Abstract
Chromosomal rearrangements involving the EVI1 proto-oncogene are a recurrent finding in myeloid leukemias and are indicative of a poor prognosis. Rearrangements of the EVI1 locus are often associated with monosomy 7 or cytogenetic detectable deletions of part of 7q. As EVI1 overexpression alone is not sufficient to induce leukemia, loss of a 7q tumour suppressor gene might be a required cooperating event. To test this hypothesis, we performed high-resolution array comparative genomic hybridization analysis of twelve EVI1 overexpressing patients and three EVI1 deregulated cell lines to search for 7q submicroscopic deletions. This analysis lead to the delineation of two critical regions, one of 0.39 Mb on 7q35 containing the CNTNAP2 gene and one of 1.33 Mb on chromosome bands 7q35–q36 comprising nine genes in EVI1 deregulated cell lines. These findings open the way to further studies aimed at identifying the culprit EVI1 implicated tumour suppressor genes on 7q.
Introduction
Chromosomal rearrangements involving chromosome band 3q26, such as translocations with various partner chromosomes or inversions of chromosome 3 are a recurrent finding in myeloid leukemias [1]. These aberrations contribute to the ectopic expression of the EVI1 proto-oncogene. EVI1 transcriptional activation has been reported in up to 10% of myeloid leukemia patients, even in the absence of 3q26 rearrangements, and is an independent indicator of adverse prognosis [2].
Retroviral integration experiments have shown that EVI1 overexpression alone is not sufficient to cause leukemia, indicating that cooperative effects are necessary for malignant transformation [3], [4]. Interestingly, over fifty percent of the 3q26 rearranged leukemias also display chromosomal abnormalities involving chromosome 7, such as monosomy 7 or deletion of part of 7q [5]. This association alludes to the existence of a 7q tumour suppressor gene, which when deleted acts in concert with EVI1 overexpression to induce malignant transformation. We performed high-resolution array comparative genomic hybridization (CGH) analysis to search for 7q submicroscopic deletions in EVI1 deregulated leukemia patients in order to identify candidate 7q tumour suppressor genes.
Materials and Methods
Patients and Cell Lines
Diagnostic bone marrow samples of twelve myeloid leukemia samples and remission bone marrow samples of two patients (case 7 and case 8) were included in the study. Fluorescence in situ hybridization (FISH) for detection of EVI1 rearrangement and reverse transcription quantitative PCR (RT-qPCR) for detection of EVI1 ectopic expression was performed as previously described [6]. The study was approved by the ethics committee of the Ghent University Hospital (2003/273).
Three EVI1 rearranged cell lines, Kasumi-3, MUTZ-3 and UCSD-AML1 were also included in the study [7], [8]. For the cell lines culture conditions were as follows, for Kasumi-3 RPMI-1640 medium (Invitrogen, Belgium) was supplemented with 15% foetal calf serum, 1% penicillin/streptomycin, 1% kanamycin, 1% glutamine, 2% HEPES (1 M), 1% sodiumpyruvate (100 nM) and 0.1% beta-mercapto ethanol (50 nM). For MUTZ-3, the used medium contained an extra 10% of the supernatant of the 5637 urinary bladder carcinoma cell line [9] and 10 ng/ml of GM-CSF (Promocell, UK). For the UCSD-AML1 cell line, the used medium contained an additional 10 ng/ml of GM-CSF (Promocell, UK). Patient and cell line characteristics and EVI1 FISH and RT-qPCR results are described in Table 1.
Table 1. Patient and cell line characteristics: diagnosis, karyotype and EVI1 FISH and RT-qPCR results.
