Abstract
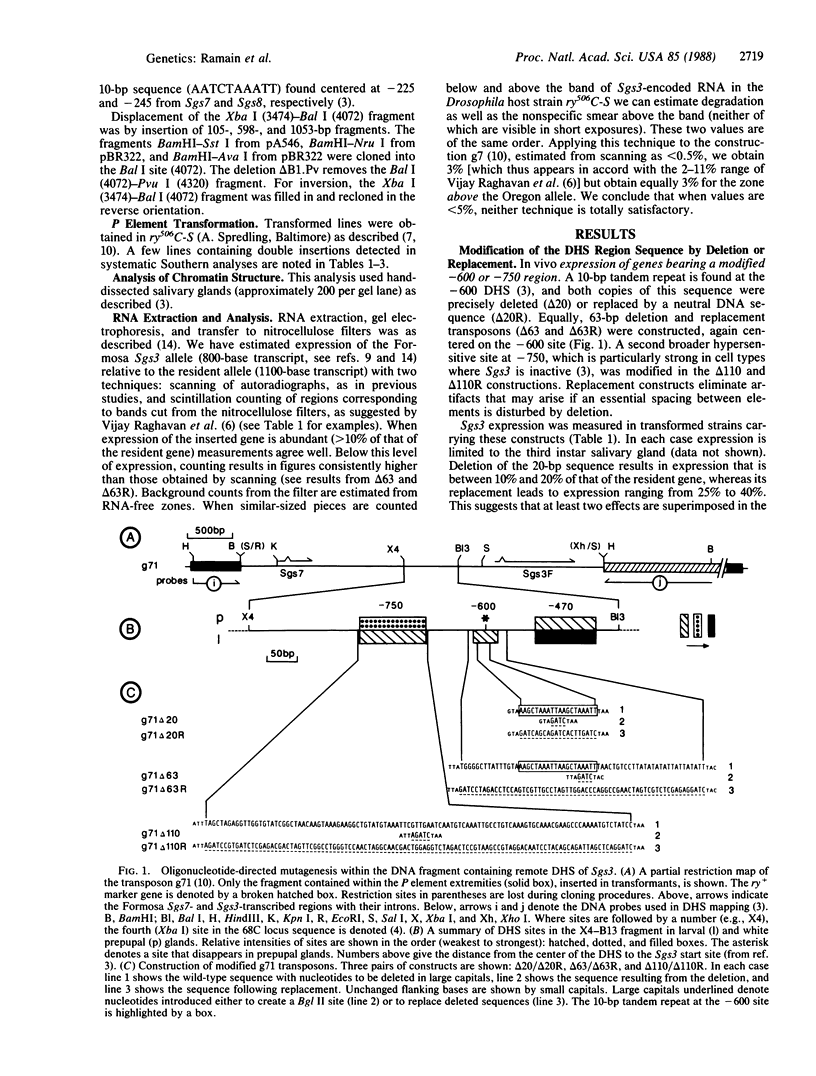
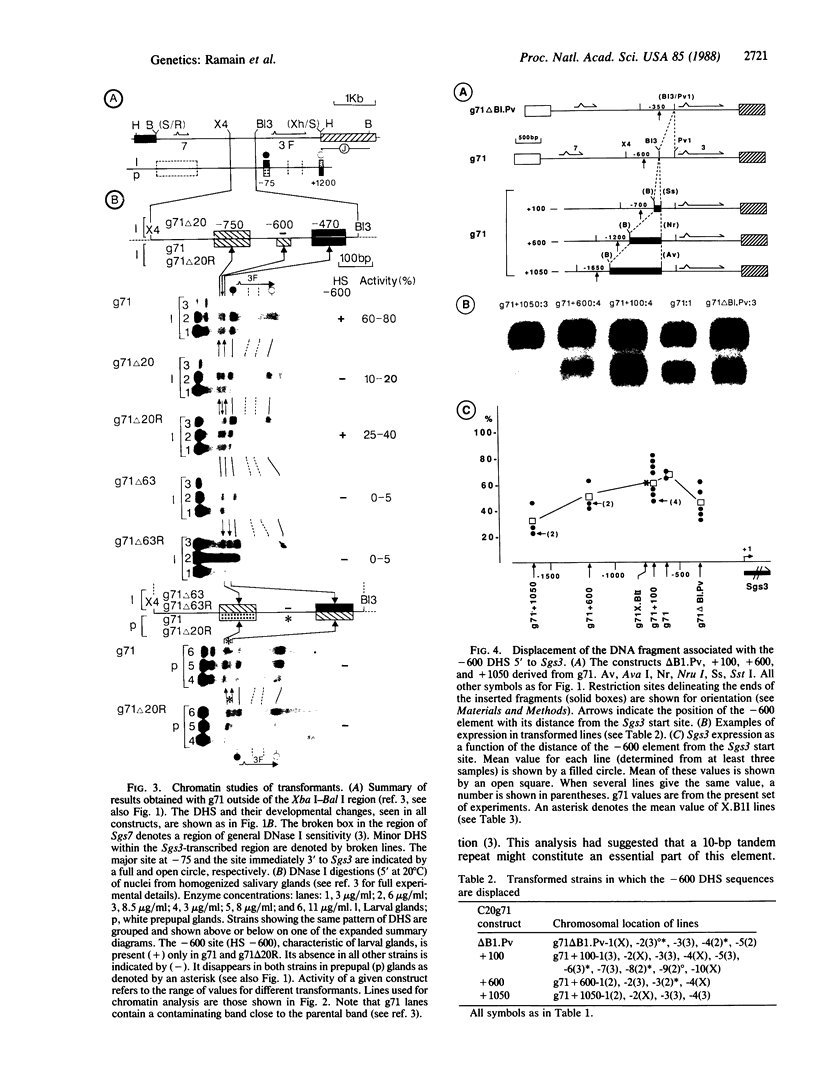
We have undertaken chromatin studies on transformed Drosophila strains carrying DNA sequences modified in the region of the DNase I (EC 3.1.4.5)-hypersensitive sites -750 and -600 base pairs upstream from the Sgs3 start site. Although both sites are developmentally specific, modifications in the -750 site have little or no effect on Sgs3-encoded transcript levels, whereas either deletion or replacement of sequences at the -600 site causes an important reduction in transcript levels. The element associated with the -600 site enhances Sgs3 transcription when displaced with respect to the start site. This combined approach has defined sequence elements necessary both for normal transcript levels as well as the chromatin structure characteristic of Sgs3 activity in vivo.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Boshart M., Weber F., Jahn G., Dorsch-Häsler K., Fleckenstein B., Schaffner W. A very strong enhancer is located upstream of an immediate early gene of human cytomegalovirus. Cell. 1985 Jun;41(2):521–530. doi: 10.1016/s0092-8674(85)80025-8. [DOI] [PubMed] [Google Scholar]
- Bourouis M., Richards G. Remote regulatory sequences of the Drosophila glue gene sgs3 as revealed by P-element transformation. Cell. 1985 Feb;40(2):349–357. doi: 10.1016/0092-8674(85)90149-7. [DOI] [PubMed] [Google Scholar]
- Costlow N. A., Simon J. A., Lis J. T. A hypersensitive site in hsp70 chromatin requires adjacent not internal DNA sequence. Nature. 1985 Jan 10;313(5998):147–149. doi: 10.1038/313147a0. [DOI] [PubMed] [Google Scholar]
- Crowley T. E., Meyerowitz E. M. Steroid regulation of RNAs transcribed from the Drosophila 68c polytene chromosome puff. Dev Biol. 1984 Mar;102(1):110–121. doi: 10.1016/0012-1606(84)90179-9. [DOI] [PubMed] [Google Scholar]
- Dente L., Cesareni G., Cortese R. pEMBL: a new family of single stranded plasmids. Nucleic Acids Res. 1983 Mar 25;11(6):1645–1655. doi: 10.1093/nar/11.6.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkner F. G., Zachau H. G. Correct transcription of an immunoglobulin kappa gene requires an upstream fragment containing conserved sequence elements. Nature. 1984 Jul 5;310(5972):71–74. doi: 10.1038/310071a0. [DOI] [PubMed] [Google Scholar]
