Abstract
Using a newly developed dual-cell quadrupole linear ion trap-orbitrap hybrid mass spectrometer (dcQLT-orbitrap), we demonstrate the utility of collecting high-resolution tandem mass spectral data for large-scale shotgun proteomics. Multiple nanoLC-MS/MS experiments on both an older generation quadrupole linear ion trap-orbitrap hybrid (QLT-orbitrap) and the dcQLT-orbitrap, using both resonant-excitation CAD and beam-type CAD (HCD) were performed. Resulting from various technological advances (e.g., a stacked ring ion guide AP inlet, a dual cell QLT, etc.), the dcQLT-orbitrap exhibited increased duty cycle (~1.5–2×) and sensitivity for both CAD (ion trap detection) and HCD (orbitrap detection) methods. As compared to the older system, the dcQLT-orbitrap produced significantly more unique peptide identification for both methods (~30% improvement for CAD and ~115% improvement for HCD). The sizeable improvement of the HCD method on the dcQLT-orbitrap system, outperforms the current standard method of CAD with ion trap detection for large-scale analysis. Finally, we demonstrate that the increased HCD performance translates to a direct and substantial improvement in protein quantitation precision using isobaric tags.
Introduction
Ion trap mass spectrometers have undergone remarkable evolution over the past twenty years. Today this technology is among the most widely used and most effective tools for large-scale protein sequence analysis.1–2 Beginning with the coupling of three-dimensional quadrupole ion traps (QIT) to atmospheric pressure (AP) inlets,3–8 the technology continued to progress with the introduction of the linear quadrupole ion trap (QLT)9–11, and most recently with the hybridization to high-resolving power Fourier transform mass analyzers (FTMS); first, with ion cyclotron resonance (ICR)12 and, more recently, the orbitrap13–14. Each of these generations has likewise spurred research aimed at discovering how to best apply the technology to analyze the highly complex mixtures of proteins present in biological systems.
The use of high-resolution and accuracy mass-to-charge (m/z) analysis of precursors (i.e., MS1) with subsequent isolation, activation, and detection in the QLT (i.e., MS2) is one such transformative method.15-16 The pairing of the fast and sensitive product ion detection in the QLT with high-resolution and accuracy FTMS1 analysis facilitates reduction of the peptide search space, yielding greatly improved specificity.17-18 The relatively long scanning time and reduced sensitivity of FTMS analysis, in general, has left the utility of high resolution MS2 data up for debate.19-22
Recently, a next-generation hybrid QLT-orbitrap mass spectrometer has been described and made commercially available. This system has been updated in three main ways: (1) the AP inlet was modified to utilize a stacked-ring ion guide (SRIG) for increased ion injection efficiency (2–10×)23, (2) a dual-cell QLT (dcQLT) was installed that affords higher ion trapping efficiency (~1.2×) and scanning rates (~2×), among other benefits24, and (3) ion transmission elements between the c-trap and orbitrap were improved25. The system also incorporates a collision cell that is integrated into the c-trap for high efficiency beam-type collision-activated dissociation (HCD)26 prior to orbitrap m/z analysis. Given the significant performance enhancements of this new system, we surmised that new opportunities to extract more information from a proteomic analyses may accompany this device. Consequently, we report here a comprehensive comparison of a dcQLT-orbitrap to the conventional QLT-orbitrap instrument for large-scale protein identification and quantitation.
Experimental
Mass spectrometry
Experiments were performed on two generations of hybrid QLT-orbitrap mass spectrometers (Thermo Fisher Scientific, Bremen, Germany). The older system, an LTQ Orbitrap XL (referred to herein as QLT-orbitrap), consists of a single QLT, which is interfaced to a heated capillary/skimmer AP inlet and is coupled to an orbitrap mass analyzer. The second generation instrument, an LTQ Orbitrap Velos (dcQLT-orbitrap), begins with a stacked-ring ion guide (SRIG) AP inlet, has a dual-cell QLT, and an integrated c-trap/collision cell (as opposed to two separate components as in the older generation).
Sample preparation
Human embryonic stem cells were lysed via sonication in 8 M urea, 75 mM NaCl, 50 mM tris (pH 8.0), 1 mM sodium orthovanadate, 10 mM sodium pyrophosphate, and a complete mini ETDA-free protease inhibitor tablet (Roche Diagnostics, Indianapolis, IN). Samples were spun at 18,000 × g for ten minutes and the supernatant was removed for further processing. The resulting protein mixture was diluted in 50 mM tris to a urea concentration of 1.5 M, reduced with dithiothreitol at 37 ºC, and alkylated with iodoacetamide for 30 minutes in the dark at room temperature. Samples were digested overnight with trypsin (Promega, Madison, WI) at 37 ºC, desalted using SepPak cartridges (Waters, Milford, MA), and labeled with the iTRAQ reagent as previously described.27–28 Samples were then desalted using SepPak cartridges prior to MS analysis.
