Abstract
IFN-γR1 deficiency is a genetic etiology of Mendelian susceptibility to mycobacterial diseases, and includes two forms of complete recessive deficiency, with or without cell surface expression, and two forms of partial deficiency, dominant or recessive. We report here a novel form of partial and recessive Interferon γ receptor 1 (IFN-γR1) deficiency, which is almost as severe as complete deficiency. The patient is homozygous for a mutation of the initiation codon (M1K). No detectable expression and function of IFN-γR1 were found in the patient's fibroblasts. However, IFN-γR1 expression was found to be impaired, but not abolished, on the EBV-transformed B cells, which could respond weakly to IFN-γ. The mechanism underlying this weak expression involves leaky translation initiation at both non-AUG codons and the third AUG codon at position 19. It results in the residual expression of IFN-γR1 protein of normal molecular weight and function. The residual IFN-γ signaling documented in this novel form of partial IFN-γR1 deficiency was not ubiquitous and was milder than that seen in other forms of partial IFN-γR1 deficiency, accounting for the more severe clinical phenotype of the patient, which was almost as severe as that of patients with complete deficiency.
INTRODUCTION
Mendelian susceptibility to mycobacterial diseases (MSMD, MIM 209950) is a rare congenital syndrome that confers predisposition to poorly virulent mycobacterial species, such as Bacillus Calmette–Guérin (BCG) and environmental mycobacteria, in otherwise healthy children (1–4). Until now, five MSMD-causing autosomal genes have been identified, including IFNGR1, which encodes the IFN-γ receptor ligand-binding chain (3–7); IFNGR2, which encodes the accessory chain of the IFN-γ receptor (8–12); IL12B, which encodes the p40 subunit shared by IL-12 and IL-23 (13,14); IL12RB1, which encodes the β1 chain shared by the receptors for IL-12 and IL-23 (15–17); signal transducer and activator of transcription 1 (STAT1) (18–21) and one X-linked gene, NF-κB essential modulator (NEMO), which mediates signaling in the NF-κB pathway (22). Interferon γ receptor 1 (IFN-γR1) deficiency was the first identified and is second most common etiology of MSMD. Until now, IFN-γR1 deficiency has been identified worldwide in 118 patients from 32 countries with 33 different mutations (Fig. 1A and unpublished data). Two major forms of IFN-γR1 deficiency have been described: complete and partial (23,24).
Figure 1.
Descriptions of the kindred, phenotype and genotype. (A) Schematic representation of the IFNGR1 gene with all previously described mutations and the M1K mutation described here (in purple italics). IFNGR1 exons are indicated by vertical bars and designated by roman numerals. Mutations in red are recessive loss-of-function mutations associated with complete defects and undetectable expression of the protein on the cell surface. Mutations in blue are recessive loss-of-function mutations associated with complete defects and surface expression of a non-functional molecule. Mutations in purple are recessive mutations associated with partial deficiency. Mutations in green are dominant mutations associated with partial deficiency. The red region indicates signal peptide. The initiation codon mutation (M1K) caused a novel partial IFN-γR1 deficiency with remarkably low levels of IFN-γR1 expression and a very week IFN-γ response. (B) Familial segregation of the M1K mutation. Both parents were heterozygous for M1K mutation and healthy, as well as the younger brother who is homozygous for the wild-type (WT) allele. The patient (filled circle) suffered from mycobacterial infections (see case report). (C) Electropherogram showing the ATG->AAG (M1K) mutation (underlined) in the patient P, as compared with a healthy control (WT/WT).
Two forms of complete IFN-γR1 deficiency have been defined on the basis cell surface expression of the receptor or not. Both forms show an abolished response to IFN-γ with regards to receptor binding, STAT1 homodimers known as gamma-activating factors (GAF) activation and HLA-DR induction. Mutations causing complete IFN-γR1 deficiencies without cell surface expression have often been found to be nonsense mutations, deletions or insertions in the coding regions for extracellular domain of IFN-γR1, which result in frameshift and a subsequent premature stop codon. The IFN-γR1 protein was not detectable on the cell surface, probably due to the degradation of the corresponding mRNA by the nonsense mediated surveillance system (3,4,25–33). The mutations identified in complete IFN-γR1 deficiency with cell surface expression were missense mutations or inframe deletions. IFNGR1 mRNA is translated to mature protein that can be transported to the cell surface but is unable to bind with IFN-γ (7,34). Patients with complete IFN-γR1 deficiency have severe clinical phenotypes, generally presenting with disseminated BCG or non-virulent mycobacterial infection early in life. High plasma concentrations of IFN-γ have frequently been observed in these patients (35). Bone marrow transplantation is currently the only curative treatment available for patients with complete IFN-γR1 deficiency (36–42). This solution remains difficult, however, due to a high rate of graft rejection resulting largely from the high concentrations of circulating IFN-γ (38).
Partial, as opposed to complete, IFN-γR1 deficiency is characterized by impaired but not abolished IFN-γ responses. Two forms of partial IFN-γR1 deficiency have been defined on the basis of differences in their characteristics and the recessive or dominant nature of defects. The recessive form is caused by a single mutation which changes from isoleucine to threonine at amino acid 87 (I87T). This single amino acid substitution decreases IFN-γR1 expression on the cell surface and results in an impaired response to IFN-γ (5,50). Dominant IFN-γR1 mutations principally affect exon 6 and include one hotspot mutation (818del4) (6,43–46). The dominant mutations give rise to a premature stop codon in the proximal intracellular domain, resulting in the production of a truncated protein lacking the intra-cellular receptor trafficking sites (6,47–49). The truncated proteins therefore accumulate on the cell surface and impede the normal signal transduction by exerting dominant-negative effects on the normal IFN-γR1 molecules. Patients with partial IFN-γR1 deficiencies generally have mild susceptibility to environmental mycobacterial disease or BCG-osis that were treatable with IFN-γ and antibiotics (36). We report here the characterization of a novel form of partial recessive IFN-γR1 deficiency, more severe than the partial forms described in previous studies.
RESULTS
Identification and segregation of the M1K mutation
We investigated a patient (P) presenting severe BCG and Mycobacterium avium infections in childhood, born to consanguineous parents in Finland (Fig. 1B). We assessed the response of whole blood from the patient to BCG and BCG + IFN-γ/IL-12, as previously described (51). We found that levels of IL-12p40 and IL-12p70 production in response to stimulation with BCG plus IFN-γ were no higher than those after BCG alone. In contrast, IFN-γ production in response to BCG plus IL-12 was normal (Supplementary Material, Fig. S1 and data not shown). Plasma IFN-γ concentration was very high (370 ng/ml, undetectable in controls, data not shown), as reported in patients with complete IFN-γR1 or IFN-γR2 deficiency (35). The IFNGR1 gene was considered the most likely candidate gene on the basis of the patient's clinical phenotype. Sequencing of the coding regions of IFNGR1 revealed a homozygous nucleotide substitution resulting in the replacement of the first methionine-encoding codon by a lysine-encoding codon (M1K) (Fig. 1C). Both parents were found to be heterozygous for this mutation and the patient's sibling carried only the wild-type allele. All of family members were healthy, with no clinical signs of mycobacterial diseases. The observed segregation pattern is therefore consistent with this mutation being recessive and pathogenic, causing MSMD when present in the homozygous state. We excluded the possibility of this mutation being an irrelevant polymorphism by sequencing 200 matched controls from the same ethnic group.
