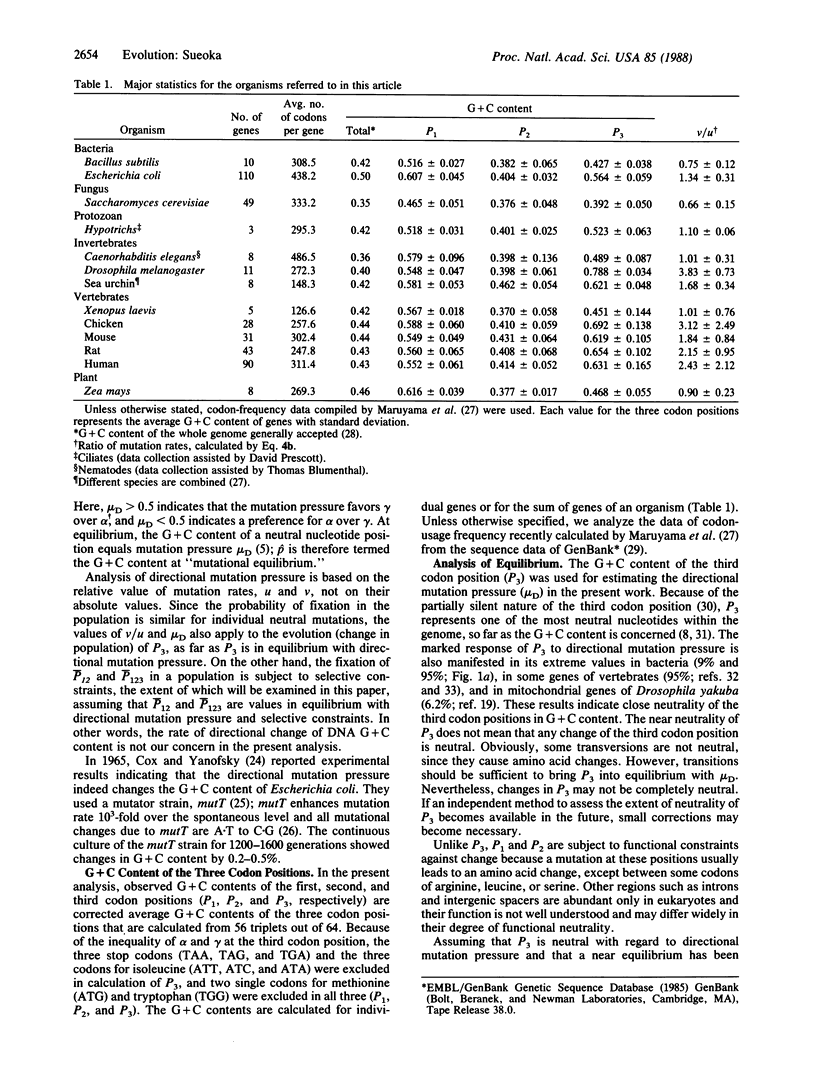
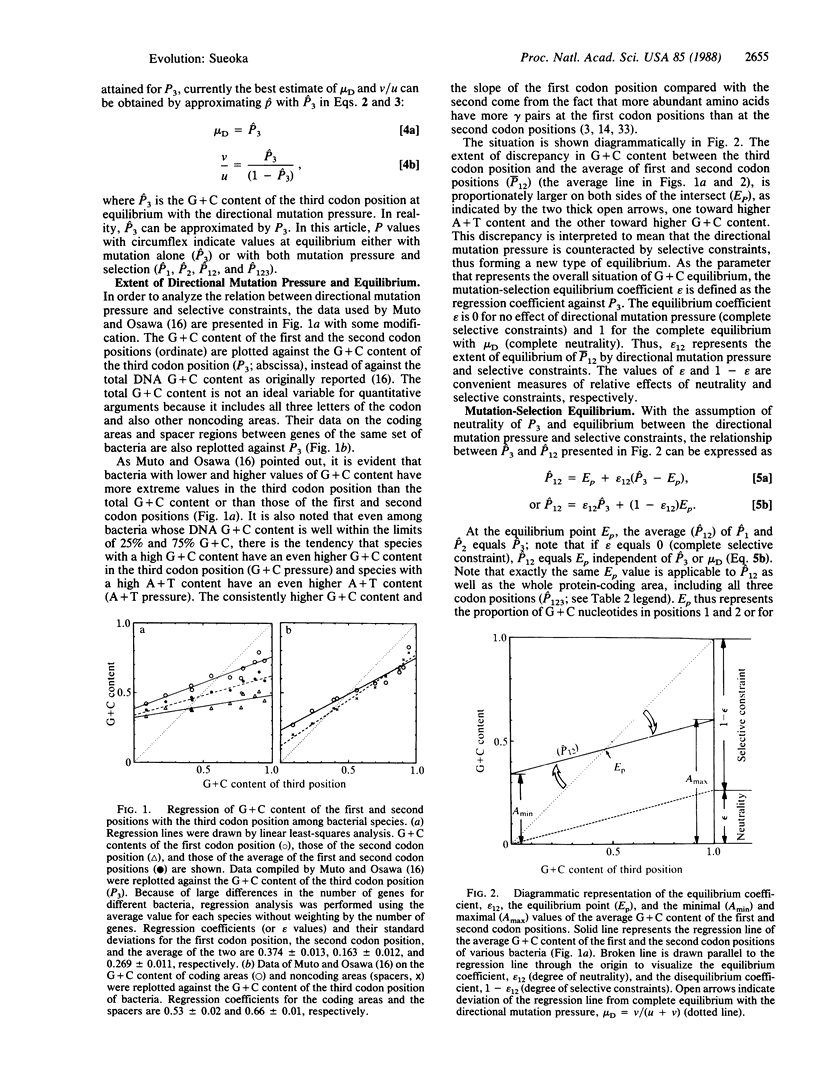
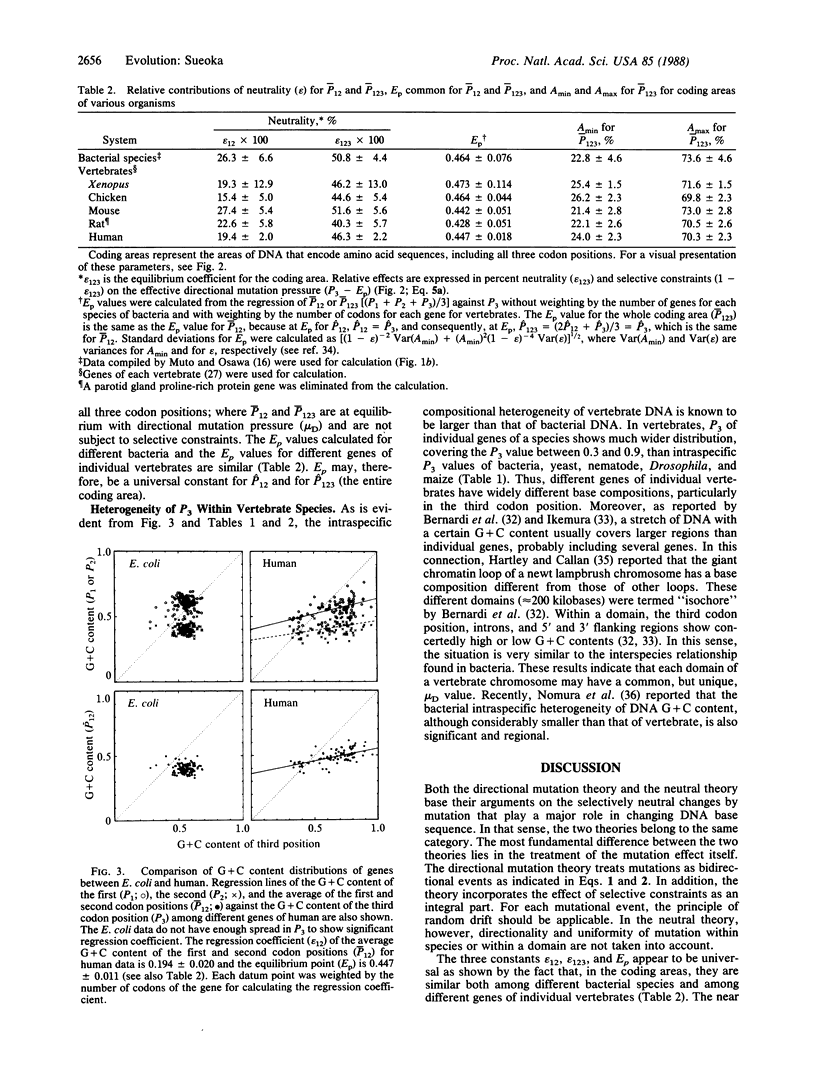
Abstract
A quantitative theory of directional mutation pressure proposed in 1962 explained the wide variation of DNA base composition observed among different bacteria and its small heterogeneity within individual bacterial species. The theory was based on the assumption that the effect of mutation on a genome is not random but has a directionality toward higher or lower guanine-plus-cytosine content of DNA, and this pressure generates directional changes more in neutral parts of the genome than in functionally significant parts. Now that DNA sequence data are available, the theory allows the estimation of the extent of neutrality of directional mutation pressure against selection. Newly defined parameters were used in the analysis, and two apparently universal constants were discovered. Analysis of DNA sequence has revealed that practically all organisms are subject to directional mutation pressure. The theory also offers plausible explanations for the large heterogeneity in guanine-plus-cytosine content among different parts of the vertebrate genome.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- BARBU E., LEE K. Y., WAHL R. Contenu en bases puriques et pyrimidiques des acides désoxyribonucléiques des bactéries. Ann Inst Pasteur (Paris) 1956 Aug;91(2):212–224. [PubMed] [Google Scholar]
- BELOZERSKY A. N., SPIRIN A. S. A correlation between the compositions of deoxyribonucleic and ribonucleic acids. Nature. 1958 Jul 12;182(4628):111–112. doi: 10.1038/182111a0. [DOI] [PubMed] [Google Scholar]
- Bak A. L., Atkins J. F., Meyer S. A. Evolution of DNA base compositions in microorganisms. Science. 1972 Mar 24;175(4028):1391–1393. doi: 10.1126/science.175.4028.1391. [DOI] [PubMed] [Google Scholar]
- Bernardi G., Olofsson B., Filipski J., Zerial M., Salinas J., Cuny G., Meunier-Rotival M., Rodier F. The mosaic genome of warm-blooded vertebrates. Science. 1985 May 24;228(4702):953–958. doi: 10.1126/science.4001930. [DOI] [PubMed] [Google Scholar]
