reaction volume. No syntaxin 1A binding to GST-C-CFTR was observed (■). Results are representative of four different experiments. (C) syn1AΔC binds to GST-N-CFTR with an approximately 1:1 stoichiometry. syn1AΔC (350 pmol) was mixed with 0.3 pmol of GST-N-CFTR, and the amount of syn1AΔC that bound was estimated by immunoblotting using known amounts of syn1AΔC as standards. (D) Munc-18a blocks the binding of GST-N-CFTR to syn1AΔC. GST-N-CFTR at the indicated concentrations was mixed with 0.1 μg of syn1AΔC in the absence or presence of 0.2 μg of Munc-18a in a 0.2-ml reaction volume. (E) GST-N-CFTR peptide blocks the inhibition of CFTR currents by membrane-anchored syntaxin 1A. Oocytes (10–12 oocytes per condition) that were expressing CFTR with or without full-length syntaxin 1A were microinjected with the indicated GST-CFTR fusion proteins (units: μM) 30 min prior to current recording (see text and ref. 6 for details). Error bars represent SEMs. The dose dependence of the block of the CFTR–syntaxin 1A interaction by GST-N-CFTR is shown in the Inset (five oocytes per concentration).
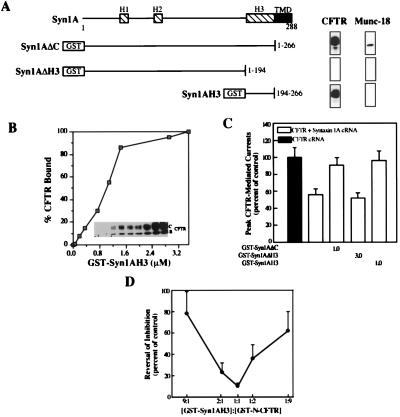
Figure 3.
The H3 domain of syntaxin 1A is necessary and sufficient for binding CFTR. (A) Schematic view of the major domains of syntaxin 1A and locations of GST fusion proteins used in this study. Right shows binding to these fusion proteins (50 μg each) of native CFTR or native Munc-18 in pull-down assays performed on lysates of HT29-CL19A colonic epithelial cells. (B) Dose-dependent binding of CFTR to isolated H3 domain (GST-syn1AH3) in pull-down assay performed on COS 7 cell lysates expressing recombinant wild-type CFTR. (C) Block of the CFTR–syntaxin 1A interaction in oocytes (8–14 oocytes per condition) by GST-syn1AH3 (units: μM). (D) The blocking effects of the GST-syn1AH3 and GST-N-CFTR peptides on the CFTR–syntaxin 1A interaction in oocytes (6 oocytes per data point) are canceled by premixing these fusion proteins at equimolar ratios (total fusion protein concentration: 2 μM).
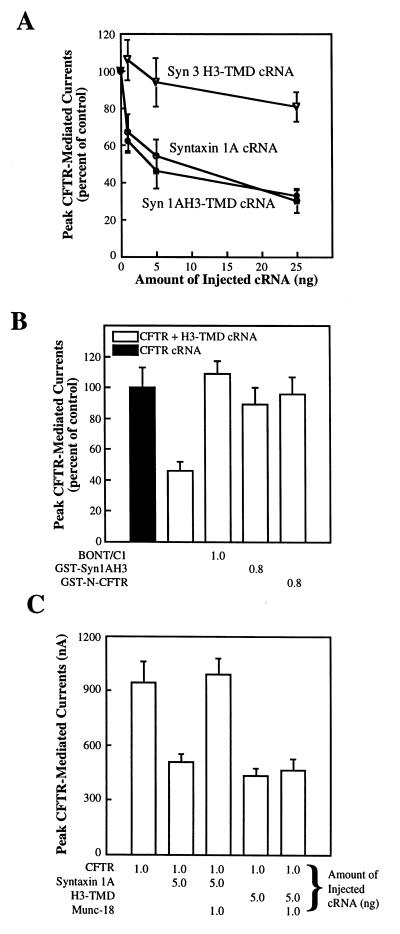
Figure 4.