Complete IFN-γR1 deficiency in fibroblasts from the patient
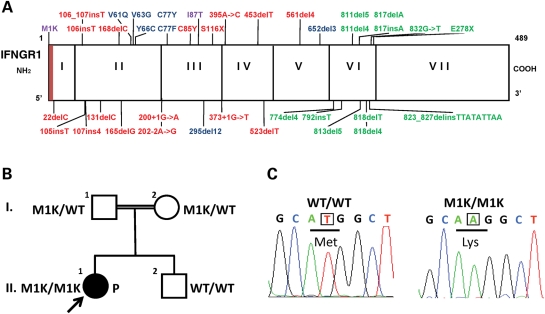
No IFN-γR1 expression was detected by FACS or immunoprecipitation and blotting (Supplementary Material, Figs S2 and S3) in the patient's fibroblasts (P fibroblast). We assessed the GAF activation by IFN-γ in fibroblasts. Unlike to I87T/I87T (5) and 818del4/WT (6) cells, P fibroblast, like the negative control (131delC/131delC) (4), displayed no GAF activation despite the normal response of all these cells to IFN-α (Supplementary Material, Fig. S4). Levels of HLA-DR expression on fibroblasts are low, but increase in response to IFN-γ. After 48 h of stimulation with 1000 or 100 000 IU/ml IFN-γ, P fibroblasts displayed no increase in HLA-DR expression (Fig. 2). Consistent with our previous study (5), I87T/I87T fibroblasts displayed only a very small increase in HLA-DR expression in response to IFN-γ stimulation. HLA-DR induction was normal in 818del4/WT fibroblasts with a high level of IFN-γ stimulation, but these cells nonetheless displayed an impairment of HLA-DR induction at an IFN-γ concentration of 10 IU/ml (data not shown). However, the observed difference in GAF activation and HLA-DR induction between the I87T/I87T and 818del4/WT fibroblasts was unexpected. This difference might plausibly be due to the accumulation of premature IFN-γR1 protein in the I87T/I87T fibroblasts (lower molecular weight, Supplementary Material, Fig. S3) in the endoplasmic reticulum or Golgi apparatus hindering the HLA-DR secretion. Further studies are required to test this hypothesis.
Figure 2.
HLA-DR induction in SV40 fibroblasts: WT/WT, I87T/I87T, 818del4/WT, P and 131delC/131delC SV40-transformed fibroblasts from were stimulated with the indicated dose of IFN-γ for 48 h. HLA-DR induction was determined by FACS analysis. Gray area: no stimulation (NS); thin line: 103 IU/ml IFN-γ; bold line: 105 IU/ml IFN-γ.
IFN-γR1 expression on the patient's EBV-B cells
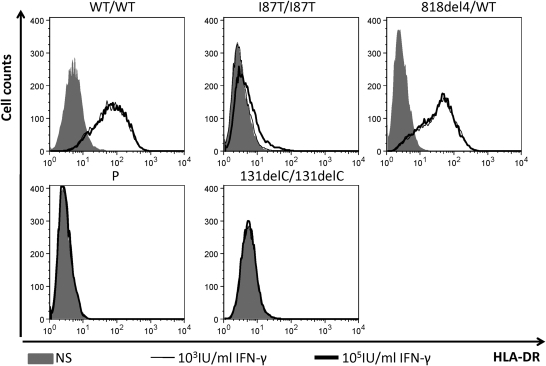
We carried out FACS analysis to investigate the impact of the M1K mutation on IFN-γR1 expression on EBV-B cells. EBV-B cells from a healthy control (WT/WT), a patient with the recessive I87T/I87T mutation (5), a patient with the 818del4/WT genotype (6), our patient (P) and a patient with complete recessive IFN-γR1 deficiency without cell surface expression (131delC/131delC) (4), were stained with two specific mouse antibodies against IFN-γR1, GIR-94 and GIR208, which recognize the extracellular part of human IFN-γR1 (Fig. 3A). We found no IFN-γR1 expression on 131delC/131delC cells (Fig. 3A), high levels of IFN-γR1 expression on the 818del4/WT cells and low levels of expression on the I87T/I87T cells, as expected. In four independent experiments, residual specific IFN-γR1 signals were systematically observed on the EBV-B cells of the patient with M1K/M1K, consistent with the expression of at least small amounts of detectable IFN-γR1 on the EBV-B cells of the patient. We also carried out scatchard assay to assess the IFN-γ binding ability on the patient's EBV-B cells. No obvious binding was observed in the M1K/M1K cells as well as I87T/I87T and negative control cell lines (Supplementary Material, Fig. S5).
Figure 3.
Faint IFN-γR1 expression in P cells. (A) FACS analysis of IFN-γR1 on EBV-B cells from a healthy control (WT/WT), a patient with partial recessive IFN-γR1 deficiency (I87T/I87T), a patient with partial dominant IFN-γR1 deficiency (818del4/WT), patient P and a patient with complete recessive IFN-γR1 deficiency with no cell surface expression (131delC/131delC). Gray area: Isotypic control antibody; bold dark line: specific extracellular IFN-γR1 antibody (GIR-94 or GIR208). (B) Immunoprecipitation of IFN-γR1 from WT/WT, I87T/I87T, 818del4/WT, P and 131delC/131delC EBV-B cells, using a specific antibody recognizing the intracellular part of IFN-γR1 (C-20), with or without subsequent PNGase F treatment. The same antibody was used to detect IFN-γR1. Left panel: without PNGase F treatment; right panel: with PNGase F treatment.
Assessment of IFN-γR1 expression by immunoprecipitation and western blotting
We investigated the characteristics of the IFN-γR1 protein with the M1K/M1K mutation, further, by carrying out immunoprecipitation with equal amounts of protein from the patient's EBV-B cells with both extracellular (GIR-94) and intracellular (C-20) antibodies against IFN-γR1 (Fig. 2B and data not shown). Western blotting resulted in the detection of a smear-like signal for IFN-γR1 in WT/WT cells, this signal having a molecular weight of around 90kD. IFNGR1 818del4/WT cells contained a smaller amount of a protein of the same size as the wild-type protein, whereas IFNGR1 I87T/I87T cells contained a smaller protein. No obvious difference was observed between P and 131delC/131delC control with complete recessive deficiency (Fig. 3B). Because IFN-γR1 belongs to the type I receptor, glycosylation in the endoplasmic reticulum and Golgi apparatus is an important event of post-translational modification stabilizing the protein and ensuring correct folding and ligand binding. Glycosylation is responsible for the diversity of molecular weights of the membrane proteins. Removal of the glycans from membrane protein leads to condensation of nascent proteins, increasing the sensitivity of detection (11). The precipitated products from EBV-B cells were treated with PNGase F to remove all the N-glycan. After digestion, a band of about 60 kDa in size was observed in WT/WT, I87T/I87T and 818del4/WT cells. A much fainter band of the same size, absent from the negative control was observed in P's EBV-B cells (Fig. 3B). However, this fainter band was not detected with P fibroblasts (Supplementary Material, Fig. S3). These data strongly support the hypothesis that the M1K/M1K mutation results in residual levels of detectable IFN-γR1 expression in EBV-B cells, but not in fibroblasts.