Saccharomyces cerevisiae (strain BY4741) was grown in YPD (yeast extract, peptone, and dextrose) media at 30 ºC to mid-log phase. Cells were harvested by centrifugation at 4 ºC for five minutes at 8000 × g and washed twice with sterile water before storage of the cell pellets at −80 ºC. Frozen pellets were thawed and washed three times prior to lysis with Y-PER (Pierce, Rockford, IL), 0.1 M DTT, complete mini ETDA-free protease inhibitor (Roche Diagnostics, Indianapolis, IN), and phosSTOP phosphatase inhibitor (Roche Diagnostics, Indianapolis, IN). Samples were pelleted and supernatant was collected. Proteins were precipitated by addition of chilled acetone and resuspended in 50 mM HEPES pH 7.5/4 M urea. To extract nuclear proteins, 8 M urea/0.4 N H2SO4 was added to the pellet. Extracted proteins from both fractions were mixed and subsequently reduced and alkylated prior to overnight digestion with trypsin (Promega, Madison, WI) at 37 ºC. Following digestion, the sample was labeled with the iTRAQ reagent as previously described, mixed in known ratios (1:1:1 and 6:2:1) and analyzed via LC-MSMS using a dcQLT-orbitrap.27–28 Separate LC-MS/MS analyses were performed using either HCD or PQD.
Liquid chromatography-tandem mass spectrometry
Liquid chromatography-tandem mass spectrometry was performed using a NanoAcquity UPLC system (Waters, Milford, MA) coupled to either a QLT-orbitrap or a dcQLT-orbitrap. Samples were loaded onto a precolumn (75 μm ID, packed with 5 cm C18 particles, Alltech, Nicholasville, KY) for ten minutes at a flow rate of 1 μm/min. Samples were then eluted over an analytical column (50 μm ID, packed with 15 cm C18 particles, Alltech, Nicholasville, KY) using a 90-minute linear gradient from 1% to 35% acetonitrile with 0.2% formic acid and a flow rate of 300 nL/min. The column-making procedure was previously described.29 An additional 30 minutes were used for column washing and equilibration.
All mass spectrometery instrument methods consisted of an MS1 analysis in the orbitrap, followed by data-dependent MS2 scans of the ten most intense precursors. Precursors were subject to dynamic exclusion for 60 s using a window of −0.5 Th to 2.5 Th. Precursors with unassigned charges states or charge states of one were also excluded. AGC target values were 1,000,000 for MS1 analysis, 10,000 for QLT MS2 analysis, and 100,000 for orbitrap MS2 analysis. Orbitrap resolution settings were 60,000 forMS1and 7500 for HCD MS 2.
Database searching and data analysis
Data reduction was performed by DTA Generator30 using a signal-to-noise ratio (S/N) threshold of 1.5 on fragment ions. The Open Mass Spectrometry Search Algorithm31 (OMSSA; version 2.1.4) was used to search spectra against the International Protein Index32 (IPI; http://www.ebi.ac.uk/IPI/) human database version 3.53 with fully tryptic enzyme specificity. An average mass tolerance of ±4.5 Da was used for the precursor, while a monoisotopic mass tolerance of ±0.01 Da was used for fragments with orbitrap detection and ±0.5 Da with QLT detection. Carbamidomethylation of cysteine, iTRAQ 4-plex on the N-terminus, and iTRAQ 4-plex on lysine were set as fixed modifications, while oxidation of methionine and iTRAQ 4-plex on tyrosine were set as variable modifications. False discovery rate (FDR) analysis was performed with custom software that iteratively checked combinations of expectation value (e-value) score and precursor mass error to find thresholds that maximize unique peptide identifications while satisfying 1% FDR. Mass accuracy and protein quantitation was evaluated with custom software.
Results and Discussion
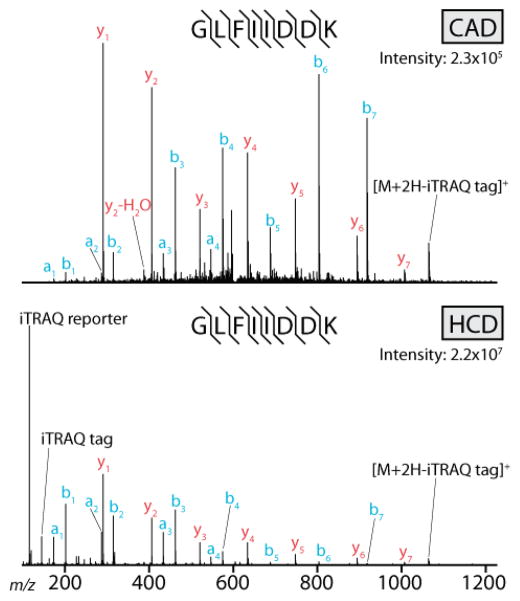
Our experiments were aimed at evaluating how effective these two generations of hybrid QLT-orbitrap mass spectrometers were at interrogating a highly complex mixture of peptides. To accomplish this, we obtained and lysed human ES cells, harvested protein, and digested the sample with trypsin. The resulting mixture was separated by nano-HPLC over a 90-minute gradient and analyzed using the QLT-orbitrap and dcQLT-orbitrap systems. On each system we interrogated the sample multiple times using separate methods that employed either ion trap CAD33–35 with QLT m/z analysis or HCD (beam-type CAD)36–38 with orbitrap m/z analysis. In total, the sample was interrogated twelve times: triplicate analyses using HCD and CAD on both systems. Example CAD and HCD tandem mass spectra, collected using the dcQLT-orbitrap system, are shown in Figure 1. The two methods produce similar, but not identical, fragmentation patterns. In both cases a complete series of b- and y-type fragments is observed. They differ slightly in that the b-type fragment ions are more intense than the y-type in the HCD spectrum. Note there is no low-mass cutoff in the HCD spectrum; the intense iTRAQ reporter ions are easily detected.
