Abstract
Unlike mammals, teleost fish can regenerate an injured retina, restoring lost visual function. Little is known of the molecular events that underlie retina regeneration. We previously found that in zebrafish, retinal injury stimulates Müller glia to generate multipotent α1-tubulin (α1T) and pax6-expressing progenitors for retinal repair. Here, we report the identification of a critical E-box in the α1T promoter that mediates transactivation by achaete-scute complex-like 1a (ascl1a) during retina regeneration. More importantly, we show that ascl1a is essential for retina regeneration. Within 4 h after retinal injury, ascl1a is induced in Müller glia. Knockdown of ascl1a blocks the induction of α1T and pax6 as well as Müller glial proliferation, consequently preventing the generation of retinal progenitors and their differentiated progeny. These data suggest ascl1a is required to convert quiescent Müller glia into actively dividing retinal progenitors, and that ascl1a is a key regulator in initiating retina regeneration.
Keywords: ascl1a, Müller glia, stem cells, regeneration, retina, zebrafish
Introduction
Recent studies suggest that after retinal injury, Müller glia can dedifferentiate and function as retinal stem cells in mammals, birds, and fish (Fischer and Reh, 2001; Ooto et al., 2004; Fausett and Goldman, 2006; Bernardos et al., 2007; Fimbel et al., 2007). However, unlike mammals and birds, where the capacity of Müller glia to regenerate new neurons is extremely limited (Fischer and Reh, 2001; Ooto et al., 2004), teleost fish mount a robust regenerative response that not only regenerates all damaged retinal neurons (Vihtelic and Hyde, 2000; Fausett and Goldman, 2006; Bernardos et al., 2007; Fimbel et al., 2007), but also results in restoration of visual function (Mensinger and Powers, 2007). Therefore, teleost fish, such as zebrafish, provide an ideal model system for identifying the molecular mechanisms underlying a robust regenerative response after retinal injury and may suggest new strategies for repairing the damaged mammalian retina. Toward this goal, global changes in gene expression have been documented in the injured zebrafish retina (Cameron et al., 2005; Kassen et al., 2007), and these studies provide a list of candidate genes that may participate in specific events associated with regeneration. Although the temporal and spatial expression pattern of some of these candidate genes suggests a role in various stages of regeneration (Yurco and Cameron, 2007), they do not inform us of the significance of these gene expression patterns.
Another approach for uncovering mechanisms of regeneration is to use promoters of regeneration-associated genes as reporters for transcription factors and other signaling molecules that mediate gene induction during retina regeneration. We have taken this approach using the α1-tubulin (α1T) promoter as a reporter for the regenerative response. In this case, transgenic zebrafish were created that harbor the α1T promoter driving enhanced green fluorescent protein (EGFP) expression (α1T:GFP) (Goldman et al., 2001). Using these fish, we recently showed that transgene-expressing Müller glia are a source of multipotent progenitors that contribute newborn cells toward retina regeneration in zebrafish (Fausett and Goldman, 2006). Here, we report the identification of an E-box within the α1T promoter that is required for its induction after retinal injury and provide evidence that the proneural basic helix-loop-helix (bHLH) transcription factor ascl1a regulates the α1T promoter via this E-box. Most importantly, we found that ascl1a is induced within 4 h after retinal injury, and knockdown of ascl1a prevents induction of regeneration-associated genes, such as α1T and pax6, and also inhibits Müller glia proliferation in response to injury. These data suggest ascl1a is a key transcription factor that is involved in initiating regeneration in the injured retina.
Materials and Methods
Animals.
The animals used in this study were treated in accordance with the guidelines of the University Committee on Use and Care of Animals at the University of Michigan. Fish were obtained from our breeding colony and raised with a 14/10 h light/dark cycle at 28°C.
Transgenic zebrafish.
1696α1TIpEGFP, del1046–846α1TIpEGFP, and 1016α1TIpEGFP transgenic fish have been described previously (Goldman and Ding, 2001; Goldman et al., 2001; Fausett and Goldman, 2006). −907α1TIpEGFP and TG-954CAα1TIpEGFP constructs were resuspended in injection buffer, and single-cell zebrafish embryos were injected, raised to adulthood, and screened for transgenic progeny as described previously (Goldman and Ding, 2001; Goldman et al., 2001).
Optic nerve lesions, eye lesions, and Morpholino-mediated gene knockdown.