| Name | Diagnosis* | Karyotype† | FISH EVI1 ‡ | RT-qPCR EVI1 ‡ |
| Kasumi-3 | AML | 46,XY,t(2;5)(p13;q33),t(3;7)(q26;q22),del(5)(q15),−8,del(9)(q32),add(12)(p11),add(16)(q13),+mar[20] | + | + |
| MUTZ-3 | AML | 46,XY,t(1;3)(q43;q13),t(2;7)(q36;q36),inv(3)(q21q26),inv(7)(p15q36),t(12;22)(p13;q12)[20] | + | + |
| UCSD-AML1 | AML | 45,XX,t(2;22)(p13;q12),t(3;3)(q21;q26),−7[20] | + | + |
| case 1 | MDS | 46,XX,t(3;8)(q26;q23) [13]/46,XX[1] | + | + |
| case 2 | AML | 47,XY,+8,inv(3)(q21q26),t(13;22)(q10;p10)[20] | + | + |
| case 3 | AML | 46,XY,t(3;21)(q26;q22),add(17)(p11.2)[12]/46,XY[5] | + | + |
| case 4 | MDS | 45,XY,add(1)(q13),t(3;21)(q26;q22),add(5)(q13),−13,−18,+22[10]/46,XY[6] | + | + |
| case 5 | AML | 46,XY,inv(3)(q21q26) [8]/46,XY[2] | + | + |
| case 6 | AML | 47,XX,t(9;11)(p22;q23),del(11)(p15),+21[8]/46,XX[12] | − | + |
| case 7 | AML | 46,XX,t(9;11)(p22;q23)[8]/46,XX[12] | − | + |
| case 8 | AML | 45,X,−X,t(9;11)(p22;q23)[12]/46,XX[6]/45,X, −X[3] | − | + |
| case 9 | AML | 45,XY,inv(3)(q21q26), −7[17] | + | + |
| case 10 | AML | 46,XY,t(3;3)(q21;q26) [15] | + | + |
| case 11 | AML | 45,XX,add(3)(q26), −7[15] | + | + |
| case 12 | MDS | 45,XY,t(3;21)(q26;q22), −7[20] | + | + |
Bold formatting indicates the 3q26 rearrangement.
AML = acute myeloid leukemia and MDS = myelodysplastic syndrome.
The chromosomal aberration implicating the EVI1 locus is indicated with bold formatting.
Positive for EVI1 FISH/qRT-PCR = +and negative for EVI1 FISH/qRT-PCR = −.
Array CGH
DNA of patient samples and cell lines was isolated using the QIAamp DNA mini kit (Qiagen, Belgium) or the Puregene Cell kit (Gentra Systems, Belgium) according to the manufacturer's descriptions. Array CGH analysis for submicroscopic 7q deletions was performed on a 44K custom array (Agilent technologies, Belgium) covering the entire chromosome 7 with a probe spacing of 1 oligonucleotide every 3 kb, according to the manufacturer's descriptions. In brief, DNA (400 ng) was labelled using the BioPrime Array CGH genomic labelling system (Invitrogen, Belgium) using Cy5 (control DNA; Promega, Belgium) and Cy3 (patient sample or cell line) labelled dCTPs (GE Healthcare, Belgium). Following labelling, hybridization, and washing of the slides, arrays were scanned using an Agilent DNA Microarray Scanner, quantified with Feature Extraction software 10.1 and data were further analyzed with arrayCGHbase [10] using a circular binary segmentation (CBS) algorithm taken into consideration log2 ratios of neighbouring probes [11]. Deletions were called heterozygous when CBS ratios were ≤−0.5 and a deletion was considered putatively homozygous when the CBS ratios were ≤−1.2. Common copy number variations (CNVs) present in the Database of Genomic Variants (www.projects.tcag.ca/variation) were excluded from analysis.
Fluorescence In Situ Hybridization
To confirm the overlapping deletions found in the cell lines Kasumi-3, UCSD-AML1 and MUTZ-3 at 7q35-q36, FISH analysis was performed as previously described [6]. FISH probes were selected from the UCSC genome browser database (http://genome.ucsc.edu) (Table 2).
Table 2. Name and position of FISH probes.
| Probe name | Position start* | Position end* |
| RP11−114L10 | 142486383 | 142604687 |
| RP11−106C6 | 148240833 | 148415979 |
| RP11−171N15 | 141216856 | 141779639 |
| RP11−728K20 | 149582837 | 149731647 |
| RP11−418B24 | 146512768 | 146667113 |
| RP11−1123J16 | 146673416 | 146811717 |
| RP4−803F10 | 146336163 | 146439692 |
| RP4−777G9 | 146949516 | 147038612 |
| RP11−79O14 | 147671295 | 147818484 |
| RP11−452J20 | 151938144 | 152144780 |
| RP11−312C1 | 152459779 | 152639208 |
UCSC genome browser database (http://genome.ucsc.edu).