CFTR currents are inhibited by the membrane-anchored H3 domain of syntaxin 1A. (A) The syntaxin 1A H3-TMD construct inhibits CFTR currents in oocytes (5–9 oocytes per data point) as effectively as does full-length syntaxin 1A (CFTR cRNA: 1 ng throughout). (B) The inhibition of CFTR currents by the syntaxin 1A H3-TMD construct in oocytes (6–12 oocytes per condition) is acutely blocked by microinjecting botulinum neurotoxin C1, GST-syn1AH3, or GST-N-CFTR into coexpressing oocytes 30 min before current recording (units: μM). (C) The inhibition of CFTR by the syntaxin 1A H3-TMD construct in oocytes (6 oocytes per condition) cannot be reversed by coexpression with Munc-18a (amounts of the injected cRNAs (ng) are shown as indicated).
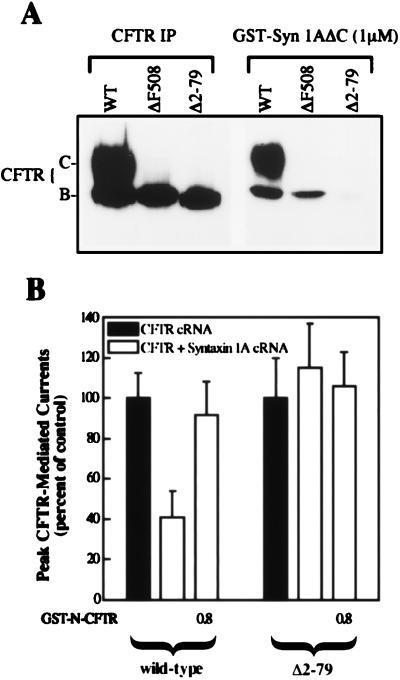
Figure 2.
The N-terminal tail of CFTR is required for the regulation of chloride currents by syntaxin 1A. (A) CFTR deletion mutant that lacks the N-terminal tail (Δ2–79) exhibits poor binding to syntaxin 1A. The indicated CFTR constructs were expressed in COS-7 cells and assayed for binding to GST-syn1AΔC at a nearly saturating concentration for the binding of wild-type band C (1 μM; ref. 6). Equal volumes of lysates were assayed for CFTR protein amount by immunoprecipitation (IP) using a monoclonal antibody to the C terminus (Genzyme). (B) Δ2–79 CFTR Cl− currents are insensitive to the coexpression of membrane-anchored syntaxin 1A in oocytes (13–18 oocytes per condition). Currents are normalized to those measured in the absence of syntaxin 1A. Absolute cAMP-stimulated currents in the absence of syntaxin 1A were 875 ± 67 nA and 180 ± 23 nA for wild-type CFTR and Δ2–79 CFTR, respectively (see Materials and Methods for technical details). Note that the Δ2–79 CFTR currents were similar in absolute magnitude to the ΔF508 currents shown in Fig. 5C, which were inhibited by syntaxin 1A. A subset of oocytes were injected with 0.8 μM GST-N-CFTR 30 min before current recording. Error bars represent SEM.
Figure 5.
Syntaxin 1A binds to and limits the functional activities of disease-associated CFTR mutants. (A) Binding of CFTR constructs expressed in COS-7 cells at subsaturating concentrations of GST-syn1AΔC. Equal volumes of lysates were assayed for CFTR protein amount by immunoprecipitation (IP). The efficiency of binding of band B (i.e., binding normalized to the IP signal) at 0.13 μM syn1AΔC was 38.4%, 36.1%, and 5.7% for wild-type CFTR, G551D CFTR, and ΔF508 CFTR, respectively. (B) ΔF508 CFTR binds efficiently to GST-syn1AΔC when this mutant is processed at reduced temperature (see text for details). (C) ΔF508 CFTR is reversibly inhibited by coexpression with full-length syntaxin 1A in oocytes (8–14 oocytes per condition). Absolute cAMP-stimulated ΔF508 CFTR currents in the absence of syntaxin 1A were 230 ± 17 nA. (D) ΔF508 CFTR currents in LLC-PK1 epithelial cells are potentiated by fusion proteins that block the CFTR–syntaxin 1A interaction. Shown are peak cAMP-activated Cl− currents 5–10 min after seal formation in the absence or presence of the indicated fusion protein in the patch pipette. Holding potential was +110 mV. GST-syn1A, cytosolic domain of syntaxin 1A (0.35 μM); GST-syn1AΔH3, truncated cytosolic domain lacking the H3 domain (0.35 μM); H3 domain, GST-syn1AH3 fusion protein (0.6 μM). cAMP-dependent currents were also potentiated by GST-syn1A in LLC-PK1 cells transfected with wild-type CFTR (results not shown).