Figure 1.
Two example single-scan tandem mass spectra collected using a dcQLT-orbitrap mass spectrometer. The same precursor was interrogated in both spectra. The top spectrum was produced using resonant-excitation CAD with QLT m/z analysis of fragment ions. The bottom spectrum was collected using beam-type CAD (HCD) with orbitrap m/z analysis of fragment ions.
Table 1 summarizes the comparison. These data reveal several surprising outcomes: first, the dcQLT-orbitrap system outperformed the older instrument in all experiments. Second, running the HCD method on the dcQLT-orbitrap produced more peptide identifications than the CAD method on the same device. The roughly equivalent performance of these two methods on the newer instrument is due to the striking gain in HCD performance between the two instruments (115%), far greater than the gain in CAD performance between the two (30%, vide infra).Using a 30-minute interval from the central portion of the gradient we calculated average values for various scan parameters, including ion injection time, MS2 scan time, number of MS2 scans acquired, and precursor peak depth. Some of the trends in this data are easily explained. For example, due to the gains in ion injection efficiency on the dcQLT-orbitrap system, ion injection times are markedly shorter (127.4 ms versus 15.1 ms for the HCD analyses). Also, as a result of employing larger AGC targets for HCD than CAD (100k and 10k, respectively), the HCD injection times are appreciably longer than the CAD times on both systems (127.4 ms versus 12.4 ms for the QLT-orbitrap, and 15.1 ms versus 10.0 ms for dcQLT-orbitrap). Scan times fit closely with what is expected based on theory and reports from the manufacturer.24 HCD analysis on the new dcQLT-orbitrap is ~150 ms faster than on the older system (265 ms versus 416 ms). This is primarily a result of the shorter ion injection times, but other improvements, such as abbreviated precursor ion isolation, contributed. The CAD scan is also faster on the newer system (191 ms versus 293 ms). This is a result of multiple factors: shorter isolation and activation times, as well as a higher scan rate in the low-pressure cell of the dcQLT.
Table 1.
Results from twelve separate analyses of a human protein sample digested with trypsin. Peptides were analyzed using two different systems and two different methods (the four combinations were run in triplicate). Here, we present the average total number of identifications and the average number of unique identifications at a 1% FDR. Also, presented is the average injection time, scan time, number of scans, and peak depth during a 30-minute interval in the middle of the LC gradient.
| dcQLT-orbitrap | all IDs | unique IDs (1%FDR) | inject time (ms) | scan time (ms) | # of scans | peak depth |
|---|---|---|---|---|---|---|
| Average FTMS-HCD | 8910 | 6830 | 15 | 265 | 5620 | 297 |
| Std Dev FTMS-HCD | 124 | 61 | 1 | 0 | 6 | 6 |
| Average ITMS-CAD | 8373 | 6161 | 10 | 191 | 7260 | 598 |
| Std Dev ITMS-CAD | 1 | 32 | 5 | 4 | 75 | 17 |
| QLT-orbitrap | all IDs | unique IDs (1%FDR) | inject time (ms) | scan time (ms) | # of scans | peak depth |
|---|---|---|---|---|---|---|
| Average FTMS-HCD | 3800 | 3169 | 127 | 416 | 3587 | 126 |
| Std Dev FTMS-HCD | 66 | 72 | 4 | 4 | 26 | 2 |
| Average ITMS-CAD | 6341 | 4751 | 12 | 293 | 5012 | 268 |
| Std Dev ITMS-CAD | 58 | 61 | 0 | 4 | 20 | 4 |
For the HCD data, the number of scans acquired per analysis is easily rationalized using the associated duty cycles of that scan type on the two instruments. As noted, HCD scan times on the newer system are ~40% faster than on the older generation, and this higher duty cycle matches exactly with the increase in the number of scans acquired by the newer system. Interestingly, the average number of scans and average peak depth for the HCD experiment analysis on the newer system match very closely to the CAD values from the older system – the main difference is that the HCD experiment resulted in 50% more high-confidence identifications (vide infra).
