Abstract
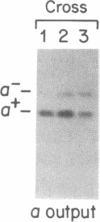
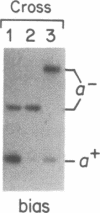
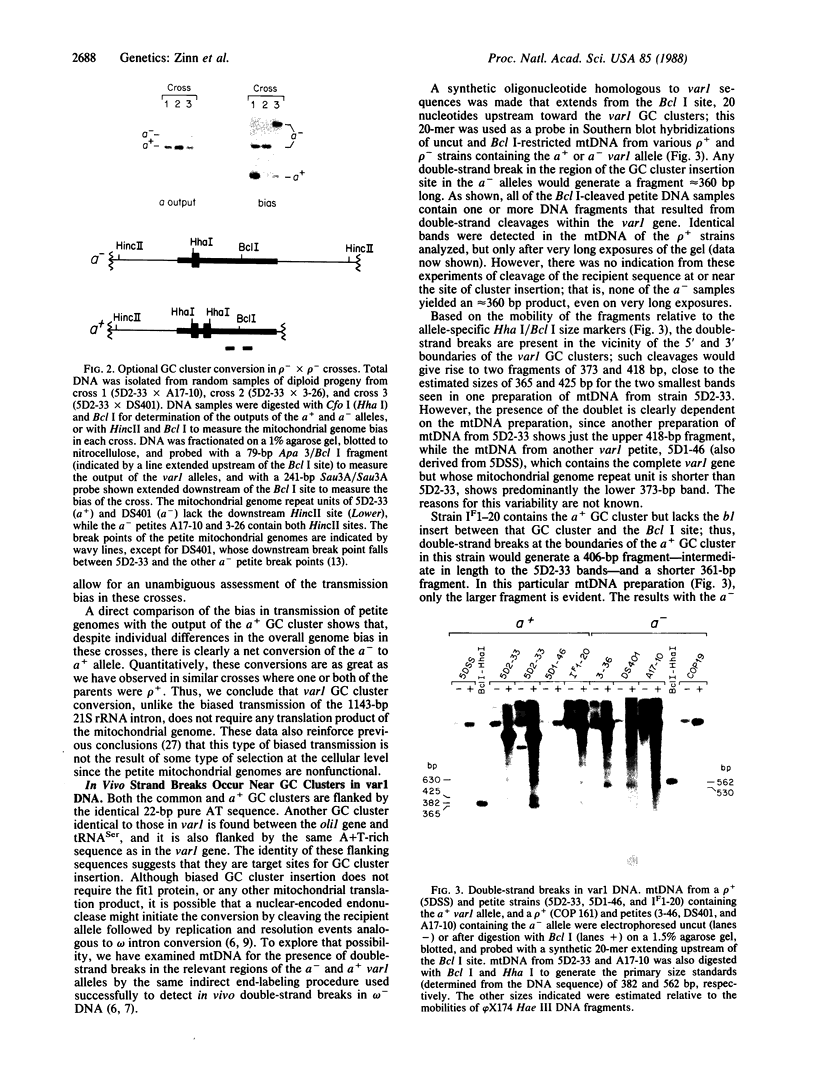
An optional 46-base-pair G + C-rich element (GC cluster) in the coding region of the yeast mitochondrial var1 gene inserts preferentially in crosses into recipient alleles that lack the sequence. Unlike a similar gene conversion event involving the insertion of an optional 1143-base-pair intron, the mitochondrial 21S rRNA gene, which requires the action of a protein encoded by a gene within that intron, conversion of the var1 GC cluster does not require any protein product of the mitochondrial genome. We have detected double-strand breaks in the var1 gene in mitochondrial DNA isolated from unmated haploid rho+ and rho- strains at or near the boundaries of the optional GC cluster, as well as at a conserved copy of that sequence 160 base pairs upstream. No double-strand breaks were detected in the recipient var1 DNA molecules in the vicinity of the optional GC cluster target sequence. This contrasts with 21S rRNA-encoding DNA (rDNA) intron conversion where the recipient, but not the donor DNA, is cleaved at the element insertion site. These results suggest that although the 21S rDNA intron and the var1 GC cluster are preferentially inserted into their respective short alleles, these conversions probably occur by different mechanisms.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ainley W. M., Hensley P., Butow R. A. Expression of GC clusters in the yeast mitochondrial var 1 gene. Translation and secondary structure implications. J Biol Chem. 1984 Jul 10;259(13):8422–8428. [PubMed] [Google Scholar]
- Ainley W. M., Macreadie I. G., Butow R. A. var1 Gene on the mitochondrial genome of Torulopsis glabrata. J Mol Biol. 1985 Aug 20;184(4):565–576. doi: 10.1016/0022-2836(85)90303-1. [DOI] [PubMed] [Google Scholar]
- Bingham C. G., Nagley P. A petite mitochondrial DNA segment arising in exceptionally high frequency in a mit- mutant of Saccharomyces cerevisiae. Biochim Biophys Acta. 1983 May 20;740(1):88–98. doi: 10.1016/0167-4781(83)90125-2. [DOI] [PubMed] [Google Scholar]
- Butow R. A., Perlman P. S., Grossman L. I. The unusual varl gene of yeast mitochondrial DNA. Science. 1985 Jun 28;228(4707):1496–1501. doi: 10.1126/science.2990030. [DOI] [PubMed] [Google Scholar]
- Colleaux L., d'Auriol L., Betermier M., Cottarel G., Jacquier A., Galibert F., Dujon B. Universal code equivalent of a yeast mitochondrial intron reading frame is expressed into E. coli as a specific double strand endonuclease. Cell. 1986 Feb 28;44(4):521–533. doi: 10.1016/0092-8674(86)90262-x. [DOI] [PubMed] [Google Scholar]
- Davis R. W., Thomas M., Cameron J., St John T. P., Scherer S., Padgett R. A. Rapid DNA isolations for enzymatic and hybridization analysis. Methods Enzymol. 1980;65(1):404–411. doi: 10.1016/s0076-6879(80)65051-4. [DOI] [PubMed] [Google Scholar]
- Dieckmann C. L., Gandy B. Preferential recombination between GC clusters in yeast mitochondrial DNA. EMBO J. 1987 Dec 20;6(13):4197–4203. doi: 10.1002/j.1460-2075.1987.tb02767.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunsmuir P., Brorein W. J., Jr, Simon M. A., Rubin G. M. Insertion of the Drosophila transposable element copia generates a 5 base pair duplication. Cell. 1980 Sep;21(2):575–579. doi: 10.1016/0092-8674(80)90495-x. [DOI] [PubMed] [Google Scholar]
- Farabaugh P. J., Fink G. R. Insertion of the eukaryotic transposable element Ty1 creates a 5-base pair duplication. Nature. 1980 Jul 24;286(5771):352–356. doi: 10.1038/286352a0. [DOI] [PubMed] [Google Scholar]
- Fonty G., Goursot R., Wilkie D., Bernardi G. The mitochondrial genome of wild-type yeast cells. VII. Recombination in crosses. J Mol Biol. 1978 Feb 25;119(2):213–235. doi: 10.1016/0022-2836(78)90435-7. [DOI] [PubMed] [Google Scholar]