Whole-cell patch-clamp studies of LLC-PK1 cells stably transfected with ΔF508 CFTR (21) were performed as described (6, 22), except that 5 mM MgATP was used in the pipette solution. Cell capacitance was measured by integrating the current during a 10-mV voltage step and subtracting a baseline established ≈15 ms after the step. Currents were activated with 500 μM 8-(4-chlorophenylthio)-cAMP (cpt-cAMP) in the pipette. Both the untransfected cells and ΔF508 CFTR-transfected LLC-PK1 cells were grown in the presence of 30 μM ZnCl2 for 18 hr prior to patch-clamping to induce recombinant protein expression under the control of the metallothionein promoter (21).
RESULTS
To test for a direct physical interaction between CFTR and syntaxin 1A, we expressed four of the major cytosolic domains of CFTR as GST fusion proteins in E. coli [i.e., the N- and C-terminal tails and the two NBDs (Fig. 1A)]. These fusion proteins were purified and tested for binding to purified recombinant cytosolic domain of syntaxin 1A (syntaxin 1AΔC, where C refers to deletion of the C-terminal membrane anchor). In these pairwise binding experiments we observed saturable and stoichiometric binding of the N-terminal tail of CFTR (GST-N-CFTR) to syntaxin 1A with an EC50 in the submicromolar range (≈400 nM; Fig. 1 B and C). The binding of syntaxin 1A to GST-N-CFTR was blocked by Munc-18a (Fig. 1D), as we observed previously for the interaction of syntaxin 1A with full-length CFTR (6). Neither of the NBDs (not shown) nor the C-terminal tail of CFTR (GST-C-CFTR) bound to syntaxin 1A (Fig. 1B); in addition, GST-N-CFTR did not bind to a syntaxin isoform that fails to inhibit CFTR currents in oocytes (i.e., syntaxin 3; ref. 6 and data not shown). The binding of syntaxin 1A to N-CFTR (but not to C-CFTR) could also be detected by using less direct binding assays such as yeast two-hybrid analysis and pull-down assays in which an N-CFTR transferrin receptor chimera was precipitated by immobilized syntaxin 1A (data not shown).
We tested the functional relevance of the binding of syntaxin 1A to the N-terminal tail of CFTR in two ways: (i) by performing peptide blocking experiments in Xenopus oocytes (Fig. 1E) and (ii) by examining a CFTR deletion mutant that lacks the N-terminal tail (Fig. 2). In the peptide blocking experiments we exploited the fact that CFTR can be acutely rescued from inhibition by membrane-anchored syntaxin 1A by microinjecting soluble GST-syntaxin fusion proteins into coexpressing oocytes within 30 min of current recording. Fig. 1E shows that microinjection of GST-N-CFTR acutely blocked the inhibition of CFTR currents by syntaxin 1A in a dose-dependent manner (IC50 of ≈300 nM), whereas the C-CFTR and NBD fusion proteins had no effect. GST-N-CFTR peptide had no effect on CFTR currents in the absence of coexpressed syntaxin 1A, which confirms that this peptide stimulates CFTR currents by disrupting the syntaxin–CFTR interaction.