Fish were anesthetized in 0.02% tricaine methane sulfonate (Sigma, St. Louis, MO) before surgery. Optic nerve crushes were performed as described previously (Hieber et al., 1998; Senut et al., 2004; Fausett and Goldman, 2006). Eye lesions were performed as described previously (Senut et al., 2004; Fausett and Goldman, 2006). Briefly, fish were anesthetized and under microscopic visualization, the right eye was gently pulled from its socket and stabbed four times (once in each quadrant) through the sclera with a 30 gauge needle. To deliver morpholinos to the injured retina, a 30 gauge needle was attached to a Hamilton syringe (Hamilton, Reno, NV) containing 1 mm morpholino (Gene Tools, Philomath, OR). Approximately 0.5 μl was injected into the vitreous after inserting the needle to the length of the bevel. We used the following lissamine-labeled morpholinos: control morpholino oligonucleotide (MO) 5′CCTCTTACCTCAGTTACAATTTATA-3′; ascl1a MO, 5′ATCTTGGCGGTGATGTCCATTTCGC-3′; ascl1a5′UTR MO, 5′AAGGAGTGAGTCAAAGCACTAAAGT-3′ [the latter two MOs have been described previously as ash1a MOs (Cau and Wilson, 2003)]. Custom tweezer-like electrodes (stainless steel, 4 mm wide, 25 mm length, 0.2 mm thickness) were then placed across the head of the fish with the cathode on the left eye and the anode on the right eye. An ECM 830 Electro Square Porator (BTX, San Diego, CA) was used to deliver five consecutive 50 ms pulses at 70 V with a 950 ms interval between pulses. The uninjected eye served as a negative control. One observer assigned letters to control and ascl1a MOs. A second observer then electroporated these MOs into fish and assigned the fish with numbers. This way, both observers could score MO-treated cells for GFP expression without any bias. For cell counts, morpholino-treated cells were scored for either GFP or bromodeoxyuridine (BrdU) labeling. The total number of morpholino-labeled cells that were labeled with GFP or BrdU was tallied for each retina to determine the percentage of MO-treated cells, which were also labeled with GFP or BrdU. The percentage of double-labeled cells for each group was calculated by taking the average from three retinas. A student's t test, assuming equal variance, was used to calculate p values.
Bromodeoxyuridine labeling.
To identify dividing cells, fish were either given a single injection of BrdU as described previously (Fausett and Goldman, 2006) or housed in 10 mm BrdU for 24 h [from 24–48 or 36–60 h postinjury (hpi)]. Fish were transferred to tanks with fresh water and killed at various times after BrdU administration to harvest the retinas.
Tissue preparation, immunohistochemistry, and in situ hybridization.
Fish were given an overdose of tricaine methane sulfonate, and the eyes from adult fish were dissected, enucleated, fixed, and sectioned as described previously (Senut et al., 2004; Fausett and Goldman, 2006). Immunohistochemistry was performed as described previously (Senut et al., 2004; Fausett and Goldman, 2006). The following primary antibodies were used: rat anti-BrdU (dividing cell marker; 1:250; Harlan, Sera-Lab, Sussex, UK); rabbit anti-GFP (1:1000; Invitrogen, Eugene, OR); and mouse anti-glutamine synthetase (GS) (glial marker; 1:500; Millipore, Temecula, CA). In situ hybridizations (ISHs) were performed with digoxigenin-labeled cRNA probes as described previously (Barthel and Raymond, 2000). Ascl1a was a gift from Eric Weinberg (University of Pennsylvania, Philadelphia, PA); Pax6 cRNA was prepared from a full-length Pax6a cDNA clone (Open Biosystems, Huntsville, AL); α1T probe was described previously (Hieber et al., 1998); notch1b and notch3 were gifts from Michael Lardelli (University of Adelaide, Adelaide, Australia); deltaA, deltaD, and olig2 were gifts from Bruce Appel (Vanderbilt University, Nashville, TN); and deltaB was a gift from Julian Lewis (UCL, London, UK). For timeline expression analysis (supplemental Table 1, available at www.jneurosci.org as supplemental material), notch3 hybridization was done individually and in combination with notch1b; similar results were obtained in each case. Delta in situ hybridization was done with deltaA individually and in combination with deltaB and deltaD; similar results were obtained in each case. Negative results were repeated twice for the 24 and 48 hpi time points except for olig2.
Imaging.