CNTNAP2 Expression Analysis
Total RNA was extracted from total bone marrow samples of thirty nine EVI1 overexpressing patients (nine with monosomy 7), five normal bone marrow samples and two CD34+ cell fractions, using the miRNeasy kit (Qiagen, Belgium). cDNA was prepared from 2 µg of total RNA with the iScript cDNA Synthesis Kit (Bio-Rad, Belgium) according to the manufacturer's descriptions and RT-qPCR for the CNTNAP2 (forward: 5′-TAGGACATGGAACCCCAATG-3′ and reverse: 5′-ATCGATTTGGCTCATCTTGG-3′) transcript was performed as previously described [12], [13]. The reference genes RPL13A (forward: 5′-CCTGGAGGAGAAGAGGAAAGAGA-3′ and reverse: 5′-TTGAGGACCTCTGTGTATTTGTCAA-3′) and YWHAZ (forward: 5′-ACTTTTGGTACATTGTGGCTTCAA-3′ and reverse: 5′-CCGCCAGGACAAACCAGTAT-3′) were used for normalisation of the RT-qPCR data.
Results and Discussion
Based on karyotyping and FISH analysis, two critical regions, one on chromosome band 7q22 [14] and one on chromosome bands 7q32–q35 [15], have been identified as the targets for 7q deletions in myeloid leukemia. In this study, array CGH was applied to search for deletions on chromosome 7 at ultra-high resolution. Using this approach, we identified several submicroscopic 7p and 7q deletions in EVI1 overexpressing patients and cell lines (Table 3 and Figure 1) including two critically deleted regions on 7q35–q36 in EVI1 deregulated cell lines. These deletions were confirmed using FISH (Figure 2).
Table 3. Array CGH results of chromosome 7 deletions.
| Name | Chromosomal position* | Start - end (Mb) | Size (Mb) | Genes† |
| Kasumi-3 | 7p22 | 2369956−6743766 | 4,37 | SDK1 (40) |
| 7p22−p15 | 7010834−26480795 | 19,47 | 63 | |
| 7q31.2 | 116412304−116584949 | 0,17 | ST7 | |
| 7q34−q36 | 142182589−148693649 | 6,51 | KEL, CNTNAP2 ‡ , CUL1 , EZH2 (48) | |
| MUTZ-3 | 7q35−q36 | 147377325−152205764 | 4,83 | CNTNAP2, CUL1, EZH2 (61) |
| UCSD-AML1 | 7q11.22 | 69266162−69318942 | 0.053 | AUTS2 ‡ |
| 7q21.11 | 78183200−78189877 | 0.007 | MAGI2 ‡ | |
| 7q35 | 146482743−146873269 | 0.391 | CNTNAP2 ‡ | |
| case 2 | 7q34 | 142351441−142361904 | 0,01 | KEL |
| case 3 | 7p22 | 4036389−4042650 | 0,01 | SDK1 |
| 7p12 | 43592619−43597672 | 0,01 | STK17A | |
| case 9 | 7q32.2 | 129076051–129078116 | 0.02 | NRF1 ‡ |
Bold formatting indicates deletions on the 7q arm.
Based upon the UCSC genome browser (http://genome.ucsc.edu/cgi-bin/hgGateway, NCBI Build 36.1).
Between brackets is the total number of genes residing in the deleted area.
Homozygous deletion.
Figure 1. Overview of deletions in cell lines and patients.
Chromosome view of chromosome 7 deletions in the cell lines Kasumi-3, MUTZ-3 and UCSD-AML1 and in cases 2, 3 and 9. Deletions are indicated using grey bars.
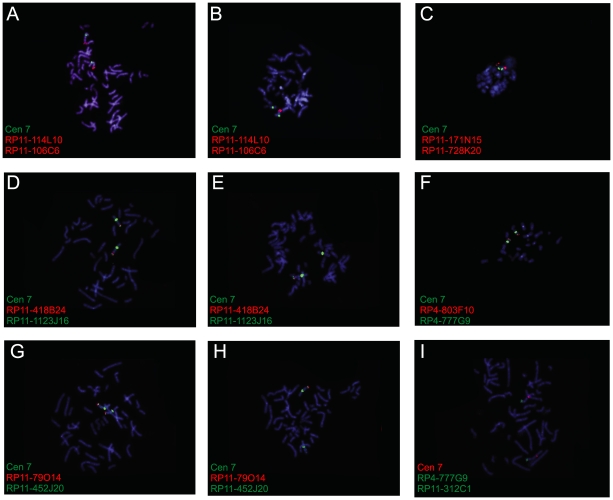
Figure 2. FISH analysis of the 7q35–q36 deletions in cell lines.