STAT1 activation and translocation by IFN-γ in the patient's EBV-B cells
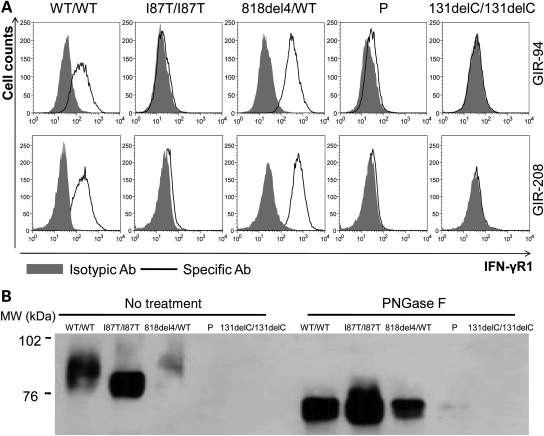
We then investigated whether the weak expression of IFN-γR1 in the patient's EBV-B cells was sufficient to mediate a cellular response to IFN-γ in terms of STAT1 phosphorylation, and the extent to which activated STAT1 homodimers (GAF) were able to bind a GAS probe corresponding to the promoter of a regulated gene. After IFN-γ stimulation, P's cells displayed detectable phosphorylated STAT1, but at much lower levels than observed in WT/WT cells and cells from other patients with partial IFN-γR1 deficiency (recessive and dominant) (Fig. 4A). The mean level of STAT1 phosphorylation in response to IFN-γ followed a gradient, as follows WT/WT > I87T/I87T > 818del4/WT > P > 131delC/131delC or STAT1−/− (18). All cell lines except STAT1 deficient cells displayed a similar level of STAT1 phosphorylation in response to IFN-α stimulation (Fig. 4A). To ascertain whether the extremely low STAT1 phosphorylation leads to functional binding to the GAS, EBV-B cells from WT/WT, 818del4/WT, I87T/I87T, P and 131delC/131delC were stimulated with various doses of IFN-γ or not and then assessed by EMSA. Unlike cells from patients with complete recessive IFN-γR1 deficiency, some GAF complexes were detectable in P cells (Fig. 4B). Consistent with the observed pattern of STAT1 phosphorylation, GAF levels in response to 105 IU/ml IFN-γ stimulation were lower in the patient with M1K/M1K mutation (3.54%) than in patients with other forms of partial IFN-γR1 deficiency I87T/I87T (69.91%), or 818del4/WT (6.42%) (Fig. 3B). Supershift assays were conducted with the nuclear extracts from P and WT/WT cells, competing the binding with non-radioactive probes, different antibodies against STAT1, STAT2, STAT3 and p48 (IRF9). Only antibody against STAT1 generated a supershift (Fig. 4C), which confirmed that the GAS probe binding proteins from P EBV-B cells was mediated by STAT1.
Figure 4.
Response of P cells to IFN-γ stimulation. (A) FACS analysis of EBV-B cells from a healthy control (WT/WT), a patient with partial recessive IFN-γR1 deficiency (I87T/I87T), a patient with partial dominant IFN-γR1 deficiency (818del4/WT), patient P and a patient with complete recessive IFN-γR1 deficiency with no cell surface expression (131delC/131delC) and a patient with complete recessive STAT1 deficiency (STAT1−/−) using specific anti-phosphorylated-Tyr-701 STAT1 in cells with and without stimulation by 105 IU/ml of IFN-γ or IFN-α for 30 min. Each experiment shown corresponds to a single representative experiment of three independent experiments. Gray area: no stimulation; bold line: 105 IU/ml IFN-γ; thin line: 105 IU/ml IFN-α. (B) EBV-B cells (10 million cells) were not stimulated or stimulated with 103 or 105 IU/ml IFN-γ or 105 IU/ml IFN-α for 20 min (upper and lower panels, respectively), EMSA shows similar levels of binding to GAS after IFN-α treatment; however, after IFN-γ stimulation, a gradient in the signal was observed as follow: WT/WT > I87T/I87T > 818del4/WT > P > 131delC/131delC. (C) WT/WT and P EBV-B cells were or were not stimulated 105 IU/ml IFN-γ. Various antibodies against STAT1, STAT2, STAT3 and p48 were added to the nuclear extract to determine the composition of the GAS-binding protein. Experiments in the presence of an excess of non-radioactive probe (Unlabeled probe) demonstrated the specificity of the WT and P complexes. Only the STAT1 antibody induced a supershift of the GAS binding protein in both the WT/WT and P cells.
The M1K mutation decreases translation efficiency
We investigated the potential mechanisms underlying the residual IFN-γR1 expression in EBV-B cells with a mutated initiation codon. We constructed a wild-type (ATG) and mutant (AAG) IFNGR1 expression vectors with V5 tags under the CMV promoter. We then transfected IFN-γR1−/− EBV-B cells, IFN-γR1−/− fibroblasts and HEK293 cells with the resulting plasmids. However, we were unable to detect the encoded protein by a western blotting with a specific anti-V5 antibody (Fig. 5A and data not show). In HEK 293T cells, proteins were detected by immunoprecipitation and western blotting, 72 h after transfection. The mutation of the initiation codon ATG to AAG greatly decreased IFN-γR1 expression, resulting in a weaker band, of the same size as wild-type IFN-γR1, on western blots (Fig. 5B). These findings suggest that the initiation codon mutation from ATG to AAG (M1K) is highly pathogenic and leads to a deficiency of translation of IFN-γR1 protein.
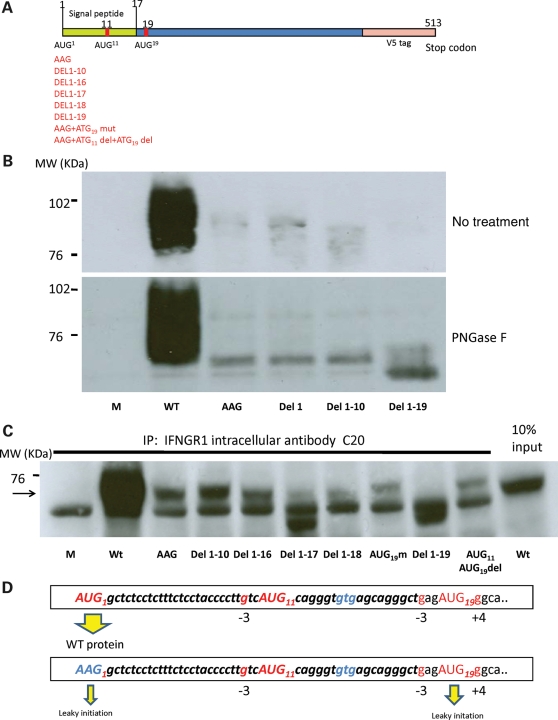
Figure 5.