Slides were examined using a Zeiss (Oberkochen, Germany) Axiophot or Olympus (Tokyo, Japan) Fluoview FV1000 laser scanning confocal microscope. Images were captured using a digital camera adapted onto the Axiophot microscope or Olympus confocal microscope. Images were processed and annotated with Adobe Photoshop CS (Adobe Systems, San Jose, CA).
Vectors.
−907α1TIpEGFP expression vector contains 907 bp of 5′ flanking α1-tubulin DNA, exon 1, and the first intron fused in frame to the GFP sequence. This promoter fragment is similar to the full-length −1696α1TIpEGFP expression vector from previous work (Goldman and Ding, 2000; Goldman et al., 2001; Senut et al., 2004), except that it is lacking 789 bp from the 5′ end. A 2 bp mutation (TG-CA) was introduced into the full-length α1T promoter by amplifying a PCR product containing the 2bp E-box mutation and cloning it into the α1TIpEGFP vector. The TG-954CAα1TIpEGFP construct is identical to the −1696 expression vector except for the TG:CA mutation at position −954. Luciferase reporter vectors are based on the pXP2 construct (Nordeen, 1998). The α1TpXP2 reporter (wild-type) contains 1696 bp of 5′ flanking α1T sequence and its first exon cloned in frame and upstream of the luciferase coding sequence. The TG-954CAα1T-pXP2 vector is identical to the wild-type vector except for a 2 bp substitution at position 954 (TG-CA). The E-box-pXP2 vector contains three copies of the α1T E-box at position 954 (5′CATGTG 3′) upstream of a minimal β-globin promoter in pXP2. The mE-box-pXP2 vector is identical to the E-box-pXP2 vector, except each E-box has a 2 bp mutation (CATGTG to CATGCA). The coding sequence of ascl1a was cloned into pCS2+ to create pCS2+ascl1a. pCS2+β-globin was a gift from Audrey Seasholtz (University of Michigan, Ann Arbor, MI).
Transactivation assays.
Human embryonic kidney 293T (HEK293T) cells were plated in 24-well plates 24 h before transfection. Cells were transfected via calcium phosphate DNA precipitation. Luciferase assays were performed in duplicate for each sample, and values were normalized to β-globin expression.
Nuclear extracts and electrophoretic mobility shift assays.
Nuclear extracts were isolated from zebrafish brain, retina, and HEK293T transfected cells using standard protocols. Oligonucleotide probes labeled with 32P-dCTP (ICN, San Diego, CA) were incubated with 5–10 μg of nuclear extracts prepared from zebrafish brain or retina, rat brain, or with in vitro synthesized proteins using TNT Coupled Transcription/Translation system (Promega, Madison, WI). Cold oligonucleotides were incubated at >50-fold excess to test binding specificity. The protein:DNA mix was then run on a nondenaturing polyacrylamide gel, and DNA migration was visualized by exposing the gel to Kodak (Rochester, NY) imaging film.
Results
An E-box is required for α1T promoter activation in proliferating Müller glia after retinal injury
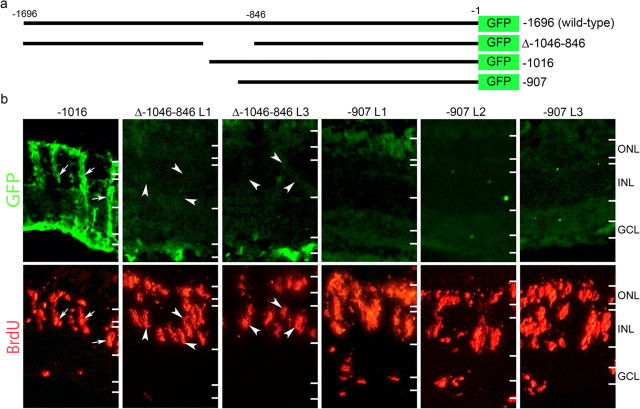
The −1696α1T:GFP and −1016α1T:GFP transgenes are induced in proliferating Müller glia after retinal injury (Senut et al., 2004; Fausett and Goldman, 2006). Identification of DNA elements mediating transgene induction may provide insight into the mechanism by which Müller glia respond to injury-induced signals and produce a dividing population of retinal stem cells. We hypothesized that these elements would lead us to transcription factors that regulate retina regeneration. Therefore, transgenic fish harboring different α1T promoter deletions driving GFP expression were screened for transgene expression in proliferating Müller glia after retinal injury (Fig. 1). This screen confirmed the −1016α1T:GFP transgene was induced after retinal injury and identified Δ-1046–846α1T:GFP and −907α1T:GFP as transgenes that no longer responded to retinal injury by inducing GFP expression in proliferating Müller glia (Fig. 1). This lack of expression is not attributable to promoter inactivation, because these promoters can drive transgene expression during development (Goldman and Ding, 2000; our unpublished observations). These results suggest that a DNA element located between nucleotides −1016 and −907 of the α1T promoter is required for transgene expression in proliferating Müller glia.