- Bibb M. J., Bibb M. J., Ward J. M., Cohen S. N. Nucleotide sequences encoding and promoting expression of three antibiotic resistance genes indigenous to Streptomyces. Mol Gen Genet. 1985;199(1):26–36. doi: 10.1007/BF00327505. [DOI] [PubMed] [Google Scholar]
- Bilofsky H. S., Burks C., Fickett J. W., Goad W. B., Lewitter F. I., Rindone W. P., Swindell C. D., Tung C. S. The GenBank genetic sequence databank. Nucleic Acids Res. 1986 Jan 10;14(1):1–4. doi: 10.1093/nar/14.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clary D. O., Wolstenholme D. R. The mitochondrial DNA molecular of Drosophila yakuba: nucleotide sequence, gene organization, and genetic code. J Mol Evol. 1985;22(3):252–271. doi: 10.1007/BF02099755. [DOI] [PubMed] [Google Scholar]
- Cox E. C. On the organization of higher chromosomes. Nat New Biol. 1972 Oct 4;239(92):133–134. doi: 10.1038/newbio239133a0. [DOI] [PubMed] [Google Scholar]
- Cox E. C., Yanofsky C. Altered base ratios in the DNA of an Escherichia coli mutator strain. Proc Natl Acad Sci U S A. 1967 Nov;58(5):1895–1902. doi: 10.1073/pnas.58.5.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley S. E., Callan H. G. RNA transcription on the giant lateral loops of the lampbrush chromosomes of the American newt Notophthalmus viridescens. J Cell Sci. 1978 Dec;34:279–288. doi: 10.1242/jcs.34.1.279. [DOI] [PubMed] [Google Scholar]
- Helftenbein E. Nucleotide sequence of a macronuclear DNA molecule coding for alpha-tubulin from the ciliate Stylonychia lemnae. Special codon usage: TAA is not a translation termination codon. Nucleic Acids Res. 1985 Jan 25;13(2):415–433. doi: 10.1093/nar/13.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikemura T. Codon usage and tRNA content in unicellular and multicellular organisms. Mol Biol Evol. 1985 Jan;2(1):13–34. doi: 10.1093/oxfordjournals.molbev.a040335. [DOI] [PubMed] [Google Scholar]