Fig. 2 shows that deletion of the N-terminal tail from CFTR greatly inhibited the physical and functional interactions between CFTR and syntaxin 1A. Like many CFTR mutants, the N-terminal deletion construct (Δ2–79 CFTR) is an endoplasmic reticulum (ER) processing mutant that inefficiently matures from the ER form (band B) to the fully processed form (band C) when expressed in mammalian cells (e.g., COS cells; Fig. 2A.) Even so, this deletion mutant generated cAMP-dependent chloride currents when expressed in Xenopus oocytes [like other CFTR processing mutants such as ΔF508 CFTR (Figs. 2B and 5; ref. 23)]. However, the currents mediated by Δ2–79 CFTR were insensitive to inhibition by syntaxin 1A, unlike those mediated by wild-type CFTR and ΔF508 CFTR (see also Fig. 5C). Consistent with our inability to detect a functional interaction between the N-terminal deletion mutant and syntaxin 1A, we observed that the binding of this mutant at a relatively high concentration of GST-syn1AΔC (1 μM) was inhibited by greater than 95% as compared with the binding of the band B forms of either wild-type CFTR or the ΔF508 CFTR processing mutant. In contrast, a CFTR deletion mutant that lacked the C-terminal tail exhibited wild-type levels of syntaxin 1A binding, and its Cl− current activity was inhibited by syntaxin 1A to the same degree as wild-type CFTR (results not shown). Collectively, our binding results and functional data indicate that syntaxin 1A binds directly and stoichiometrically to the N-terminal tail of CFTR (Fig. 1), and that this domain of CFTR is important for the negative regulation of CFTR currents by syntaxin 1A (Figs. 1E and 2).
Using a similar strategy, we identified a minimal cytoplasmic domain of syntaxin 1A that is both necessary and sufficient for CFTR binding (Fig. 3). Four major domains constitute syntaxin 1A (7): three putative helical domains (H1–H3) and a C-terminal membrane anchor (Fig. 3A). The H3 domain is necessary but not sufficient for syntaxin 1A to participate fully in the membrane fusion reaction cycle [i.e., SNARE complex assembly and disassembly (24, 25)]. Deletion of the H3 domain eliminated the binding of both CFTR (Fig. 3A) and Munc-18a (Fig. 3A and ref. 25) to syntaxin 1A. Interestingly, the H3 domain itself (GST-syn1AH3) exhibited saturable binding to CFTR (Figs. 3 A and B), although the apparent affinity of this interaction (EC50 of 1 μM) was somewhat lower than that for intact cytoplasmic syntaxin 1A (EC50 of 0.3 μM; ref. 6). In contrast, Munc-18 was unable to bind to the isolated H3 domain (Fig. 3A and ref. 25). Microinjection of GST-syn1AH3 into oocytes that were coexpressing CFTR and full-length syntaxin 1A blocked the inhibition of CFTR currents by membrane-anchored syntaxin 1A, as we observed for GST-N-CFTR (Fig. 1E) and for intact cytoplasmic syntaxin 1A (Fig. 3C and ref. 6). The syntaxin 1A fusion protein that lacked the H3 domain (GST-syn1AΔH3) was unable to block the inhibition of CFTR currents by membrane-anchored syntaxin 1A (Fig. 3C), as expected on the basis of our binding data. Neither GST-syn1AH3 nor GST-syn1AΔH3 affected CFTR currents in the absence of membrane-anchored syntaxin 1A (not shown). In addition, the blocking effects of GST-syn1AH3 and of GST-N-CFTR on the CFTR–syntaxin interaction could be canceled by mixing these proteins at equimolar ratios prior to microinjection (Fig. 3D). The effects of these peptides could not be cancelled either by mixing GST-syn1AH3 with GST-C-CFTR or by mixing the GST-N-CFTR peptide with the syntaxin 1 fusion protein lacking the H3 domain (GST-syn1AΔH3; not shown). These latter results verify the specificity of the syn1AH3 and N-CFTR fusion proteins in disrupting the CFTR–syntaxin interaction in oocytes, and they support our biochemical evidence for a 1:1 stoichiometric interaction between these protein domains (Fig. 1).
Given that the isolated H3 domain of syntaxin 1A is capable of binding CFTR, we examined the possibility that CFTR currents could be inhibited by a minimal syntaxin construct that is composed solely of the membrane-anchored H3 domain (syn1AH3-TMD). Fig. 4A shows that, like wild-type syntaxin 1A, expression of this minimal construct from cRNA effectively inhibited CFTR currents in a dose-dependent manner. As a control for specificity we also coinjected CFTR cRNA with the cRNA encoding the corresponding region of syntaxin 3 (syn3 H3-TMD). This latter construct had negligible effects on CFTR current activity. The inhibition of CFTR currents by the membrane-anchored H3 domain of syntaxin 1A could be acutely reversed by microinjecting either botulinum neurotoxin C1 (i.e., a protease that cleaves syntaxin from the membrane; ref. 12) or the GST-syn1AH3 and GST-N-CFTR fusion proteins (Fig. 4B), just as we observed for full-length syntaxin 1A (6). However, CFTR currents could not be rescued from this inhibition by Munc-18a, which completely blocks the inhibition of CFTR currents by full-length syntaxin 1A when coexpressed with these molecules in oocytes (Fig. 4C and ref. 6). The results of our experiments with membrane-anchored H3 domain indicate that (i) this minimal syntaxin 1A construct can effectively regulate CFTR currents, even though it cannot replace wild-type syntaxin 1A in the membrane fusion reaction cycle (25), and (ii) the regulation of CFTR by this construct is Munc-18 independent, which is consistent with the biochemical evidence that the N-terminal region of syntaxin 1A is required for Munc-18 binding (Fig. 3A and ref. 25).