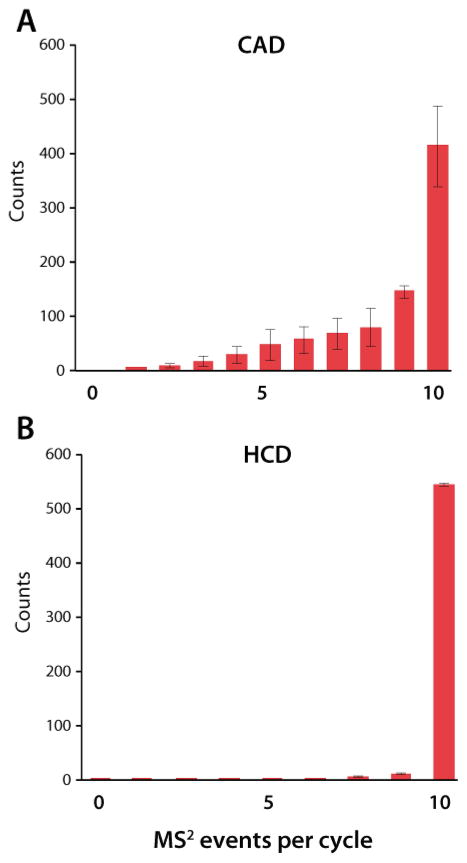
Unlike the HCD method, the increase in the number of MS2 spectra acquired is not proportional to the improved duty cycle of the dcQLT-orbitrap CAD method (i.e., resonant-excitation CAD with QLT m/z analysis). Whereas the predicted number of HCD scans matched within 0.2% of the number actually acquired (based on the increase in duty cycle and the number of scans acquired using the older generation instrument), the CAD experiment acquired 6% fewer spectra than one would have expected (i.e,. one would expect the CAD method to collect ~500 more spectra). Irrespective of this disproportionate increase in the number of precursors interrogated, the average peak depth for those precursors increased twofold. To explain this we examined the average number of MS2 events that followed the MS1 survey scans (Figure 2). Following the MS1 survey scan, the instrument generates a list of viable precursors – taking into account dynamic exclusion, charge state exclusion, etc. If, however, the instrument is unable to generate a sufficiently long list of precursor targets to satisfy the number of data -dependent scans requested, it will default back to acquiring another MS1 scan. During the HCD analysis using the dcQLT-orbitrap system (Figure 2A), nearly every MS1 scan is followed by ten data-dependent MS2 scans. There were always enough precursors detected by the instrument to fulfill the requested number of data-dependent scans. However, during the CAD analysis on this system, quite often, fewer than ten data-dependent scans were acquired (Figure 2B). There were simply not enough precursors available to satisfy the fast duty cycle of the CAD experiment on the dcQLT-orbitrap. The insufficient number of precursors propagated the low peak depth for CAD experiment on the dcQLT-orbitrap – whenever a viable precursor was detected in an MS1 scan it was immediately selected and activated (i.e., every peak was activated very early on in its elution such that they were always of low relative abundance). From these data we conclude there are two primary reasons that the extremely fast duty cycle of the CAD scan on the dcQLT-orbitrap instrument did not result in the expected increase in peptide identifications: first, the instrument interrogated precursors too early in their elution profile, which consequently lowered the probability a of successful identification. Second, there were too few available precursors, based on our relatively standard exclusion settings, to consistently satisfy the instrument. Doubtless a reevaluation of optimal dynamic exclusion settings would improve the number of spectral identifications resulting from this method. Improved peak-picking algorithms could also help this situation. Furthermore, the speed of this method on the dcQLT-orbitrap might permit routine data-independent sampling.39–42
Figure 2.
The average number of MS 2events that followed the MS1survey scan during a 30-minute interval in the middle of the LC gradient.
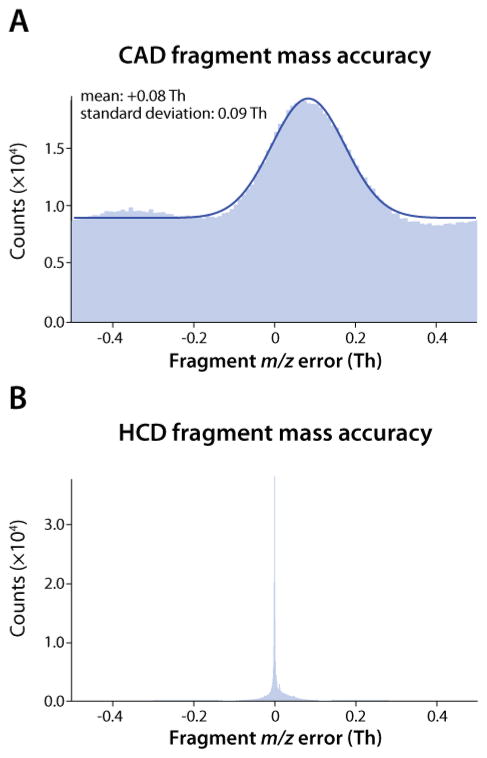
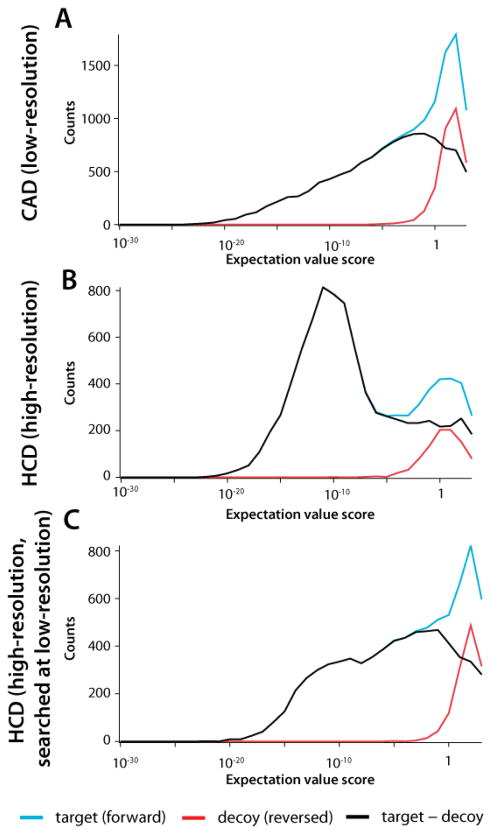
The role of MS2 spectral mass accuracy in these experiments is quite significant. As noted above, the sampling statistics of the CAD experiment performed on the QLT-orbitrap system and the HCD experiment performed on the dcQLT-orbitrap system were similar (i.e., duty cycle, number of scans acquired, and peak depth). With that said, the HCD experiment yielded ~50% more high-confidence unique identifications. Even at a resolving power of 7500, orbitrap mass accuracy is high (Figure 3B), which enabled use of a tight fragment mass tolerance (±0.01 Da) when performing database searching. This improved mass accuracy led to