A), D) and G) FISH analysis on cytogenetically normal controls with probes located within the 7q35–q36 deleted regions. B), E) and H) FISH analysis with probes located within the 7q35–q36 deleted regions on the cell lines Kasumi-3 (B), UCSD-AML1 (E) and MUTZ-3 (H). C), F) and I) FISH analysis with probes just outside of the 7q35–q36 deleted regions on the cell lines Kasumi-3 (C), UCSD-AML1 (F) and MUTZ-3 (I).
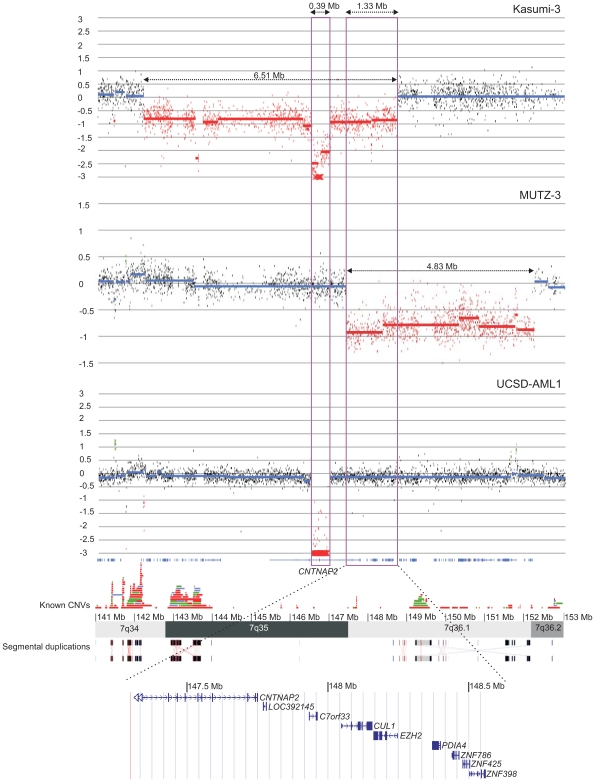
Interestingly, in the Kasumi-3 cell line a 680 kb homozygous deletion on 7q encompassing the CNTNAP2 gene was identified, located within a 6.51 Mb heterozygous deleted region at 7q34–q36. The observation of homozygous deletions has been of great importance in tumour genetics as the deleted region often contains putative tumour suppressor genes. In the MUTZ-3 cell line, a 4.83 Mb deletion was present within the same chromosomal region, delineating a shortest region of overlap (SRO) of 1.33 Mb (Figure 3) encompassing nine genes (Table 4). Interestingly, in the UCSD-AML1 cell line a homozygous deletion of 0.39 Mb was detected encompassing the CNTNAP2 gene, which delineated a second SRO of 0.39 Mb on 7q35 (Figure 3).
Figure 3. Array CGH profile of chromosome 7 (141 Mb–153 Mb) for Kasumi-3, MUTZ-3 and UCSD-AML1.
A) Array CGH profile of Kasumi-3, B) MUTZ-3 and C) UCSD-AML1 indicating the 0.39 Mb SRO on 7q35 and the 1.33 Mb SRO on chromosome bands 7q35–q36. Log2-ratios of the clones are depicted by vertical dots corresponding to the respective genomic position (NCBI build 36). Deletions are shown in red. The SROs are indicated with a purple box. The bottom of the figure shows the genomic position with an indication of known CNVs as present in the Database of Genomic Variants (http://projects.tcag.ca/variation/). Regions of segmental duplication are displayed using black and grey boxes indicating identical genomic regions. A screenshot of the UCSC Genome Browser (NCBI build 36, http://genome.ucsc.edu) shows an overview of the known RefSeq genes in the 1.33 Mb critical region.
Table 4. Genes located in the 1.33 Mb SRO on 7q35−q36.
| Gene name | Annotation |
| CNTNAP2 | contactin associated protein-like 2 isoform b |
| LOC392145 | hypothetical protein LOC392145 |
| C7orf33 | hypothetical protein LOC202865 |
| CUL1 | cullin 1 |
| EZH2 | enhancer of zeste homolog 2 |
| PDIA4 | protein disulfide isomerase associated 4 |
| ZNF786 | zinc finger protein 786 |
| ZNF425 | zinc finger protein 425 |
| ZNF398 | zinc finger 398 isoform a |
CNTNAP2 is located in a common fragile site which is inactivated in different types of cancers such as brain, ovarian and breast tumours [16], and methylation of the promoter of this gene has been described in pancreatic adenocarcinoma [17]. However, upon RT-qPCR analysis of the CNTNAP2 gene no expression could be detected in normal bone marrow samples, CD34+ cell fractions or EVI1 overexpressing bone marrow samples (data not shown), indicating that this gene is not a straightforward candidate 7q tumour suppressor gene cooperating with EVI1 overexpression.