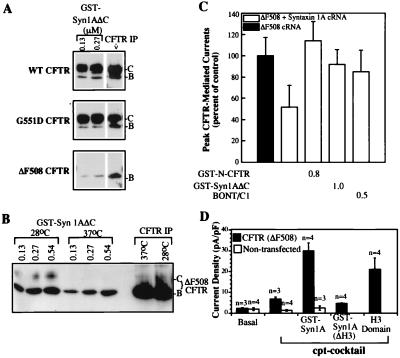
We next determined whether syntaxin 1A binds to and inhibits the activities of disease-associated mutants of CFTR, many of which are partial-loss-of-function mutants whose residual activity may be limited by this interaction. Those CFTR mutants that produce full-length translation products can be classified into three categories (2): (i) ER processing mutants that inefficiently traffic to the Golgi apparatus (e.g., the most common allele, ΔF508); (ii) regulation mutants that mature normally but are refractory to activation by ATP (e.g., G551D); and (iii) conduction mutants that also mature normally but exhibit reduced single-channel conductances (e.g., R117H). We observed that CFTR mutants from each of these classes are capable of binding syntaxin 1A, as determined in pull-down assays performed on lysates prepared from transfected COS cells (Fig. 5A and results not shown). Interestingly, however, the ER glycosylated forms (band B) of the processing mutants consistently exhibited less efficient binding at subsaturating concentrations of syntaxin 1A (0.13 and 0.27 μM) than the ER forms of wild-type CFTR and the nonprocessing mutants (Fig. 5 A and B and legend). This decrease in the efficiency of binding is probably because of altered protein conformation or because of protein–protein interactions within the ER that hinder binding to exogenous syntaxin 1A, rather than because of the mutations per se. In this regard, we could recover wild-type levels of binding for both the band B and band C forms of ΔF508 CFTR by promoting maturation of this temperature-sensitive mutant by growing the COS fibroblasts at reduced temperature (Fig. 5B and ref. 26).
Because the most common disease-associated mutant of CFTR (ΔF508) is capable of binding syntaxin 1A, we asked whether the functional activity of this mutant is limited by its interaction with syntaxin 1A. This is an important question, given the great interest in devising strategies to improve the functional activity of this CFTR mutant. Fig. 5C shows that syntaxin 1A inhibited the chloride current activity of ΔF508 CFTR in oocytes. This inhibition was blocked by GST-N-CFTR and GST-syn1AΔC fusion proteins and by botulinum neurotoxin C1, as we observed for wild-type CFTR (6). We next determined whether the functional activity of ΔF508 CFTR in epithelial cells could be enhanced by disrupting its interaction with endogenous syntaxin 1A. For this purpose we introduced syntaxin fusion proteins that block the syntaxin–CFTR interaction through a whole-cell patch pipette into LLC-PK1 epithelial cells transfected with recombinant ΔF508 CFTR and monitored the effects of these peptides on whole-cell chloride currents mediated by the ΔF508 mutant (Fig. 5D). LLC-PK1 cells express native syntaxin 1A at approximately the same level as colonic epithelial cell lines such as T84, as determined by immunoblotting (results not shown). The expression of recombinant ΔF508 CFTR in these cells is under the control of a zinc-inducible promoter: i.e., after zinc induction these cells produce enough mature ΔF508 CFTR protein to generate small cAMP-activated currents (ref. 21; we chose this approach to generate mature ΔF508 protein because it is more effective than growing epithelial cells at reduced temperature; unpublished observations). Fig. 5D shows that inclusion of GST-syn1AΔC or GST-syn1AH3 in the patch pipette acutely and markedly potentiated the macroscopic currents carried by ΔF508 CFTR, as we had observed previously for native CFTR in colonic epithelial cells (6). These fusion proteins had no effect on whole cell currents in untransfected LLC-PK1 cells that express no ΔF508 CFTR, nor were the ΔF508 CFTR currents affected by the syntaxin 1A fusion protein that lacks the H3 domain [GST-syn1AΔH3, which is unable to block the CFTR–syntaxin interaction in oocytes (Fig. 3)]. Thus, the functional activity of the ΔF508 mutant can be potentiated by reagents that disrupt its interaction with native syntaxin 1A in cultured epithelial cells.