Figure 1.
A 109 bp region of the α1T promoter is required for transgene expression in dedifferentiating Müller glia. a, Schematic representation of α1T promoter constructs. The bars represent promoter sequence, and the numbers indicate relative position from the start codon. −1696 is the wild-type promoter described previously (Hieber et al., 1998; Goldman et al., 2001). Δ-1046–846 has been described previously (Goldman and Ding, 2000). The −1016 promoter directs transgene expression in Müller glia (Fausett and Goldman, 2006). The −907 promoter lacks 789 bp of upstream sequence. b, Transgenic fish received retinal injuries on day 0 and were given a 4 h pulse of BrdU at 4 dpi. Transgenic fish, which carry the required DNA element, express GFP in BrdU-labeled Müller glia (-1016, arrows), whereas transgenic fish lacking the element do not (Δ-1046–846 and −907). Two independent lines of Δ-1046–846 and three independent lines of −907 transgenic fish all display a lack of GFP expression in BrdU-labeled cells (arrowheads in Δ-1046–846). The images for −1016 and Δ-1046–846 are from the same sections. Because the −907 transgenic fish display very weak GFP expression in general, we used serial sections to obtain the −907 images. GCL, Ganglion cell layer.
To further narrow-in on this element, we searched for potential transcription factor binding sites within this 109 bp region by performing gel electrophoretic mobility shift assays. Radiolabeled oligonucleotide probes spanning different parts of the 109 bp sequence were incubated with nuclear protein extracts prepared from zebrafish or rat brain (Fig. 2a) or zebrafish retina (Fig. 2d) and resolved on native polyacrylamide gels. These experiments revealed specific binding of nuclear proteins to probe 4 (Fig. 2a, arrow), which contains a single E-box sequence CATGTG (Fig. 2b). Two base pair mutations within the E-box (Fig. 2b, probes 4–3, 4–4) disrupted protein binding (Fig. 2c, lanes 3, 4), whereas mutations in surrounding nucleotides (Fig. 2b, probes 4–1, 4–2, 4–5) had no effect on binding (Fig. 2c, lanes 1, 2, 5). In addition, E-box mutations prevented unlabeled probes from competing for binding with wild-type radiolabeled probes even at 50-fold molar excess (Fig. 2c, lanes 8, 9). Although nuclear extracts prepared from brain or retina bound probe 4 (Fig. 2), those prepared from rat liver, fish gill, fish muscle, and fish kidney did not (data not shown). To further test the specificity of this binding, we used a probe (Eb), which contains an E-box (CAGATG), that is not required for transgene expression after retinal injury but is required for transgene induction in retinal ganglion cells after optic nerve crush (Senut et al., 2004). Unlike the probe harboring the E-box at position −954, probe Eb did not bind nuclear proteins from zebrafish brain (Fig. 2e, lanes 4–6). These results suggest that the binding we observe is specific to the E-box at position −954.
Figure 2.
An E-box within the 109 bp region that is required for transgene expression in Müller glia binds nuclear extracts from zebrafish and rat brain and zebrafish retina. a, Electrophoretic mobility shift assay using a probe from the 109 bp region binds nuclear extracts from zebrafish and rat brain. The arrow indicates specific binding. Cold indicates where 50-fold molar excess unlabeled probe was added as competition. Extract indicates whether zebrafish (zf) or rat brain extracts were added. b, Nucleotide sequence of the probes used for electrophoretic mobility shift assay. The E-box is outlined in probe 4 with a box. Mutations are indicated by italicized and underlined text. c, Mutations to the E-box (lanes 3 and 4) render the probe unable to bind nuclear extracts from zebrafish brain, whereas mutations to non-E-box nucleotides do not affect binding (lanes 1, 2, and 5). Probes correspond to those shown in b. Unlabeled mutant probes compete with wild-type probe binding when the E-box is intact (lanes 6, 7, and 10), but not when the E-box is mutated (lanes 8 and 9), even at 50-fold molar excess. d, Nuclear extracts from zebrafish retina bind specifically to the E-box. e, An E-box probe from a different region of the promoter (Eb) is unable to compete with the E-box from probe 4 (lane 3) and does not bind to zebrafish brain nuclear extracts (lane 4).