greatly improved search specificity (Figure 4B). By using such narrow fragment ion tolerances, the expectation value scores of true positives tended to be high,43 resulting in a distinctly bimodal distribution with well-separated target and decoy populations. By comparison, QLT mass accuracy is decidedly lower, forcing an expanded search tolerance of ±0.5 Da (Figure 3A). As a result of these search parameters and data quality, the target and decoy distributions largely overlap making it difficult to distinguish between the two populations without the utilization of additional factor(s) such as precursor mass accuracy (Figure 4A). Note that the bimodal distribution of the HCD data is a result of both the high accuracy of the orbitrap analyzer and the HCD dissociation process – if the orbitrap HCD data is searched with a mass accuracy of ±0.5 Da the resulting distribution is still bimodal, although the separation between target and decoy populations is less pronounced (Figure 4C). As an additional control, the same sample was interrogated on our dcQLT-orbitrap using a method where every precursor is activated by HCD and CAD, and, every scan utilizes orbitrap analysis. In this experiment, HCD still outperformed CAD, identifying 4,831 unique peptides as compared to 4,108 (1% FDR).
Figure 3.
The mass accuracy of all the fragment ions detected using the dcQLT -orbitrap during the three (A) CAD-QLT and (B) the three HCD-orbitrap analyses.
Figure 4.
Plots of identifications from the target database (blue), decoy database (red) and the difference between the two (black) as a function of identification expectation value. Plots are broken down into the (A) CAD -QLT experiment, (B) HCD-orbitrap experiment, and (C) the HCD-orbitrap experiment searched with a product tolerance of ±0.5Th.
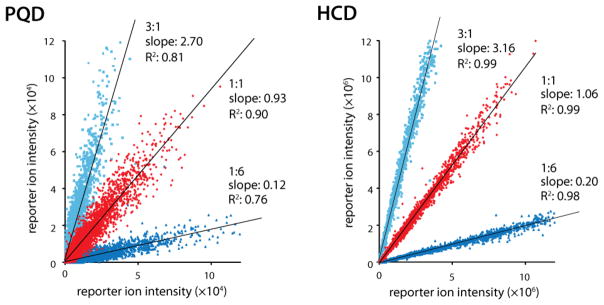
Prior to the incorporation of the HCD cell into the hybrid QLT-orbitrap, relative quantitation by isobaric tagging (e.g., iTRAQ and TMT)44–45 was not straight forward to perform on ion trap-based instrumentation. Both PQD and ETD are compatible with isobaric tags; however, limitations in robustness and reporter ion intensity have limited their mainstream adoption.27–28,46 The introduction of beam-type CAD (HCD), which eliminates the low mass cutoff imposed by resonant-excitation CAD, on the QLT-orbitrap hybrid has been a boon for the use of isobaric tag -based quantitation strategies.29,47 Still, the method had experienced a sluggish uptake as HCD requires orbitrap m/z analysis – a method that on previous instruments was less sensitive as compared to resonant-excitation CAD. We reasoned, however, that the new dcQLT-orbitrap system, with its excellent HCD performance, would allow routine use of isobaric tagging methods. To test this hypothesis we analyzed peptide mixtures from yeast that were labeled with isobaric tags using PQD and HCD fragmentation on the dcQLT-orbitrap. Here the HCD method identified 36% more peptides than PQD (7,607 versus 5,584) and produced much more reliable quantitative information (Figure 5). Although the average (log2) ratios produced by both methods were close to the expected values (ideal 1:6 = −2.58, 1:1 = 0, 3:1 = 1.58; PQD 1:6 = −2.38, 1:1 = −0.01, 3:1 = 1.59; HCD 1:6 = −2.39, 1:1 = 0.05, 3:1 = 1.75) the standard deviations were much smaller for HCD (PQD = 0.30, HCD = 0.07). The respective AGC targets of the two methods (100k for HCD versus 10k for PQD) likely influenced the quality of the quantitation results produced by each technique; however, independent of the AGC target values used, the contribution of the iTRAQ reporter ions to the overall product ion intensity was substantially higher in HCD spectra than in PQD.