- Grindley N. D., Sherratt D. J. Sequence analysis at IS1 insertion sites: models for transposition. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 2):1257–1261. doi: 10.1101/sqb.1979.043.01.142. [DOI] [PubMed] [Google Scholar]
- Hudspeth M. E., Ainley W. M., Shumard D. S., Butow R. A., Grossman L. I. Location and structure of the var1 gene on yeast mitochondrial DNA: nucleotide sequence of the 40.0 allele. Cell. 1982 Sep;30(2):617–626. doi: 10.1016/0092-8674(82)90258-6. [DOI] [PubMed] [Google Scholar]
- Hudspeth M. E., Vincent R. D., Perlman P. S., Shumard D. S., Treisman L. O., Grossman L. I. Expandable var1 gene of yeast mitochondrial DNA: in-frame insertions can explain the strain-specific protein size polymorphisms. Proc Natl Acad Sci U S A. 1984 May;81(10):3148–3152. doi: 10.1073/pnas.81.10.3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquier A., Dujon B. An intron-encoded protein is active in a gene conversion process that spreads an intron into a mitochondrial gene. Cell. 1985 Jun;41(2):383–394. doi: 10.1016/s0092-8674(85)80011-8. [DOI] [PubMed] [Google Scholar]
- Macreadie I. G., Scott R. M., Zinn A. R., Butow R. A. Transposition of an intron in yeast mitochondria requires a protein encoded by that intron. Cell. 1985 Jun;41(2):395–402. doi: 10.1016/s0092-8674(85)80012-x. [DOI] [PubMed] [Google Scholar]
- Orr-Weaver T. L., Szostak J. W., Rothstein R. J. Yeast transformation: a model system for the study of recombination. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6354–6358. doi: 10.1073/pnas.78.10.6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prunell A., Bernardi G. The mitochondrial genome of wild-type yeast cells. VI. Genome organization. J Mol Biol. 1977 Feb 15;110(1):53–74. doi: 10.1016/s0022-2836(77)80098-3. [DOI] [PubMed] [Google Scholar]
- Sander M., Hsieh T. S. Drosophila topoisomerase II double-strand DNA cleavage: analysis of DNA sequence homology at the cleavage site. Nucleic Acids Res. 1985 Feb 25;13(4):1057–1072. doi: 10.1093/nar/13.4.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker C., Goff S., Gilboa E., Paskind M., Mitra S. W., Baltimore D. Structure of a cloned circular Moloney murine leukemia virus DNA molecule containing an inverted segment: implications for retrovirus integration. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3932–3936. doi: 10.1073/pnas.77.7.3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strausberg R. L., Butow R. A. Gene conversion at the var1 locus on yeast mitochondrial DNA. Proc Natl Acad Sci U S A. 1981 Jan;78(1):494–498. doi: 10.1073/pnas.78.1.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strausberg R. L., Vincent R. D., Perlman P. S., Butow R. A. Asymmetric gene conversion at inserted segments on yeast mitochondrial DNA. Nature. 1978 Dec 7;276(5688):577–583. doi: 10.1038/276577a0. [DOI] [PubMed] [Google Scholar]
- Zinn A. R., Butow R. A. Kinetics and intermediates of yeast mitochondrial DNA recombination. Cold Spring Harb Symp Quant Biol. 1984;49:115–121. doi: 10.1101/sqb.1984.049.01.015. [DOI] [PubMed] [Google Scholar]
- Zinn A. R., Butow R. A. Nonreciprocal exchange between alleles of the yeast mitochondrial 21S rRNA gene: kinetics and the involvement of a double-strand break. Cell. 1985 Apr;40(4):887–895. doi: 10.1016/0092-8674(85)90348-4. [DOI] [PubMed] [Google Scholar]
- de Zamaroczy M., Bernardi G. The GC clusters of the mitochondrial genome of yeast and their evolutionary origin. Gene. 1986;41(1):1–22. doi: 10.1016/0378-1119(86)90262-3. [DOI] [PubMed] [Google Scholar]
- de Zamaroczy M., Faugeron-Fonty G., Bernardi G. Excision sequences in the mitochondrial genome of yeast. Gene. 1983 Mar;21(3):193–202. doi: 10.1016/0378-1119(83)90002-1. [DOI] [PubMed] [Google Scholar]