Subsequent screening of twelve EVI1 overexpressing patient samples with (3 cases) or without (9 cases) monosomy 7, revealed no submicroscopic alterations within the above described SROs. However, focal deletions in other genomic regions on chromosome arms 7p and 7q were detected.
Of interest is that both the Kasumi-3 and the MUTZ-3 cell line display chromosomal rearrangements involving chromosome 7. It is well known that small deletions can occur at chromosomal breakpoints [18]. Therefore, the observed 4.83 Mb deletion at 7q36 in MUTZ-3 might have originated as a consequence of the t(2;7)(q36;q36) and/or the inv(7)(p15q36) present in this cell line.
Of the nine genes located within the 1.33 Mb SRO CUL1 and EZH2 are the most promising candidates due to known function in and association with cancer. The CUL1 gene encodes a protein that, when associated with other proteins such as Skp1, forms the Skp2 E3 ubiquitin ligase E3 complex involved in degradation of different proteins [19]. In AML, the Skp2 complex has been described to target the MEF transcription factor which induces G1/S transition. Impairment of the Skp2 complex leads to a decrease in MEF degradation thereby promoting cell proliferation [20]. Deletion and mutation of other E3 ubiquitin ligase complex members such as the F-box containing FBXW7 gene has already been described in T-cell acute lymphoblastic leukemia (T-ALL) where it is associated with poor prognosis [21]. Moreover, in AML loss-of-heterozygosity (LOH) of the EZH2 (polycomb group) gene (enhancer of zest) was detected in 5 out of 21 patients [22]. However, further analyses such as mutation screening and functional RNA interference screens [23] are needed to identify the genes contributing to EVI1 leukemogenesis.
In addition to the regions described above, other 7q deletions in the EVI1 overexpressing patients as well as in the UCSD-AML1 cell line were detected. In UCSD-AML1 two additional small 7q deletions were observed, a 53 kb (7q11.22) and 7 kb (7q21.11) deletion and containing the AUTS2 and MAGI2 genes respectively. In case 2, a small deletion of 10 kb on 7q34 was observed encompassing the KEL blood group gene. In Kasumi-3, this gene was located in a larger deleted region of 6.51 Mb on 7q34–q36. In case 9, a small homozygous deletion of 20 kb on 7q32.2 was detected. Located in this region is the NRF1 gene for which inactivating mutations have already been described in liver cancer [24].
Besides the above mentioned 7q deletions, several 7p deletions were also detected, some of which were common between different patients and/or cell lines. In case 3, a small deletion of 10 kb on 7p22 was observed encompassing the SDK1 sphingosine dependent protein kinase gene. In Kasumi-3, this gene was located in a larger deleted region of 4.37 Mb on 7p22. As involvement of the SDK1 gene in apoptosis has already been described [25], deletion of this gene could lead to a decreased rate of cell death. For patient 3 deletion of the STK17A gene at 7p12 was detected. Deletion of this pro-apoptotic gene had already been described in laryngeal squamous cell carcinoma [26]. It should be noted that it is possible that the above described small 7q and 7p deletions in patient samples are not leukemogenic as no constitutional material of these patients was available. In the remaining patient samples no chromosome 7 deletions could be detected.
In conclusion, using high-resolution array CGH, we were able to delineate two critical regions for 7q deletions in myeloid leukemias that were not detectable using standard karyotyping. Besides a 0.39 Mb SRO on 7q35 containing the CNTNAP2 gene, we identified a 1.33 Mb region on chromosome bands 7q35–q36 containing nine putative tumour suppressor genes in EVI1 deregulated cell lines. Further analysis of these candidates is warranted to investigate their role in EVI1 mediated malignant transformation. Identification of an EVI1 cooperating 7q tumour suppressor gene opens perspectives for novel treatment strategies selectively restoring the expression of the deleted genes.