Figure 5.
The measured iTRAQ product ion ratios of samples that were mixed in known amounts. The samples were analyzed using PQD and HCD.
Conclusion
The dcQLT-orbitrap mass spectrometer incorporates several hardware modifications that enhance scan speed and sensitivity. In particular, improvements in ion injection efficiency (AP inlet) and orbitrap injection efficiency make rapid and sensitive tandem mass spectra collection in the FT analyzer possible. These improvements, combined with beam-type CAD (HCD) allow for the widespread use of isobaric tags for large-scale quantification of QLT hybrid mass spectrometers. Here we demonstrate that the achieved mass accuracy of the orbitrap-analyzed MS/MS spectra results in a distinctly bimodal distribution with well-separated target and decoy populations and, ultimately, greatly improved database search specificity. We predict that such capabilities may stimulate a paradigm shift in mass spectrometry-based proteomics – i.e., the routine acquisition of high-resolution tandem mass spectra for large -scale protein sequencing and quantitation.
Using conventional dynamic exclusion settings the very fast scanning dcQLT instrument did secure genuine gains (CAD method) over the older generation single QLT device (6161 versus 4751 unique IDs); however, the HCD experiment (with orbitrap detection) bested both with 6830. We conclude a thorough evaluation of data-dependent selection parameters may indeed propel the CAD method to the top. Nevertheless, the remarkably strong performance of the HCD method, as compared to previous generations, and its compatibility with isobaric tagging, provides a viable method that will doubtless offer new opportunities. One such opportunity, which is beyond the scope of the current study, is the acquisition of high-resolution ETD tandem mass spectra.48–50 Recent work by our laboratory demonstrated that c- and z -type fragment ions can be readily distinguished from one another on the basis of mass alone with sufficient mass measurement accuracy.51 The dcQLT-orbitrap instrument will likewise allow for fast and sensitive acquisition of ETD tandem mass spectra, data that can be directly annotated for fragment ion type. Finally, for ETD and CAD, high-resolution tandem mass spectra will be of obvious benefit for those wishing to directly interpret tandem mass spectra (de novo).52–58
Acknowledgments
We thank Mark Tervo, Danielle Swaney, and Jason Russell of the University of Wisconsin - Madison and Jens Griep-Raming, Dirk Nolting, Alexander Makarov, Vlad Zabrouskov, Jae Schwartz, John Syka, and George Stafford of Thermo Fisher Scientific for helpful discussions. Mark Levenstein kindly provided us with the stem cells. This work was funded by the Beckman Foundation, the National Science Foundation (NSF 0701846 and 0747990 to JJC), and the National Institutes of Health (NIH R01GM080148 and PO1GM081629 to JJC). GCM and DHP acknowledge support from an NIH predoctoral fellowships (Biotechnology Training Program - 5T32GM08349, and Genetics Training Program - 5T32HG002760).
References
- 1.Washburn MP, Wolters D, Yates JR. Nat Biotechnol. 2001;19:242. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 2.de Godoy LMF, Olsen JV, Cox J, Nielsen ML, Hubner NC, Frohlich F, Walther TC, Mann M. Nature. 2008;455:1251. doi: 10.1038/nature07341. [DOI] [PubMed] [Google Scholar]
- 3.Mcluckey SA, Glish GL, Asano KG. Anal Chim Acta. 1989;225:25. [Google Scholar]
- 4.Vanberkel GJ, Glish GL, Mcluckey SA. Analytical Chemistry. 1990;62:1284. [Google Scholar]
- 5.Vanberkel GJ, Mcluckey SA, Glish GL. Analytical Chemistry. 1991;63:1098. doi: 10.1021/ac00018a014. [DOI] [PubMed] [Google Scholar]
- 6.Mcluckey SA, Vanberkel GJ, Glish GL, Huang EC, Henion JD. Analytical Chemistry. 1991;63:375. [Google Scholar]
- 7.Bruins AP, Covey TR, Henion JD. Anal Chem. 1987;59:2642. [Google Scholar]
- 8.Fenn JB, Mann M, Meng CK, Wong SF, Whitehouse CM. Science. 1989;246:64. doi: 10.1126/science.2675315. [DOI] [PubMed] [Google Scholar]
- 9.Schwartz JC, Senko MW, Syka JE. J Am Soc Mass Spectrom. 2002;13:659. doi: 10.1016/S1044-0305(02)00384-7. [DOI] [PubMed] [Google Scholar]