Identification of potential initiation codons. (A) Schematic representation of the IFNGR1 gene cloned in a pcDNA3-V5 vector. The positions of various ATG codons close to the signal peptide and mutations are indicated. (B) HEK 293T cells were transfected with mock, wild-type (WT), first ATG->AAG mutant (AAG), first ATG deletion (Del1), first 10 codons deletion (Del1-10) and first 19 codons deletion (Del1-19) plasmids. We extracted proteins and carried out immunoprecipitation with intracellular antibody against IFN-γR1 (C20). Revelation was done with anti-V5 antibody. The mutation of the first ATG codon greatly decreased IFN-γR1 expression. No residual IFN-γR1 expression was observed if the first 19 codons were deleted. In the lower panel, the same extracts were subjected to PNGase F treatment: the construct lacking the first 19 codons did not generate a protein of similar molecular weight to the construct with AAG mutant or with the first 10 codons deletion. (C) The proteins precipitated by C20 were subjected to PNGase F treatment and then electrophoresis. Transfection with a mock plasmid (Lane 1), a wild-type IFNGR1 plasmid (Lane 2); an ATG mutated to AAG IFNGR1 plasmid (Lane 3); a plasmid lacking the first 10 codons (Lane 4) or the first 16 codons (Lane 5) resulted in similar levels of expression as transfection with a plasmid carrying the AAG mutation alone. Lanes 6 and 7: deletion of the first 17 codons or the first 18 codons decreased residual expression. Lane 9: deletion of the first 19 codons abolished residual expression of the normal-sized IFN-γR1. Lanes 8 and Lane 10: the mutation of ATG19 or deletion of ATG11 and ATG19 did not decrease the residual expression. Lane 11: as a control, 10% of the inputs for immunoprecipitation from wild-type IFNGR1 transfection were loaded on the gel. Arrow indicates the bands of residual IFN-γR1 expression. (D) Diagram of leaky scanning, highlighting the mechanisms of the residual expression in the M1K mutation. Upper panel, initiation takes place at the first AUG in the favorable context and generates sufficient amounts of full-length protein. Lower panel, leaky scanning occurs when the first AUG is mutated to AAG. In these conditions, initiation may take place on a non-AUG codon (AAG1) and at AUG19.
Leaky initiation leads to partial expression
We tried to identify the amino acid which is responsible for the initiation of the residual IFN-γR1 expression observed in cells from the patient with the M1K mutation, by constructing several vectors with deletion of the first 10 codons and subsequently deletion of first 19 codons. Deletion of the first 10 codons nonetheless resulted in a weak band corresponding to a protein of the same size as the wild-type IFN-γR1 (Fig. 5B). However, this residual expression was abolished by deletion of the first 19 codons. Treatment of the precipitated products with PNGaseF confirmed that deletion of the first 19 codons abolished the residual IFN-γR1 expression (Fig. 5B). We therefore conclude that codons between codon 11 and codon 19 were responsible for the residual levels of expression of a protein of similar size to the wild-type protein observed. Two ATG codons are present in this region: ATG11 and ATG19. Similar levels of residual expression were observed if the first 13 codons or the first 16 codons of the coding sequence were deleted (Supplementary Material, Fig. S6). Deletion of first 17 and 18 codons of the coding sequence resulted in lower levels of residual expression than deletion the first 16 codons, but expression remained detectable (Fig. 5C). In contrast, no expression was detected if the first 19 codons were deleted (Fig. 5C). These data suggests that codons 17, 18 and 19 of the coding sequence were essential for the residual expression of IFN-γR1 expression (Fig. 5D). The decrease in translation associated with the deletion of codons 17 and 18 may be accounted for by these codons constituting a favorable context for translation initiation from the AUG19 codon of the transcript.
We then investigate whether the AUG19 was the only translation initiation site responsible for the residual levels of M1K IFN-γR1 expression observed. The mutation of codon 19 alone on the M1K IFNGR1 did not prevent residual expression, even if both ATG11 and ATG19 were deleted (Fig. 5C). As the 5′-UTR of the IFNGR1 mRNA contained no other AUG codons, our findings suggest that non-AUG mediated initiation may also lead to residual IFN-γR1 expression in patients with the M1K mutation.
DISCUSSION
We report here a novel form of IFN-γR1 deficiency due to a homozygous M1K mutation. Unlike other forms of partial IFN-γR1 deficiency, IFN-γR1 expression was expressed weakly in EBV-B cells and not at all in fibroblasts, due to mutation of the first ATG codon, resulting in a decrease in translation initiation efficiency. The M1K/M1K mutation caused a much more severe impairment of the phosphorylation of STAT1 and GAF-binding proteins than observed in other partial forms of IFN-γR1 deficiency, whether dominant or recessive, in EBV-B cells. In addition, M1K/M1K fibroblasts displayed a loss of function for GAF activation and HLA-DR induction in response to IFN-γ stimulation. The cellular phenotype is therefore more severe than that of any of the other known forms of partial IFN-γR1 deficiency, but less severe than that of complete IFN-γR1 deficiency. Like patients with the complete IFN-γR1 deficiency and unlike other patients with partial IFN-γR1 deficiency, this patient had a high plasma concentration of IFN-γ and showed no granulomatous reaction. This patient presented BCG disease after BCG vaccination at birth, and disseminated environmental mycobacterial disease affecting at multiple organs at the age of seven, similar to that observed in patients with complete IFN-γR1 deficiency. The clinical phenotype of this patient was too severe to be cured by treatment with IFN-γ and antibiotics. The patient was successfully treated with HSCT, a treatment generally considered for patients with complete IFN-γR1 deficiency but not for those with partial deficiency (39).
During translation initiation in eukaryotic cells, the small (40S) subunit of the eukaryotic ribosome binds to the capped 5′-end of the mRNA. It then migrates, stopping at the first AUG codon in a favorable context for translation initiation (52). If the first ATG is mutated, translation may occur through re-initiation or context dependent leaky scanning (53–56). In our case, re-initiation was impossible because there was no small open reading frame (ORF) upstream from the main ORF. The only possible explanation was therefore leaky scanning. In the IFNGR1 mRNA, mutation of the first AUG to an AAG prevents efficient translation initiation at this position. However, leaky ribosomes continue scanning to find the optimal downstream AUG START codon for translation initiation. In mammals, the optimal context for AUGSTART codon recognition is −3 A or G and +4 G. Two downstream AUG codons (AUG11 and AUG19) have a −3 G residue, but only the AUG19 codon has a +4 G residual, and therefore a better context for translation initiation. Some translation might be initiated at AUG11, but most of the ribosomes are likely to continue scanning until they reach the AUG19, the GCU17GAG18aug19G context of which mimics the consensus Kozak sequence (GCCRCCaugG). Our results highlight the importance for translation initiation of the nucleotides in position -6 to -1, because the deletion of codon 17 or of codons 17 and 18 decreased the efficiency of translation initiation from AUG19.
Residual expression was observed even when both ATG11 and ATG19 were deleted, consistent with the additional involvement of an inefficient non-AUG mediated mechanism in leaky scanning. In a favorable context, codons with a two nucleotides in common with AUG (e.g. ACG, AUU, CUG etc.) have been shown to initiate translation to various degrees in vivo and in vitro (57). Three such non-AUGs are present among the first 19 codons of the IFNGR1 M1K allele: AAG1, GUG14 and AGG16. These codons in favorable contexts may have different initiation effects, with GUG theoretically the strongest. However, only AAG1 is located in a good context for initiation, so non-AUGs-mediated initiation probably occurred at the AAG1.
We have defined a novel form of partial IFN-γR1 deficiency that is more severe immunologically and clinically that the known forms of partial IFN-γR1 deficiency, whether recessive or dominant. The causal mutation affects the initiating translation codon and leaky initiation at other AUG and non-AUG codons account for the residual expression and function of IFN-γR1 in some, but not all cell types. There may be several reasons for the observed pattern of cellular specificity. First, different cell types have different amounts of endoplasmic reticulum and post-translational modification procedure. Second, differences in the tissue specific profile of t-RNA may affect the efficiencies of non-AUG mediated initiation. As the patient was treated by hematopoietic stem cell transplantation (HSCT), it is not possible to investigate further the specific phenotypes of different immune cells, such as macrophages, T cells and NK cells. However, this study neatly highlights the tight correlation between the cellular and the clinical phenotype in patients with IFN-γR1 deficiency (58). A careful experimental investigation must be made along with clinical considerations when making therapeutic decisions in these patients.