DISCUSSION
We have provided evidence for a direct interaction between syntaxin 1A and CFTR that limits the functional activities of both normal and disease-associated forms of CFTR. Syntaxin 1A binds directly and stoichiometrically to the N-terminal cytoplasmic tail of CFTR. The interaction between syntaxin 1A and this region of CFTR is blocked by Munc-18, which itself binds to syntaxin 1A and prevents the physical and functional interactions between CFTR and syntaxin 1A (6). Our functional data indicate that the binding of membrane-anchored syntaxin 1A to the N-terminal tail of CFTR is essential for the inhibition of CFTR currents by this molecule. However, our results do not exclude additional interactions between syntaxin 1A and other CFTR domains that may modulate this mode of CFTR regulation. Conceivably, syntaxin 1A regulates CFTR activity by influencing interactions between the N-terminal tail and domains that are involved in channel activation and gating (i.e., the NBDs and regulatory domain; refs. 4 and 5). It is also possible that syntaxin 1A modulates the numbers of CFTR molecules at the cell surface, although our results indicate that such an effect would be mediated by the physical interaction between these proteins rather than by a generic effect of syntaxin 1A on membrane traffic. These different modes of CFTR channel regulation are not mutually exclusive. Clarifying the mechanistic details of the syntaxin 1A–CFTR interaction will require detailed studies of the single channel properties and surface expression of CFTR in the presence and absence of membrane-anchored syntaxin 1A.
Our results also indicate that CFTR binds to the H3 domain of syntaxin 1A and that this domain is important for the syntaxin-dependent regulation of CFTR currents. Since the H3 domain is essential for the participation of syntaxin 1A in membrane fusion reactions (25), the binding of CFTR to this domain raises the possibility that the CFTR–syntaxin 1A interaction could influence the functional properties of syntaxin as well [i.e., the functional interaction may be bidirectional, as argued for the interaction between syntaxin 1A and calcium channels in brain (16–18, 27)]. For example, the CFTR chloride channel may mask this domain and thereby regulate the availability of syntaxin 1A for participating in membrane fusion reactions in certain cell types and intracellular compartments. This could explain in part the emerging evidence that CFTR can fine tune membrane traffic in epithelial cells (28–30).
We have also shown that syntaxin 1A physically and functionally interacts with disease-associated mutants of CFTR including the most common mutant, ΔF508 CFTR. Interestingly, we consistently observed that the ER forms of the processing mutants (e.g., ΔF508) bound to syntaxin 1A with somewhat lower efficiency than did the ER forms of wild-type CFTR or nonprocessing mutants. The physiological significance of this observation is unclear, since the acute regulation of CFTR currents in oocytes and epithelial cells by syntaxin 1A indicates that the functional interaction between these molecules occurs at least in part beyond the biosynthetic pathway (as is consistent with the primarily apical location of syntaxin 1A in colonic epithelial cells; data not shown). The apparently reduced affinity for syntaxin 1A that was exhibited by the ER processing mutants most likely reflects altered protein conformation or stable interactions with ER resident proteins (e.g., chaperones such as calnexin or Hsp70; refs. 31 and 32) that hinder in vitro binding to exogenous syntaxin 1A. The more physiologically significant finding was that the functional activity of ΔF508 CFTR is limited by its interaction with syntaxin 1A when this mutant is allowed to escape the ER and proceed to the cell surface. Moreover, ΔF508 currents in oocytes and cultured epithelial cells can be acutely and markedly potentiated by fusion proteins that block the syntaxin–CFTR interaction. Whether or not syntaxin 1A also limits the function of wild-type and mutant forms of CFTR in vivo remains to be determined. If so, maneuvers that relieve CFTR from syntaxin 1A inhibition may be therapeutically beneficial for those many cystic fibrosis patients with partial-loss-of-function mutants that traffic normally to the cell surface or, in the case of patients with ER processing mutants, when used in combination with methods to release these mutant proteins from the ER.