- Garfinkel M. D., Pruitt R. E., Meyerowitz E. M. DNA sequences, gene regulation and modular protein evolution in the Drosophila 68C glue gene cluster. J Mol Biol. 1983 Aug 25;168(4):765–789. doi: 10.1016/s0022-2836(83)80074-6. [DOI] [PubMed] [Google Scholar]
- Giangrande A., Mettling C., Richards G. Sps-3 transcript levels are determined by multiple remote sequence elements. EMBO J. 1987 Oct;6(10):3079–3084. doi: 10.1002/j.1460-2075.1987.tb02615.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbomel P., Bourachot B., Yaniv M. Two distinct enhancers with different cell specificities coexist in the regulatory region of polyoma. Cell. 1984 Dec;39(3 Pt 2):653–662. doi: 10.1016/0092-8674(84)90472-0. [DOI] [PubMed] [Google Scholar]
- Jongstra J., Reudelhuber T. L., Oudet P., Benoist C., Chae C. B., Jeltsch J. M., Mathis D. J., Chambon P. Induction of altered chromatin structures by simian virus 40 enhancer and promoter elements. Nature. 1984 Feb 23;307(5953):708–714. doi: 10.1038/307708a0. [DOI] [PubMed] [Google Scholar]
- Jowett T. The regulatory domain of a larval serum protein gene in Drosophila melanogaster. EMBO J. 1985 Dec 30;4(13B):3789–3795. doi: 10.1002/j.1460-2075.1985.tb04149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis W., Shermoen A. W., Beckendorf S. K. A transposable element inserted just 5' to a Drosophila glue protein gene alters gene expression and chromatin structure. Cell. 1983 Aug;34(1):75–84. doi: 10.1016/0092-8674(83)90137-x. [DOI] [PubMed] [Google Scholar]
- Norris K., Norris F., Christiansen L., Fiil N. Efficient site-directed mutagenesis by simultaneous use of two primers. Nucleic Acids Res. 1983 Aug 11;11(15):5103–5112. doi: 10.1093/nar/11.15.5103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkert C. A., Ornitz D. M., Brinster R. L., Palmiter R. D. An albumin enhancer located 10 kb upstream functions along with its promoter to direct efficient, liver-specific expression in transgenic mice. Genes Dev. 1987 May;1(3):268–276. doi: 10.1101/gad.1.3.268. [DOI] [PubMed] [Google Scholar]