MATERIALS AND METHODS
Case report
Patient P is a 9-year-old Finnish girl whose parents are second degree cousins. BCG vaccination as a newborn led to severe inguinal lymphadenitis, treated by surgery and followed-up with 6 months of treatment with isoniazid/rifampicin/ethambutol. At the age of seven, the patient suffered hip and leg pain, weight loss, fatigue, fever and respiratory distress. She presented pulmonary infiltration and pleural fluid on chest X-ray, an enlarged spleen with multiple lesions on ultra-sound scan, a mediastinal mass on computed tomography scan, and pelvic and femoral bone lesions on magnetic resonance imaging. Histological samples from bone, lung and mediastinum showed acid-fast bacilli with staining, but no granulomatous reaction. Mycobacterium avium intracellulare was cultured from multiple samples. No evidence of any recognized immune deficiency was found in this patient. The patient's parents and younger brother are healthy. The patient survived HSCT and detailed clinical information was reported elsewhere (39). All members of the family agreed to participate in this study, which was approved by the respective hospital's ethics committee.
Cell culture and stimulation, DNA extraction, PCR and sequencing
EBV-transformed B lymphocytes (EBV-B cells), SV40-transformed fibroblasts (fibroblasts) and HEK 293T cells were cultured as previously described (5–7,55). EBV-B cells and fibroblasts were stimulated with the indicated doses of IFN-γ (Imukin, Boehringer Ingelheim) and IFN-α2b (IntronA, Schering Plough). Genomic DNA was extracted from fresh blood cells, and PCR amplification and sequencing were carried out as previously described (4). Primers and PCR conditions are available upon request. Sequencing was carried out on an ABI 3130x (Applied Biosystems) sequencer.
Expression vectors and cell transfection
The wild-type IFNGR1 allele was inserted into the V5-topo-pcDNA3 (Invitrogen) according to the manufacturer's instructions. The ATG>AAG mutant was generated and various parts of the nucleotides from the first AUG to the third AUG were deleted by site-directed mutagenesis (Stratagene, Quickchange site-directed mutagenesis kit) according to the manufacturer's instructions. Primers are available upon request. We transfected HEK 293T cells with one of the various IFNGR1 V5-tagged pcDNA3.1 vectors or an insert-less V5-tagged pcDNA3 vector (mock), using a phosphate calcium transfection kit (Invitrogen) according to the manufacturer's instructions.
Electrophoretic mobility shift assay
Electrophoretic mobility shift assay (EMSA) was carried out as previously described (5–7). Briefly, cells were stimulated for 20 min with IFN-γ or IFN-α at the indicated doses. We incubated 10 µg (stimulated with IFN-γ) or 1 µg (stimulated with IFN-α) of nuclear extract with 32P-labeled (α-dATP) GAS (from the FCGR1 promoter) probe and subjected the mixture to electrophoresis in a polyacrylamide gel.
Immunoprecipitation and western blotting
EBV-B cells (50 millions cells) or HEK293T cells were lysed in lysis buffer containing 20 mm Tris–HCl pH 7.4, 140 mm NaCl, 2 mm EDTA, 50 mm NaF, 0.5% sodium deoxycholate and 1% NP-40 together with 100 mm orthovanadate, 200 mm PMSF, 1% aprotinin, 1 mg/ml pepstatine, 1 mg/ml leupeptine and 1 mg/ml antipain. We then subjected 1.5 mg of cell lysate to immunoprecipitation on SigmaPrep spin columns (Sigma MC1000) with 2 µg of specific antibody against IFN-γR1, either C-20 (Santa Cruz Biotechnology) or GIR-94 (BD biosciences Pharmingen), and protein G (P-3296, Sigma). Immunoprecipitates were left untreated or were treated with PNGase F (Biolabs, P0704L) before western blotting, which was carried out as previously described (20). The following antibodies were used: Anti-V5 antibody (Invitrogen, 46-0705), C-20 antibody (Santa Cruz Biotechnology), ECL™ horseradish peroxidase-conjugated donkey anti-rabbit IgG antibody (NA934V, GE Health Care UK Limited) and ECL™ horseradish peroxidase-conjugated sheep anti-mouse IgG antibody (NA931V, GE Health Care UK Limited).
Flow cytometry
The methods for detecting cell surface expression of IFN-γR1 and HLA-DR have been described elsewhere (5,6). STAT1 phosphorylation were assessed by activating cells with IFN-γ or IFN-α for 30 min, washing them in cold 1× PBS, incubating them with 4% paraformaldehyde (PFA) for 10 min at room temperature, washing them with 1× PBS, and incubating them with 100% methanol for 10 min at 4°C. Cells were washed twice and incubated with PBS 1× + 1% SAB + 0.1% saponin for 10 min at 4°C. Cells were then washed and incubated for 1 h at 4°C with either an antibody against phosphorylated STAT1 (612132, BD Transduction Laboratories) or with the corresponding isotype antibody (554121,BD Transduction Laboratories). Cells were then washed and incubated with Alexa G488 (Molecular Probe, Invitrogen) for 20 min at 4°C. Cells were washed three times, and signals were analyzed with a FACScanTM machine, using CELLQuestTM software (Becton Dickinson).
Whole-blood assay of the IL-12-IFN-γ circuit
Whole-blood assays were performed as previously described (51). Heparin-treated blood samples from a healthy control and P were stimulated in vitro with BCG alone or with BCG plus IFN-γ or IL-12 (R&D). Supernatants were collected after 48 h of stimulation and ELISA were performed with specific antibodies directed against IFN-γ, IL-12p70, or IL-12p40, using the human Quantikine HS kits for IL-12p70 and IL-12p40 from R&D and the human Pelipair IFN-γ kit from Sanquin, according to the manufacturer's instructions.