- Jukes T. H., Bhushan V. Silent nucleotide substitutions and G + C content of some mitochondrial and bacterial genes. J Mol Evol. 1986;24(1-2):39–44. doi: 10.1007/BF02099949. [DOI] [PubMed] [Google Scholar]
- Jukes T. H., Osawa S., Muto A. Divergence and directional mutation pressures. Nature. 1987 Feb 19;325(6106):668–668. doi: 10.1038/325668b0. [DOI] [PubMed] [Google Scholar]
- Jukes T. H. The genetic code. II. Am Sci. 1965 Dec;53(4):477–487. [PubMed] [Google Scholar]
- Kimura M. Evolutionary rate at the molecular level. Nature. 1968 Feb 17;217(5129):624–626. doi: 10.1038/217624a0. [DOI] [PubMed] [Google Scholar]
- King J. L., Jukes T. H. Non-Darwinian evolution. Science. 1969 May 16;164(3881):788–798. doi: 10.1126/science.164.3881.788. [DOI] [PubMed] [Google Scholar]
- Kramer J. M., Cox G. N., Hirsh D. Comparisons of the complete sequences of two collagen genes from Caenorhabditis elegans. Cell. 1982 Sep;30(2):599–606. doi: 10.1016/0092-8674(82)90256-2. [DOI] [PubMed] [Google Scholar]
- Kuchino Y., Hanyu N., Tashiro F., Nishimura S. Tetrahymena thermophila glutamine tRNA and its gene that corresponds to UAA termination codon. Proc Natl Acad Sci U S A. 1985 Jul;82(14):4758–4762. doi: 10.1073/pnas.82.14.4758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama T., Gojobori T., Aota S., Ikemura T. Codon usage tabulated from the GenBank genetic sequence data. Nucleic Acids Res. 1986;14 (Suppl):r151–r197. doi: 10.1093/nar/14.suppl.r151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preer J. R., Jr, Preer L. B., Rudman B. M., Barnett A. J. Deviation from the universal code shown by the gene for surface protein 51A in Paramecium. Nature. 1985 Mar 14;314(6007):188–190. doi: 10.1038/314188a0. [DOI] [PubMed] [Google Scholar]