Acknowledgments
We would like to thank Dr. S Meyer and D. White (University of Manchester) for their help in optimizing the culture conditions of the MUTZ-3 cell line and L. Vantomme and S. Baute for the excellent technical assistance.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: A. De Weer is the recipient of a BOF grant (Bijzonder Onderzoeksfonds UGent, grant no. 01D28905). B. Poppe is a senior clinical investigator of Fonds voor Wetenschappelijk Onderzoek (FWO) Vlaanderen, P. Van Vlierberghe is a post-doc of FWO-Vlaanderen and S. Vergult is an aspirant of FWO-Vlaanderen. This study was supported by the FWO-Vlaanderen, grant no. G.0106.05, by GOA-UGent, grant no. 12051203 and by IWT-Vlaanderen SBO grant no. 060848. This text presents research results of the Belgian Program of Interuniversity Poles of Attraction initiated by the Belgian State, Prime Minister's Office, Science Policy Programming. The scientific responsibility is assumed by the authors. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Nucifora G. The EVI1 gene in myeloid leukemia. Leukemia. 1997;11:2022–2031. doi: 10.1038/sj.leu.2400880. [DOI] [PubMed] [Google Scholar]
- 2.Lugthart S, Drunen EV, Norden YV, Hoven AV, Erpelinck CA, et al. High EVI1 levels predict adverse outcome in acute myeloid leukemia: prevalence of EVI1 overexpression and chromosome 3q26 abnormalities underestimated. Blood. 2008 doi: 10.1182/blood-2007-10-119230. [DOI] [PubMed] [Google Scholar]
- 3.Calmels B, Ferguson C, Laukkanen MO, Adler R, Faulhaber M, et al. Recurrent retroviral vector integration at the Mds1/Evi1 locus in nonhuman primate hematopoietic cells. Blood. 2005;106:2530–2533. doi: 10.1182/blood-2005-03-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Du Y, Jenkins NA, Copeland NG. Insertional mutagenesis identifies genes that promote the immortalization of primary bone marrow progenitor cells. Blood. 2005;106:3932–3939. doi: 10.1182/blood-2005-03-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lopingco MC, Perkins AS. Molecular analysis of Evi1, a zinc finger oncogene involved in myeloid leukemia. Curr Top Microbiol Immunol. 1996;211:211–222. doi: 10.1007/978-3-642-85232-9_21. [DOI] [PubMed] [Google Scholar]
- 6.De Weer A, Speleman F, Cauwelier B, Van Roy N, Yigit N, et al. EVI1 overexpression in t(3;17) positive myeloid malignancies results from juxtaposition of EVI1 to the MSI2 locus at 17q22. Haematologica. 2008;93:1903–1907. doi: 10.3324/haematol.13192. [DOI] [PubMed] [Google Scholar]
- 7.Asou H, Suzukawa K, Kita K, Nakase K, Ueda H, et al. Establishment of an undifferentiated leukemia cell line (Kasumi-3) with t(3;7)(q27;q22) and activation of the EVI1 gene. Japanese journal of cancer research. 1996;87:269–274. doi: 10.1111/j.1349-7006.1996.tb00216.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu ZB, Ma W, Zaborski M, MacLeod R, Quentmeier H, et al. Establishment and characterization of two novel cytokine-responsive acute myeloid and monocytic leukemia cell lines, MUTZ-2 and MUTZ-3. Leukemia. 1996;10:1025–1040. [PubMed] [Google Scholar]
- 9.Welte K, Platzer E, Lu L, Gabrilove JL, Levi E, et al. Purification and biochemical characterization of human pluripotent hematopoietic colony-stimulating factor. Proc Natl Acad Sci U S A. 1985;82:1526–1530. doi: 10.1073/pnas.82.5.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Menten B, Pattyn F, De Preter K, Robbrecht P, Michels E, et al. arrayCGHbase: an analysis platform for comparative genomic hybridization microarrays. BMC Bioinformatics. 2005;6:124. doi: 10.1186/1471-2105-6-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Venkatraman ES, Olshen AB. A faster circular binary segmentation algorithm for the analysis of array CGH data. Bioinformatics. 2007;23:657–663. doi: 10.1093/bioinformatics/btl646. [DOI] [PubMed] [Google Scholar]