SUPPLEMENTARY MATERIAL
FUNDING
The Laboratory of Human Genetics of Infectious Diseases is supported by grants from The Rockefeller University Center for Clinical and Translational Science grant number 5UL1RR024143-03 and The Rockefeller University, the Schlumberger Foundation, the BNP-Paribas Foundation, the ‘Institut Universitaire de France,’ and the EU-grant QLK2-CT-2002-00846. X.-F.K. is supported by a Choh-Hao Li Memorial Fund Scholar award and the Shanghai Educational Development Foundation.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Bertrand Boisson, Emmanuelle Jouanguy, Svetlana Mazel, Horst von Bernuth and Ludovic de Beaucoudrey for helpful discussions. We thank Ron Liebman, Tatiana Kochetkov, Erin Kirk, Yelena Nemirovskaya, Brooke Delaney, Lucile Janiere, Yoann Rose, Martine Courat, Tony Leclerc and Guy Brami for technical and secretarial assistance and all members of the Laboratory of Human Genetics of Infectious Diseases for helpful discussions.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Casanova J.L., Abel L. Primary immunodeficiencies: a field in its infancy. Science. 2007;317:617–619. doi: 10.1126/science.1142963. [DOI] [PubMed] [Google Scholar]
- 2.Casanova J.L., Abel L. Genetic dissection of immunity to mycobacteria: the human model. Annu. Rev. Immunol. 2002;20:581–620. doi: 10.1146/annurev.immunol.20.081501.125851. [DOI] [PubMed] [Google Scholar]
- 3.Newport M.J., Huxley C.M., Huston S., Hawrylowicz C.M., Oostra B.A., Williamson R., Levin M. A mutation in the interferon-gamma-receptor gene and susceptibility to mycobacterial infection. N. Engl. J. Med. 1996;335:1941–1949. doi: 10.1056/NEJM199612263352602. [DOI] [PubMed] [Google Scholar]
- 4.Jouanguy E., Altare F., Lamhamedi S., Revy P., Emile J.F., Newport M., Levin M., Blanche S., Seboun E., Fischer A., et al. Interferon-gamma-receptor deficiency in an infant with fatal bacille Calmette–Guerin infection. N. Engl. J. Med. 1996;335:1956–1961. doi: 10.1056/NEJM199612263352604. [DOI] [PubMed] [Google Scholar]
- 5.Jouanguy E., Lamhamedi-Cherradi S., Altare F., Fondaneche M.C., Tuerlinckx D., Blanche S., Emile J.F., Gaillard J.L., Schreiber R., Levin M., et al. Partial interferon-gamma receptor 1 deficiency in a child with tuberculoid bacillus Calmette–Guerin infection and a sibling with clinical tuberculosis. J. Clin. Invest. 1997;100:2658–2664. doi: 10.1172/JCI119810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jouanguy E., Lamhamedi-Cherradi S., Lammas D., Dorman S.E., Fondaneche M.C., Dupuis S., Doffinger R., Altare F., Girdlestone J., Emile J.F., et al. A human IFNGR1 small deletion hotspot associated with dominant susceptibility to mycobacterial infection. Nat. Genet. 1999;21:370–378. doi: 10.1038/7701. [DOI] [PubMed] [Google Scholar]
- 7.Jouanguy E., Dupuis S., Pallier A., Doffinger R., Fondaneche M.C., Fieschi C., Lamhamedi-Cherradi S., Altare F., Emile J.F., Lutz P., et al. In a novel form of IFN-gamma receptor 1 deficiency, cell surface receptors fail to bind IFN-gamma. J. Clin. Invest. 2000;105:1429–1436. doi: 10.1172/JCI9166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doffinger R., Jouanguy E., Dupuis S., Fondaneche M.C., Stephan J.L., Emile J.F., Lamhamedi-Cherradi S., Altare F., Pallier A., Barcenas-Morales G., et al. Partial interferon-gamma receptor signaling chain deficiency in a patient with bacille Calmette–Guerin and Mycobacterium abscessus infection. J. Infect. Dis. 2000;181:379–384. doi: 10.1086/315197. [DOI] [PubMed] [Google Scholar]
- 9.Dorman S.E., Holland S.M. Mutation in the signal-transducing chain of the interferon-gamma receptor and susceptibility to mycobacterial infection. J. Clin. Invest. 1998;101:2364–2369. doi: 10.1172/JCI2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rosenzweig S.D., Dorman S.E., Uzel G., Shaw S., Scurlock A., Brown M.R., Buckley R.H., Holland S.M. A novel mutation in IFN-gamma receptor 2 with dominant negative activity: biological consequences of homozygous and heterozygous states. J. Immunol. 2004;173:4000–4008. doi: 10.4049/jimmunol.173.6.4000. [DOI] [PubMed] [Google Scholar]
- 11.Vogt G., Chapgier A., Yang K., Chuzhanova N., Feinberg J., Fieschi C., Boisson-Dupuis S., Alcais A., Filipe-Santos O., Bustamante J., et al. Gains of glycosylation comprise an unexpectedly large group of pathogenic mutations. Nat. Genet. 2005;37:692–700. doi: 10.1038/ng1581. [DOI] [PubMed] [Google Scholar]
- 12.Vogt G., Bustamante J., Chapgier A., Feinberg J., Boisson Dupuis S., Picard C., Mahlaoui N., Gineau L., Alcais A., Lamaze C., et al. Complementation of a pathogenic IFNGR2 misfolding mutation with modifiers of N-glycosylation. J. Exp. Med. 2008;205:1729–1737. doi: 10.1084/jem.20071987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Altare F., Lammas D., Revy P., Jouanguy E., Doffinger R., Lamhamedi S., Drysdale P., Scheel-Toellner D., Girdlestone J., Darbyshire P., et al. Inherited interleukin 12 deficiency in a child with bacille Calmette–Guerin and Salmonella enteritidis disseminated infection. J. Clin. Invest. 1998;102:2035–2040. doi: 10.1172/JCI4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Picard C., Fieschi C., Altare F., Al-Jumaah S., Al-Hajjar S., Feinberg J., Dupuis S., Soudais C., Al-Mohsen I.Z., Genin E., et al. Inherited interleukin-12 deficiency: IL12B genotype and clinical phenotype of 13 patients from six kindreds. Am. J. Hum. Genet. 2002;70:336–348. doi: 10.1086/338625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Altare F., Durandy A., Lammas D., Emile J.F., Lamhamedi S., Le Deist F., Drysdale P., Jouanguy E., Doffinger R., Bernaudin F., et al. Impairment of mycobacterial immunity in human interleukin-12 receptor deficiency. Science. 1998;280:1432–1435. doi: 10.1126/science.280.5368.1432. [DOI] [PubMed] [Google Scholar]
- 16.Fieschi C., Dupuis S., Catherinot E., Feinberg J., Bustamante J., Breiman A., Altare F., Baretto R., Le Deist F., Kayal S., et al. Low penetrance, broad resistance, and favorable outcome of interleukin 12 receptor beta1 deficiency: medical and immunological implications. J. Exp. Med. 2003;197:527–535. doi: 10.1084/jem.20021769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Jong R., Altare F., Haagen I.A., Elferink D.G., Boer T., van Breda Vriesman P.J., Kabel P.J., Draaisma J.M., van Dissel J.T., Kroon F.P., et al. Severe mycobacterial and Salmonella infections in interleukin-12 receptor-deficient patients. Science. 1998;280:1435–1438. doi: 10.1126/science.280.5368.1435. [DOI] [PubMed] [Google Scholar]