- Raschke W. C. Cloned murine T200 (Ly-5) cDNA reveals multiple transcripts within B- and T-lymphocyte lineages. Proc Natl Acad Sci U S A. 1987 Jan;84(1):161–165. doi: 10.1073/pnas.84.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolfe R., Meselson M. THE RELATIVE HOMOGENEITY OF MICROBIAL DNA. Proc Natl Acad Sci U S A. 1959 Jul;45(7):1039–1043. doi: 10.1073/pnas.45.7.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SUEOKA N. Compositional correlation between deoxyribonucleic acid and protein. Cold Spring Harb Symp Quant Biol. 1961;26:35–43. doi: 10.1101/sqb.1961.026.01.009. [DOI] [PubMed] [Google Scholar]
- SUEOKA N., MARMUR J., DOTY P., 2nd Dependence of the density of deoxyribonucleic acids on guanine-cytosine content. Nature. 1959 May 23;183(4673):1429–1431. doi: 10.1038/1831429a0. [DOI] [PubMed] [Google Scholar]
- SUEOKA N. On the genetic basis of variation and heterogeneity of DNA base composition. Proc Natl Acad Sci U S A. 1962 Apr 15;48:582–592. doi: 10.1073/pnas.48.4.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sueoka N. A STATISTICAL ANALYSIS OF DEOXYRIBONUCLEIC ACID DISTRIBUTION IN DENSITY GRADIENT CENTRIFUGATION. Proc Natl Acad Sci U S A. 1959 Oct;45(10):1480–1490. doi: 10.1073/pnas.45.10.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sueoka N. CORRELATION BETWEEN BASE COMPOSITION OF DEOXYRIBONUCLEIC ACID AND AMINO ACID COMPOSITION OF PROTEIN. Proc Natl Acad Sci U S A. 1961 Aug;47(8):1141–1149. doi: 10.1073/pnas.47.8.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treffers H. P., Spinelli V., Belser N. O. A Factor (or Mutator Gene) Influencing Mutation Rates in Escherichia Coli. Proc Natl Acad Sci U S A. 1954 Nov;40(11):1064–1071. doi: 10.1073/pnas.40.11.1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky C., Cox E. C., Horn V. The unusual mutagenic specificity of an E. Coli mutator gene. Proc Natl Acad Sci U S A. 1966 Feb;55(2):274–281. doi: 10.1073/pnas.55.2.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky C., vanCleemput M. Nucleotide sequence of trpE of Salmonella typhimurium and its homology with the corresponding sequence of Escherichia coli. J Mol Biol. 1982 Mar 5;155(3):235–246. doi: 10.1016/0022-2836(82)90003-1. [DOI] [PubMed] [Google Scholar]