- 10.Hager JW. Rapid Communications in Mass Spectrometry. 2002;16:512. doi: 10.1002/rcm.1061. [DOI] [PubMed] [Google Scholar]
- 11.Welling M, Schuessler HA, Thompson RI, Walther H. International Journal of Mass Spectrometry. 1998;172:95. [Google Scholar]
- 12.Syka JEP, Marto JA, Bai DL, Horning S, Senko MW, Schwartz JC, Ueberheide B, Garcia B, Busby S, Muratore T, Shabanowitz J, Hunt DF. J Proteome Res. 2004;3:621. doi: 10.1021/pr0499794. [DOI] [PubMed] [Google Scholar]
- 13.Makarov A, Denisov E, Kholomeev A, Baischun W, Lange O, Strupat K, Horning S. Analytical Chemistry. 2006;78:2113. doi: 10.1021/ac0518811. [DOI] [PubMed] [Google Scholar]
- 14.Yates JR, Cociorva D, Liao LJ, Zabrouskov V. Anal Chem. 2006;78:493. doi: 10.1021/ac0514624. [DOI] [PubMed] [Google Scholar]
- 15.Haas W, Faherty BK, Gerber SA, Elias JE, Beausoleil SA, Bakalarski CE, Li X, Villen J, Gygi SP. Mol Cell Proteomics. 2006;5:1326. doi: 10.1074/mcp.M500339-MCP200. [DOI] [PubMed] [Google Scholar]
- 16.Bakalarski CE, Haas W, Dephoure NE, Gygi SP. Anal Bioanal Chem. 2007;389:1409. doi: 10.1007/s00216-007-1563-x. [DOI] [PubMed] [Google Scholar]
- 17.Boyne MT, Garcia BA, Li M, Zamdborg L, Wenger CD, Babai S, Kelleher NL. J Proteome Res. 2009;8:374. doi: 10.1021/pr800635m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elias JE, Gygi SP. Nature Methods. 2007;4:207. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 19.Mann M, Kelleher NL. P Natl Acad Sci USA. 2008;105:18132. doi: 10.1073/pnas.0800788105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scherl A, Shaffer SA, Taylor GK, Hernandez P, Appel RD, Binz PA, Goodlett DR. Journal of the American Society for Mass Spectrometry. 2008;19:891. doi: 10.1016/j.jasms.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parks BA, Jiang L, Thomas PM, Wenger CD, Roth MJ, Boyne MT, 2nd, Burke PV, Kwast KE, Kelleher NL. Anal Chem. 2007;79:7984. doi: 10.1021/ac070553t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wenger CD, Boyne MT, 2nd, Ferguson JT, Robinson DE, Kelleher NL. Anal Chem. 2008;80:8055. doi: 10.1021/ac8010704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wouters ER, Splendore M, Mullen C, Schwartz JC, Senko MW, Dunyach J-J. 57th Conference on Mass Spectrometry and Allied Topics; Philadelphia, Pennsylvania. 2009. [Google Scholar]
- 24.Pekar Second T, Blethrow JD, Schwartz JC, Merrihew GE, Maccoss MJ, Swaney DL, Russell JD, Coon JJ, Zabrouskov V. Anal Chem. 2009 doi: 10.1021/ac901278y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Damoc E, Denisov E, Blethrow J, Second TP, Zabrouskov V, Griep-Raming J, Makarov A, Moehring T. 57th Conference on Mass Spectrometry and Allied Topics; Philadelphia, Pennsylvania. 2009. [Google Scholar]
- 26.Olsen JV, Macek B, Lange O, Makarov A, Horning S, Mann M. Nat Methods. 2007;4:709. doi: 10.1038/nmeth1060. [DOI] [PubMed] [Google Scholar]
- 27.Phanstiel D, Unwin R, McAlister GC, Coon JJ. Anal Chem. 2009;81:1693. doi: 10.1021/ac8019202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Phanstiel D, Zhang Y, Marto JA, Coon JJ. Journal of the American Society for Mass Spectrometry. 2008;19:1255. doi: 10.1016/j.jasms.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Y, Ficarro SB, Li SJ, Marto JA. Journal of the American Society for Mass Spectrometry. 2009;20:1425. doi: 10.1016/j.jasms.2009.03.019. [DOI] [PubMed] [Google Scholar]
- 30.Good DM, Wenger CD, McAlister GC, Bai DL, Hunt DF, Coon JJ. Journal of the American Society for Mass Spectrometry. 2009;20:1435. doi: 10.1016/j.jasms.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Geer LY, Markey SP, Kowalak JA, Wagner L, Xu M, Maynard DM, Yang XY, Shi WY, Bryant SH. J Proteome Res. 2004;3:958. doi: 10.1021/pr0499491. [DOI] [PubMed] [Google Scholar]
- 32.Kersey PJ, Duarte J, Williams A, Karavidopoulou Y, Birney E, Apweiler R. Proteomics. 2004;4:1985. doi: 10.1002/pmic.200300721. [DOI] [PubMed] [Google Scholar]
- 33.Mcluckey SA, Glish GL, Kelley PE. Analytical Chemistry. 1987;59:1670. [Google Scholar]
- 34.Louris JN, Brodbeltlustig JS, Cooks RG, Glish GL, Vanberkel GJ, Mcluckey SA. Int J Mass Spectrom. 1990;96:117. [Google Scholar]
- 35.March RE, McMahon AW, Allinson ET, Londry FA, Alfred RL, Todd JFJ, Vedel F. Int J Mass Spectrom. 1990;99:109. [Google Scholar]