- 18.Chapgier A., Kong X.F., Boisson-Dupuis S., Jouanguy E., Averbuch D., Feinberg J., Zhang S.Y., Bustamante J., Vogt G., Lejeune J., et al. A partial form of recessive STAT1 deficiency in humans. J. Clin. Invest. 2009;119:1502–1514. doi: 10.1172/JCI37083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dupuis S., Dargemont C., Fieschi C., Thomassin N., Rosenzweig S., Harris J., Holland S.M., Schreiber R.D., Casanova J.L. Impairment of mycobacterial but not viral immunity by a germline human STAT1 mutation. Science. 2001;293:300–303. doi: 10.1126/science.1061154. [DOI] [PubMed] [Google Scholar]
- 20.Dupuis S., Jouanguy E., Al-Hajjar S., Fieschi C., Al-Mohsen I.Z., Al-Jumaah S., Yang K., Chapgier A., Eidenschenk C., Eid P., et al. Impaired response to interferon-alpha/beta and lethal viral disease in human STAT1 deficiency. Nat. Genet. 2003;33:388–391. doi: 10.1038/ng1097. [DOI] [PubMed] [Google Scholar]
- 21.Chapgier A., Boisson-Dupuis S., Jouanguy E., Vogt G., Feinberg J., Prochnicka-Chalufour A., Casrouge A., Yang K., Soudais C., Fieschi C., et al. Novel STAT1 alleles in otherwise healthy patients with mycobacterial disease. PLoS Genet. 2006;2:e131. doi: 10.1371/journal.pgen.0020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Filipe-Santos O., Bustamante J., Haverkamp M.H., Vinolo E., Ku C.L., Puel A., Frucht D.M., Christel K., von Bernuth H., Jouanguy E., et al. X-linked susceptibility to mycobacteria is caused by mutations in NEMO impairing CD40-dependent IL-12 production. J. Exp. Med. 2006;203:1745–1759. doi: 10.1084/jem.20060085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Filipe-Santos O., Bustamante J., Chapgier A., Vogt G., de Beaucoudrey L., Feinberg J., Jouanguy E., Boisson-Dupuis S., Fieschi C., Picard C., et al. Inborn errors of IL-12/23- and IFN-gamma-mediated immunity: molecular, cellular, and clinical features. Semin. Immunol. 2006;18:347–361. doi: 10.1016/j.smim.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 24.Fortin A., Abel L., Casanova J.L., Gros P. Host genetics of mycobacterial diseases in mice and men: forward genetic studies of BCG-osis and tuberculosis. Annu Rev Genomics Hum Genet. 2007;8:163–192. doi: 10.1146/annurev.genom.8.080706.092315. [DOI] [PubMed] [Google Scholar]
- 25.Noordzij J.G., Hartwig N.G., Verreck F.A., De Bruin-Versteeg S., De Boer T., Van Dissel J.T., De Groot R., Ottenhoff T.H., Van Dongen J.J. Two patients with complete defects in interferon gamma receptor-dependent signaling. J. Clin. Immunol. 2007;27:490–496. doi: 10.1007/s10875-007-9097-8. [DOI] [PubMed] [Google Scholar]
- 26.Altare F., Jouanguy E., Lamhamedi-Cherradi S., Fondaneche M.C., Fizame C., Ribierre F., Merlin G., Dembic Z., Schreiber R., Lisowska-Grospierre B., et al. A causative relationship between mutant IFNgR1 alleles and impaired cellular response to IFNgamma in a compound heterozygous child. Am. J. Hum. Genet. 1998;62:723–726. doi: 10.1086/301750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsolia M.N., Chapgier A., Taprantzi P., Servitzoglou M., Tassios I., Spyridis N., Papageorgiou F., Santos O.F., Casanova J.L., Spyridis P. Disseminated nontuberculous mycobacterial infection in a child with interferon-gamma receptor 1 deficiency. Eur. J. Pediatr. 2006;165:458–461. doi: 10.1007/s00431-006-0110-7. [DOI] [PubMed] [Google Scholar]
- 28.Holland S.M., Dorman S.E., Kwon A., Pitha-Rowe I.F., Frucht D.M., Gerstberger S.M., Noel G.J., Vesterhus P., Brown M.R., Fleisher T.A. Abnormal regulation of interferon-gamma, interleukin-12, and tumor necrosis factor-alpha in human interferon-gamma receptor 1 deficiency. J. Infect. Dis. 1998;178:1095–1104. doi: 10.1086/515670. [DOI] [PubMed] [Google Scholar]
- 29.Rosenzweig S., Dorman S.E., Roesler J., Palacios J., Zelazko M., Holland S.M. 561del4 defines a novel small deletion hotspot in the interferon-gamma receptor 1 chain. Clin. Immunol. 2002;102:25–27. doi: 10.1006/clim.2001.5135. [DOI] [PubMed] [Google Scholar]
- 30.Pierre-Audigier C., Jouanguy E., Lamhamedi S., Altare F., Rauzier J., Vincent V., Canioni D., Emile J.F., Fischer A., Blanche S., et al. Fatal disseminated Mycobacterium smegmatis infection in a child with inherited interferon gamma receptor deficiency. Clin. Infect. Dis. 1997;24:982–984. doi: 10.1093/clinids/24.5.982. [DOI] [PubMed] [Google Scholar]
- 31.Roesler J., Kofink B., Wendisch J., Heyden S., Paul D., Friedrich W., Casanova J.L., Leupold W., Gahr M., Rosen-Wolff A. Listeria monocytogenes and recurrent mycobacterial infections in a child with complete interferon-gamma-receptor (IFNgammaR1) deficiency: mutational analysis and evaluation of therapeutic options. Exp. Hematol. 1999;27:1368–1374. doi: 10.1016/s0301-472x(99)00077-6. [DOI] [PubMed] [Google Scholar]
- 32.Cunningham J.A., Kellner J.D., Bridge P.J., Trevenen C.L., McLeod D.R., Davies H.D. Disseminated bacille Calmette–Guerin infection in an infant with a novel deletion in the interferon-gamma receptor gene. Int. J. Tuberc. Lung. Dis. 2000;4:791–794. [PubMed] [Google Scholar]
- 33.Koscielniak E., de Boer T., Dupuis S., Naumann L., Casanova J.L., Ottenhoff T.H. Disseminated Mycobacterium peregrinum infection in a child with complete interferon-gamma receptor-1 deficiency. Pediatr. Infect. Dis. J. 2003;22:378–380. [PubMed] [Google Scholar]
- 34.Allende L.M., Lopez-Goyanes A., Paz-Artal E., Corell A., Garcia-Perez M.A., Varela P., Scarpellini A., Negreira S., Palenque E., Arnaiz-Villena A. A point mutation in a domain of gamma interferon receptor 1 provokes severe immunodeficiency. Clin. Diagn. Lab. Immunol. 2001;8:133–137. doi: 10.1128/CDLI.8.1.133-137.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fieschi C., Dupuis S., Picard C., Smith C.I., Holland S.M., Casanova J.L. High levels of interferon gamma in the plasma of children with complete interferon gamma receptor deficiency. Pediatrics. 2001;107:E48. doi: 10.1542/peds.107.4.e48. [DOI] [PubMed] [Google Scholar]
- 36.Dorman S.E., Picard C., Lammas D., Heyne K., van Dissel J.T., Baretto R., Rosenzweig S.D., Newport M., Levin M., Roesler J., et al. Clinical features of dominant and recessive interferon gamma receptor 1 deficiencies. Lancet. 2004;364:2113–2121. doi: 10.1016/S0140-6736(04)17552-1. [DOI] [PubMed] [Google Scholar]
- 37.Chantrain C.F., Bruwier A., Brichard B., Largent V., Chapgier A., Feinberg J., Casanova J.L., Stalens J.P., Vermylen C. Successful hematopoietic stem cell transplantation in a child with active disseminated Mycobacterium fortuitum infection and interferon-gamma receptor 1 deficiency. Bone Marrow Transplant. 2006;38:75–76. doi: 10.1038/sj.bmt.1705399. [DOI] [PubMed] [Google Scholar]