- Raghavan K. V., Crosby M. A., Mathers P. H., Meyerowitz E. M. Sequences sufficient for correct regulation of Sgs-3 lie close to or within the gene. EMBO J. 1986 Dec 1;5(12):3321–3326. doi: 10.1002/j.1460-2075.1986.tb04646.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramain P., Bourouis M., Dretzen G., Richards G., Sobkowiak A., Bellard M. Changes in the chromatin structure of Drosophila glue genes accompany developmental cessation of transcription in wild type and transformed strains. Cell. 1986 May 23;45(4):545–553. doi: 10.1016/0092-8674(86)90286-2. [DOI] [PubMed] [Google Scholar]
- Richards G., Cassab A., Bourouis M., Jarry B., Dissous C. The normal developmental regulation of a cloned sgs3 'glue' gene chromosomally integrated in Drosophila melanogaster by P element transformation. EMBO J. 1983;2(12):2137–2142. doi: 10.1002/j.1460-2075.1983.tb01714.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shermoen A. W., Jongens J., Barnett S. W., Flynn K., Beckendorf S. K. Developmental regulation by an enhancer from the Sgs-4 gene of Drosophila. EMBO J. 1987 Jan;6(1):207–214. doi: 10.1002/j.1460-2075.1987.tb04740.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theisen M., Stief A., Sippel A. E. The lysozyme enhancer: cell-specific activation of the chicken lysozyme gene by a far-upstream DNA element. EMBO J. 1986 Apr;5(4):719–724. doi: 10.1002/j.1460-2075.1986.tb04273.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widen S. G., Papaconstantinou J. Liver-specific expression of the mouse alpha-fetoprotein gene is mediated by cis-acting DNA elements. Proc Natl Acad Sci U S A. 1986 Nov;83(21):8196–8200. doi: 10.1073/pnas.83.21.8196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. The 5' ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature. 1980 Aug 28;286(5776):854–860. doi: 10.1038/286854a0. [DOI] [PubMed] [Google Scholar]
- Zenke M., Grundström T., Matthes H., Wintzerith M., Schatz C., Wildeman A., Chambon P. Multiple sequence motifs are involved in SV40 enhancer function. EMBO J. 1986 Feb;5(2):387–397. doi: 10.1002/j.1460-2075.1986.tb04224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]