Acknowledgments
We thank S. Cheng for the LLC-PK1 cell line expressing ΔF508 CFTR, J. Pevsner for Munc-18 cDNA, T. Südhof for syntaxin 1A cDNA, and E. Sorscher for purified NBD1 and NBD2. We also thank M. Bennett and M. Caplan for helpful discussions, S. Reddy, Ge Li, and W. Xie for technical assistance, and M. N. Shelton and C. T. Starr for secretarial support. This work was supported by the National Institutes of Health (DK51868 and DK50830 to K.L.K.; DA10509 to M.W.Q.; DK47339 to J.F.C.), the W. M. Keck Foundation (931360 to M.W.Q.), the Cystic Fibrosis Foundation (COLLAW96PO to J.F.C.), and an American Heart Association Postdoctoral Fellowship to A.N.
ABBREVIATIONS
- CFTR
cystic fibrosis transmembrane conductance regulator
- NBD
nucleotide-binding domain
- GST
glutathione S-transferase
- ER
endoplasmic reticulum
- cRNA
complementary RNA
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Bear C E, Li C, Kartner N, Bridges R J, Jensen T J, Ramjeesingh M, Riordan J R. Cell. 1992;68:809–818. doi: 10.1016/0092-8674(92)90155-6. [DOI] [PubMed] [Google Scholar]
- 2.Welsh M J, Smith A E. Cell. 1993;73:1251–1254. doi: 10.1016/0092-8674(93)90353-r. [DOI] [PubMed] [Google Scholar]
- 3.Riordan J R, Rommens J M, Kerem B, Alon N, Rozmahel R, Grzelczak Z, Zielenski J, Lok S, Plavsic N, Chou J, Drumm M L, Iannuzzi M C, Collins F S, Tsui L C. Science. 1989;245:1066–1072. doi: 10.1126/science.2475911. [DOI] [PubMed] [Google Scholar]
- 4.Cheng S H, Rich D P, Marshall J, Gregory R D, Welsh M K, Smith A E. Cell. 1991;66:1027–1036. doi: 10.1016/0092-8674(91)90446-6. [DOI] [PubMed] [Google Scholar]
- 5.Carson M R, Travis S M, Welsh M J. J Biol Chem. 1995;270:1711–1717. doi: 10.1074/jbc.270.4.1711. [DOI] [PubMed] [Google Scholar]
- 6.Naren A P, Nelson D J, Xie W, Jovov B, Pevsner J, Bennett M K, Benos D J, Quick M W, Kirk K L. Nature (London) 1997;390:302–305. doi: 10.1038/36882. [DOI] [PubMed] [Google Scholar]
- 7.Bennett M K, Garcia-Arrarás J E, Elferink L A, Peterson K, Fleming A M, Hazuka C D, Scheller R H. Cell. 1993;74:1–20. doi: 10.1016/0092-8674(93)90466-4. [DOI] [PubMed] [Google Scholar]
- 8.Rowe J, Dascher C, Bannykh S, Plutner H, Balch W E. Science. 1998;279:696–700. doi: 10.1126/science.279.5351.696. [DOI] [PubMed] [Google Scholar]
- 9.Hata Y, Slaughter C A, Südhof T C. Nature (London) 1993;366:347–351. doi: 10.1038/366347a0. [DOI] [PubMed] [Google Scholar]
- 10.Pevsner J, Hsu S-C, Scheller R H. Proc Natl Acad Sci USA. 1994;91:1445–1449. doi: 10.1073/pnas.91.4.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Südhof T C. Nature (London) 1995;375:645–653. doi: 10.1038/375645a0. [DOI] [PubMed] [Google Scholar]
- 12.Blasi J, Chapman E R, Yamasaki S, Binz T, Niemann H, Jahn R. EMBO J. 1993;12:4821–4828. doi: 10.1002/j.1460-2075.1993.tb06171.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bennett M K, Calakos N, Scheller R H. Science. 1992;257:255–259. doi: 10.1126/science.1321498. [DOI] [PubMed] [Google Scholar]