- 38.Rottman M., Soudais C., Vogt G., Renia L., Emile J.F., Decaluwe H., Gaillard J.L., Casanova J.L. IFN-gamma mediates the rejection of haematopoietic stem cells in IFN-gammaR1-deficient hosts. PLoS Med. 2008;5:e26. doi: 10.1371/journal.pmed.0050026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moilanen P., Korppi M., Hovi L., Chapgier A., Feinberg J., Kong X.F., Boisson-Dupuis S., Arola M., Casanova J.L., Saarinen-Pihkala U.M. Successful Hematopoietic Stem Cell Transplantation From an Unrelated Donor in a Child With Interferon Gamma Receptor Deficiency. Pediatr. Infect. Dis. J. 2009;28:658–660. doi: 10.1097/INF.0b013e318195092e. [DOI] [PubMed] [Google Scholar]
- 40.Roesler J., Horwitz M.E., Picard C., Bordigoni P., Davies G., Koscielniak E., Levin M., Veys P., Reuter U., Schulz A., et al. Hematopoietic stem cell transplantation for complete IFN-gamma receptor 1 deficiency: a multi-institutional survey. J. Pediatr. 2004;145:806–812. doi: 10.1016/j.jpeds.2004.08.021. [DOI] [PubMed] [Google Scholar]
- 41.Reuter U., Roesler J., Thiede C., Schulz A., Classen C.F., Oelschlagel U., Debatin K.M., Friedrich W. Correction of complete interferon-gamma receptor 1 deficiency by bone marrow transplantation. Blood. 2002;100:4234–4235. doi: 10.1182/blood-2002-02-0433. [DOI] [PubMed] [Google Scholar]
- 42.Horwitz M.E., Uzel G., Linton G.F., Miller J.A., Brown M.R., Malech H.L., Holland S.M. Persistent Mycobacterium avium infection following nonmyeloablative allogeneic peripheral blood stem cell transplantation for interferon-gamma receptor-1 deficiency. Blood. 2003;102:2692–2694. doi: 10.1182/blood-2003-04-1268. [DOI] [PubMed] [Google Scholar]
- 43.Lee W.I., Huang J.L., Lin T.Y., Hsueh C., Wong A.M., Hsieh M.Y., Chiu C.H., Jaing T.H. Chinese patients with defective IL-12/23-interferon-gamma circuit in Taiwan: partial dominant interferon-gamma receptor 1 mutation presenting as cutaneous granuloma and IL-12 receptor beta1 mutation as pneumatocele. J. Clin. Immunol. 2009;29:238–245. doi: 10.1007/s10875-008-9253-9. [DOI] [PubMed] [Google Scholar]
- 44.Glosli H., Stray-Pedersen A., Brun A.C., Holtmon L.W., Tonjum T., Chapgier A., Casanova J.L., Abrahamsen T.G. Infections due to various atypical mycobacteria in a Norwegian multiplex family with dominant interferon-gamma receptor deficiency. Clin. Infect. Dis. 2008;46:e23–e27. doi: 10.1086/525855. [DOI] [PubMed] [Google Scholar]
- 45.Muszlak M., Chapgier A., Barry Harivelo R., Castella C., Cremades F., Goulois E., Laporte R., Casanova J.L., Ranaivoarivony V., Hebert J.C., et al. Multifocal infection due to Mycobacterium intracellulare: first case of interferon gamma receptor partial dominant deficiency in tropical French territory. Arch. Pediatr. 2007;14:270–272. doi: 10.1016/j.arcped.2006.11.023. [DOI] [PubMed] [Google Scholar]
- 46.Arend S.M., Janssen R., Gosen J.J., Waanders H., de Boer T., Ottenhoff T.H., van Dissel J.T. Multifocal osteomyelitis caused by nontuberculous mycobacteria in patients with a genetic defect of the interferon-gamma receptor. Neth. J. Med. 2001;59:140–151. doi: 10.1016/s0300-2977(01)00152-8. [DOI] [PubMed] [Google Scholar]
- 47.Okada S., Ishikawa N., Shirao K., Kawaguchi H., Tsumura M., Ohno Y., Yasunaga S., Ohtsubo M., Takihara Y., Kobayashi M. The novel IFNGR1 mutation 774del4 produces a truncated form of interferon-gamma receptor 1 and has a dominant-negative effect on interferon-gamma signal transduction. J. Med. Genet. 2007;44:485–491. doi: 10.1136/jmg.2007.049635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Storgaard M., Varming K., Herlin T., Obel N. Novel mutation in the interferon-gamma-receptor gene and susceptibility to mycobacterial infections. Scand. J. Immunol. 2006;64:137–139. doi: 10.1111/j.1365-3083.2006.01775.x. [DOI] [PubMed] [Google Scholar]
- 49.Villella A., Picard C., Jouanguy E., Dupuis S., Popko S., Abughali N., Meyerson H., Casanova J.L., Hostoffer R.W. Recurrent Mycobacterium avium osteomyelitis associated with a novel dominant interferon gamma receptor mutation. Pediatrics. 2001;107:E47. doi: 10.1542/peds.107.4.e47. [DOI] [PubMed] [Google Scholar]
- 50.Remiszewski P., Roszkowska-Sliz B., Winek J., Chapgier A., Feinberg J., Langfort R., Bestry I., Augustynowicz-Kopec E., Ptak J., Casanova J.L., et al. Disseminated Mycobacterium avium infection in a 20-year-old female with partial recessive IFNgammaR1 deficiency. Respiration. 2006;73:375–378. doi: 10.1159/000088682. [DOI] [PubMed] [Google Scholar]
- 51.Feinberg J., Fieschi C., Doffinger R., Feinberg M., Leclerc T., Boisson-Dupuis S., Picard C., Bustamante J., Chapgier A., Filipe-Santos O., et al. Bacillus Calmette Guerin triggers the IL-12/IFN-gamma axis by an IRAK-4- and NEMO-dependent, non-cognate interaction between monocytes, NK, and T lymphocytes. Eur. J. Immunol. 2004;34:3276–3284. doi: 10.1002/eji.200425221. [DOI] [PubMed] [Google Scholar]
- 52.Kozak M. Pushing the limits of the scanning mechanism for initiation of translation. Gene. 2002;299:1–34. doi: 10.1016/S0378-1119(02)01056-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kozak M. Context effects and inefficient initiation at non-AUG codons in eucaryotic cell-free translation systems. Mol. Cell. Biol. 1989;9:5073–5080. doi: 10.1128/mcb.9.11.5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kozak M. Adherence to the first-AUG rule when a second AUG codon follows closely upon the first. Proc. Natl. Acad. Sci. USA. 1995;92:2662–2666. doi: 10.1073/pnas.92.7.2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Puel A., Reichenbach J., Bustamante J., Ku C.L., Feinberg J., Doffinger R., Bonnet M., Filipe-Santos O., de Beaucoudrey L., Durandy A., et al. The NEMO mutation creating the most-upstream premature stop codon is hypomorphic because of a reinitiation of translation. Am. J. Hum. Genet. 2006;78:691–701. doi: 10.1086/501532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kozak M. Emerging links between initiation of translation and human diseases. Mamm. Genome. 2002;13:401–410. doi: 10.1007/s00335-002-4002-5. [DOI] [PubMed] [Google Scholar]
- 57.Peabody D.S. Translation initiation at non-AUG triplets in mammalian cells. J. Biol. Chem. 1989;264:5031–5035. [PubMed] [Google Scholar]
- 58.Dupuis S., Doffinger R., Picard C., Fieschi C., Altare F., Jouanguy E., Abel L., Casanova J.L. Human interferon-gamma-mediated immunity is a genetically controlled continuous trait that determines the outcome of cobacterial invasion. Immunol. Rev. 2000;178:129–137. doi: 10.1034/j.1600-065x.2000.17810.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.