- 36.McLafferty FW, Venktaraghavan R, Irving P. Biochemical and Biophysical Research Communications. 1970;39 doi: 10.1016/0006-291x(70)90789-8. [DOI] [PubMed] [Google Scholar]
- 37.Wipf HK, Irving P, McCamish M, Venkataraghavan R, McLafferty FW. Journal of the American Chemical Society. 1973;95 doi: 10.1021/ja00791a048. [DOI] [PubMed] [Google Scholar]
- 38.Hunt DF, Yates JR, Shabanowitz J, Winston S, Hauer CR. Proceedings of the National Academy of Sciences of the United States of America. 1986;83:6233. doi: 10.1073/pnas.83.17.6233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Masselon C, Anderson GA, Harkewicz R, Bruce JE, Pasa-Tolic L, Smith RD. Anal Chem. 2000;72:1918. doi: 10.1021/ac991133+. [DOI] [PubMed] [Google Scholar]
- 40.Panchaud A, Scherl A, Shaffer SA, von Haller PD, Kulasekara HD, Miller SI, Goodlett DR. Anal Chem. 2009;81:6481. doi: 10.1021/ac900888s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Purvine S, Eppel JT, Yi EC, Goodlett DR. Proteomics. 2003;3:847. doi: 10.1002/pmic.200300362. [DOI] [PubMed] [Google Scholar]
- 42.Venable JD, Dong MQ, Wohlschlegel J, Dillin A, Yates JR. Nature Methods. 2004;1:39. doi: 10.1038/nmeth705. [DOI] [PubMed] [Google Scholar]
- 43.Meng FY, Cargile BJ, Miller LM, Forbes AJ, Johnson JR, Kelleher NL. Nat Biotechnol. 2001;19:952. doi: 10.1038/nbt1001-952. [DOI] [PubMed] [Google Scholar]
- 44.Thompson A, Schafer J, Kuhn K, Kienle S, Schwarz J, Schmidt G, Neumann T, Hamon C. Anal Chem. 2003;75:1895. doi: 10.1021/ac0262560. [DOI] [PubMed] [Google Scholar]
- 45.Ross PL, Huang YLN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, Purkayastha S, Juhasz P, Martin S, Bartlet-Jones M, He F, Jacobson A, Pappin DJ. Molecular & Cellular Proteomics. 2004;3:1154. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 46.Griffin TJ, Xie HW, Bandhakavi S, Popko J, Mohan A, Carlis JV, Higgins L. Journal of Proteome Research. 2007;6:4200. doi: 10.1021/pr070291b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bantscheff M, Boesche M, Eberhard D, Matthieson T, Sweetman G, Kuster B. Molecular & Cellular Proteomics. 2008;7:1702. doi: 10.1074/mcp.M800029-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McAlister GC, Phanstiel D, Good DM, Berggren WT, Coon JJ. Analytical Chemistry. 2007:3525. doi: 10.1021/ac070020k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McAlister GC, Berggren WT, Griep-Raming J, Horning S, Makarov A, Phanstiel D, Stafford G, Swaney DL, Syka JEP, Zabrouskov V, Coon JJ. Journal of Proteome Research. 2008;7:3127. doi: 10.1021/pr800264t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kaplan DA, Hartmer R, Speir JP, Stoermer C, Gumerov D, Easterling ML, Brekenfeld A, Kim T, Laukien F, Park MA. Rapid Commun Mass Spectrom. 2008;22:271. doi: 10.1002/rcm.3356. [DOI] [PubMed] [Google Scholar]
- 51.Hubler SL, Jue A, Keith J, McAlister GC, Craciun G, Coon JJ. Journal of the American Chemical Society. 2008;130:6388. doi: 10.1021/ja7099985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ueberheide BM, Fenyo D, Alewood PF, Chait BT. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:6910. doi: 10.1073/pnas.0900745106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Spengler B. Journal of the American Society for Mass Spectrometry. 2004;15:703. doi: 10.1016/j.jasms.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 54.Spengler B. Eur J Mass Spectrom. 2007;13:83. doi: 10.1255/ejms.840. [DOI] [PubMed] [Google Scholar]
- 55.Horn DM, Zubarev RA, McLafferty FW. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:10313. doi: 10.1073/pnas.97.19.10313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Budnik BA, Olsen JV, Egorov TA, Anisimova VE, Galkina TG, Musolyamov AK, Grishin EV, Zubarev RA. Journal of Mass Spectrometry. 2004;39:193. doi: 10.1002/jms.577. [DOI] [PubMed] [Google Scholar]
- 57.Savitski MM, Nielsen ML, Kjeldsen F, Zubarev RA. Journal of Proteome Research. 2005;4:2348. doi: 10.1021/pr050288x. [DOI] [PubMed] [Google Scholar]
- 58.Frank AM, Savitski MM, Nielsen ML, Zubarev RA, Pevzner PA. Journal of Proteome Research. 2007;6:114. doi: 10.1021/pr060271u. [DOI] [PMC free article] [PubMed] [Google Scholar]