- 14.Yoshida A, Oho C, Omori A, Kywahara R, Ito T, Takahashi M. J Biol Chem. 1992;267:24925–24928. [PubMed] [Google Scholar]
- 15.Sheng Z-H, Rettig J, Takahashi M, Catterall W A. Neuron. 1994;13:1303–1313. doi: 10.1016/0896-6273(94)90417-0. [DOI] [PubMed] [Google Scholar]
- 16.Bezprovanny I, Scheller R H, Tsien R W. Nature (London) 1995;378:623–626. doi: 10.1038/378623a0. [DOI] [PubMed] [Google Scholar]
- 17.Wiser O, Bennett M K, Atlas D. EMBO J. 1996;15:4100–4110. [PMC free article] [PubMed] [Google Scholar]
- 18.Stanley E F, Mirotznik R R. Nature (London) 1997;385:340–343. doi: 10.1038/385340a0. [DOI] [PubMed] [Google Scholar]
- 19.Hartman J, Huang Z, Rado T A, Peng S, Jilling T, Muccio D D, Sorscher E J. J Biol Chem. 1992;267:6455–6458. [PubMed] [Google Scholar]
- 20.Quick M W, Lester H A. In: Methods in Neurosciences: Ion Channels of Excitable Cells. Conn P M, editor. San Diego: Academic; 1994. pp. 261–279. [Google Scholar]
- 21.Marshall J, Fang S, Ostedgaard L S, O’Riordan C R, Ferrara D, Amara J F, Hoppe H, Scheule R K, Welsh M J, Smith A E, Cheng S H. J Biol Chem. 1994;269:2987–2995. [PubMed] [Google Scholar]
- 22.Chan H-C, Kaetzel M A, Nelson D J, Hazarika P, Dedman J R. J Biol Chem. 1992;267:8411–8416. [PubMed] [Google Scholar]
- 23.Drumm M L, Wilkinson D J, Smit L S, Worrell R T, Strong T V, Frizzell R A, Dawson D C, Collins F S. Science. 1991;254:1797–1799. doi: 10.1126/science.1722350. [DOI] [PubMed] [Google Scholar]
- 24.Rothman J E. Nature (London) 1994;372:55–63. doi: 10.1038/372055a0. [DOI] [PubMed] [Google Scholar]
- 25.Kee Y, Lin R C, Hsu S-C, Scheller R H. Neuron. 1995;14:991–998. doi: 10.1016/0896-6273(95)90337-2. [DOI] [PubMed] [Google Scholar]
- 26.Denning G M, Anderson M P, Amara J F, Marshall J, Smith E, Welsh M J. Nature (London) 1992;358:761–764. doi: 10.1038/358761a0. [DOI] [PubMed] [Google Scholar]
- 27.Mochida S, Sheng Z-H, Baker C, Kobayashi H, Catterall W A. Neuron. 1995;17:781–788. doi: 10.1016/s0896-6273(00)80209-3. [DOI] [PubMed] [Google Scholar]
- 28.Bradbury N A, Jilling T, Berta G, Sorscher E, Bridges R J, Kirk K L. Science. 1992;256:530–532. doi: 10.1126/science.1373908. [DOI] [PubMed] [Google Scholar]
- 29.Schwiebert E M, Gesek F, Ercolani L, Wjasow C, Gruenert D C, Karlson K, Stanton B A. Am J Physiol. 1994;267:C272–C281. doi: 10.1152/ajpcell.1994.267.1.C272. [DOI] [PubMed] [Google Scholar]
- 30.Mergey M, Lemnaouar M, Veissiere D, Perricaudet M, Gruenert D C, Picard J, Capeau J, Brahimi H M, Paul A. Am J Physiol. 1995;269:L855–L864. doi: 10.1152/ajplung.1995.269.6.L855. [DOI] [PubMed] [Google Scholar]
- 31.Yang Y, Janich S, Cohn J A, Wilson J M. Proc Natl Acad Sci USA. 1993;90:9480–9484. doi: 10.1073/pnas.90.20.9480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pind S, Riordan J R, Williams D B. J Biol Chem. 1994;269:12784–12788. [PubMed] [Google Scholar]