Might this E-box mediate transgene induction in Muller glia after retinal injury? To investigate this question, we placed the 2 bp mutation from probe 4–4 (Fig. 2b) in the full-length −1696α1T promoter to create TG-954CAα1T:GFP transgenic zebrafish (Fig. 3a). Three independent lines of transgenic fish were identified and examined for transgene expression during development. The TG-954CA promoter directs transgene expression to the brain, spinal cord, and retina during development, which is similar to the wild-type promoter (supplemental Fig. 1, available at www.jneurosci.org as supplemental material). We reported previously that mature fish harboring the −1696α1T:GFP or −1016α1T:GFP transgene express GFP in retinal stem cells located at the circumferential germinal zone (CGZ) (Goldman et al., 2001; Fausett and Goldman, 2006), where new retinal cells are continually added as the eye grows. Interestingly, TG-954CAα1T:GFP fish do not express the transgene in the CGZ (supplemental Fig. 2, available at www.jneurosci.org as supplemental material), suggesting the E-box is required for transgene expression in retinal stem cells. Consistent with the idea that this particular E-box is essential for α1T expression in adult retinal stem cells is our observation that after retinal lesion, TG-954CAα1T:GFP fish do not express GFP in proliferating Müller glia (Fig. 3b). Occasionally we noticed GFP+ cells in the ganglion cell layer after retinal lesion (Fig. 3b, arrows). These appeared to be axotomized retinal ganglion cells, which are known to induce expression from the −1696α1T:GFP transgene (Goldman and Ding, 2000; Goldman et al., 2001; Senut et al., 2004). We confirmed this in TG-954CAα1T:GFP fish by observing robust GFP expression in retinal ganglion cells after optic nerve crush (supplemental Fig. 3, available at www.jneurosci.org as supplemental material). Therefore, the identified E-box is specifically necessary for transgene expression in adult retinal stem cells residing in either the retinal periphery (CGZ) or in the central retina after injury (proliferating Müller glia).
Figure 3.

Mutation of −954 E-box in the wild-type −1696 α1T promoter prevents transgene induction in proliferating Müller glia after retina injury. a, Schematic representation of α1T promoter:GFP constructs. The wild-type −1696α1T:GFP transgene (-1696) and the 2 bp mutation created in the E-box at position −954 in the −1696 construct (TG-954CA) are indicated. b, Three independent lines of transgenic fish received retinal injuries on day 0 and were given a 4 h pulse of BrdU at 4 dpi. Note that none of the lines harboring the 2 bp mutation induced transgene GFP expression in BrdU-labeled Müller glia (arrowheads). See Figure 1 for comparison with the wild-type −1696α1T:GFP transgene response. The arrows in the lowest right GFP panel show transgene induction in retinal ganglion cells, the axons of which were presumably damaged when we injured the retina in this fish.
Ascl1a is rapidly induced in Müller glia after retinal injury
The results described above suggest basic helix-loop-helix transcription factors may be crucial for retina regeneration. One such protein, ascl1a, attracted our attention because of the following: (1) the chick homolog of ascl1a is expressed in Müller glia-derived progenitors in the injured chick retina (Fischer and Reh, 2001); (2) ascl1a is induced in Müller glia during regeneration of the zebrafish retina (Yurco and Cameron, 2007); and (3) the mouse homolog of ascl1a is expressed in retinal (Jasoni and Reh, 1996) and neural (Torii et al., 1999; Yun et al., 2002) progenitors. To confirm ascl1a was induced in proliferating Müller glia, we examined injured retinas from −1016α1T:GFP fish for ascl1a expression. We observed that ascl1a expressing cells correspond precisely to GFP+ Müller glia at 4 d postinjury (dpi) (Fig. 4a–c), which we previously identified as proliferating Müller glia-derived retinal progenitors (Fausett and Goldman, 2006).
Figure 4.