- 12.Poppe B, Dastugue N, Vandesompele J, Cauwelier B, De Smet B, et al. EVI1 is consistently expressed as principal transcript in common and rare recurrent 3q26 rearrangements. Genes, chromosomes & cancer. 2006;45:349–356. doi: 10.1002/gcc.20295. [DOI] [PubMed] [Google Scholar]
- 13.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome biology. 2002;3:1–11. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Le Beau MM, Espinosa R, 3rd, Davis EM, Eisenbart JD, Larson RA, et al. Cytogenetic and molecular delineation of a region of chromosome 7 commonly deleted in malignant myeloid diseases. Blood. 1996;88:1930–1935. [PubMed] [Google Scholar]
- 15.Dohner K, Brown J, Hehmann U, Hetzel C, Stewart J, et al. Molecular cytogenetic characterization of a critical region in bands 7q35-q36 commonly deleted in malignant myeloid disorders. Blood. 1998;92:4031–4035. [PubMed] [Google Scholar]
- 16.McAvoy S, Ganapathiraju SC, Ducharme-Smith AL, Pritchett JR, Kosari F, et al. Non-random inactivation of large common fragile site genes in different cancers. Cytogenet Genome Res. 2007;118:260–269. doi: 10.1159/000108309. [DOI] [PubMed] [Google Scholar]
- 17.Omura N, Li CP, Li A, Hong SM, Walter K, et al. Genome-wide profiling of methylated promoters in pancreatic adenocarcinoma. Cancer Biol Ther. 2008;7:1146–1156. doi: 10.4161/cbt.7.7.6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kolomietz E, Al-Maghrabi J, Brennan S, Karaskova J, Minkin S, et al. Primary chromosomal rearrangements of leukemia are frequently accompanied by extensive submicroscopic deletions and may lead to altered prognosis. Blood. 2001;97:3581–3588. doi: 10.1182/blood.v97.11.3581. [DOI] [PubMed] [Google Scholar]
- 19.Yang X, Menon S, Lykke-Andersen K, Tsuge T, Di X, et al. The COP9 signalosome inhibits p27(kip1) degradation and impedes G1-S phase progression via deneddylation of SCF Cul1. Curr Biol. 2002;12:667–672. doi: 10.1016/s0960-9822(02)00791-1. [DOI] [PubMed] [Google Scholar]
- 20.Liu Y, Hedvat CV, Mao S, Zhu XH, Yao J, et al. The ETS protein MEF is regulated by phosphorylation-dependent proteolysis via the protein-ubiquitin ligase SCFSkp2. Mol Cell Biol. 2006;26:3114–3123. doi: 10.1128/MCB.26.8.3114-3123.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Asnafi V, Buzyn A, Le Noir S, Baleydier F, Simon A, et al. NOTCH1/FBXW7 mutation identifies a large subgroup with favourable outcome in adult T-cell acute lymphoblastic leukemia (T-ALL): a GRAALL study. Blood. 2008 doi: 10.1182/blood-2008-10-184069. [DOI] [PubMed] [Google Scholar]
- 22.Cardoso C, Mignon C, Hetet G, Grandchamps B, Fontes M, et al. The human EZH2 gene: genomic organisation and revised mapping in 7q35 within the critical region for malignant myeloid disorders. Eur J Hum Genet. 2000;8:174–180. doi: 10.1038/sj.ejhg.5200439. [DOI] [PubMed] [Google Scholar]
- 23.Ebert BL, Pretz J, Bosco J, Chang CY, Tamayo P, et al. Identification of RPS14 as a 5q- syndrome gene by RNA interference screen. Nature. 2008;451:335–339. doi: 10.1038/nature06494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu Z, Chen L, Leung L, Yen TS, Lee C, et al. Liver-specific inactivation of the Nrf1 gene in adult mouse leads to nonalcoholic steatohepatitis and hepatic neoplasia. Proc Natl Acad Sci U S A. 2005;102:4120–4125. doi: 10.1073/pnas.0500660102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suzuki E, Handa K, Toledo MS, Hakomori S. Sphingosine-dependent apoptosis: a unified concept based on multiple mechanisms operating in concert. Proc Natl Acad Sci U S A. 2004;101:14788–14793. doi: 10.1073/pnas.0406536101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Giefing M, Martin-Subero JI, Kiwerska K, Jarmuz M, Grenman R, et al. Characterization of homozygous deletions in laryngeal squamous cell carcinoma cell lines. Cancer Genet Cytogenet. 2008;184:38–43. doi: 10.1016/j.cancergencyto.2008.03.004. [DOI] [PubMed] [Google Scholar]