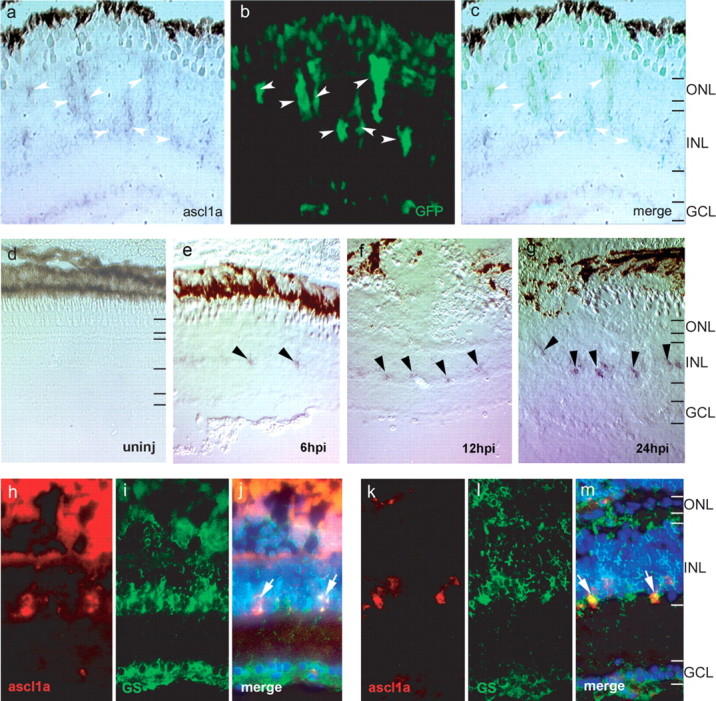
Ascl1a is induced in proliferating Müller glia after retinal injury. a–c, Ascl1a expression is detected by in situ hybridization (a) in GFP+ Müller glia (b) at 4 dpi (arrowheads). c shows the merge image of a and b. d–g, In situ hybridization for ascl1a from 6–24 hpi. Ascl1a is not expressed in the uninjured retina (d). Ascl1a is induced in cells of the INL at 6 hpi (e, arrowheads). Ascl1a expression gradually increases and is easily detected at 24 hpi (g, arrowheads). h–m, Ascl1a in situ hybridization (red, h, k) and glutamine synthetase immunostaining (green, i, l) show ascl1a is induced in Müller glia at 6 hpi (arrows in merge, j) and 24 hpi (arrows in merge, m). DAPI nuclear staining is shown in the merged panels. GCL, Ganglion cell layer.
To determine whether ascl1a expression precedes Müller glia proliferation and α1T induction, which begin at ∼24 hpi (Fausett and Goldman, 2006), we examined retinas at 6, 12, 18, and 24 hpi for ascl1a expression by ISH. We detected a low level of expression at 6, 12, and 18 hpi and stronger expression at 24 hpi (6, 12, and 24 hpi shown in Fig. 4d–g). Colocalization of ascl1a and glutamine synthetase at these early time points confirmed ascl1a-expressing cells are Müller glia (Fig. 4h–m). These results indicate that ascl1a is induced in Müller glia at least 18 h before they enter the cell cycle and begin expressing the α1T transgene.
To put ascl1a and α1T gene induction in the context of other genes induced in the injured retina, which are known to be involved in retina development and induced during retina regeneration, we compared ascl1a and α1T expression with pax6, delta, notch, and olig2 at various times after retinal injury using ISH. The results of this analysis are summarized in supplemental Table 1 (available at www.jneurosci.org as supplemental material) and show that ascl1a induction is the earliest indication of Müller glia dedifferentiation after retinal injury. Ascl1a was first detectable at 4 hpi, increasing at 24 hpi and reaching peak levels by 48 hpi. In contrast endogenous α1T expression increases at 24 hpi, whereas delta, notch, and olig2 are induced between 48 and 96 hpi. Because pax6 is expressed by amacrine cells (Hitchcock et al., 1996), it was difficult to detect its induction until ectopic expression appeared in the inner and outer nuclear layers (ONLs) at 96 hpi. This result is consistent with the observation that not all GFP+ Müller glia induce pax6 at 4 dpi (Fausett and Goldman, 2006), suggesting pax6 induction in Müller glia occurs after GFP induction. Although low levels of pax6 protein have been reported in Müller glia (Bernardos et al., 2007), we did not detect pax6 induction in transgene expressing Müller glia at 6 or 24 hpi (supplemental Fig. 4, available at www.jneurosci.org as supplemental material).
Ascl1a regulates α1T expression
Because injury-induced expression of the −1696α1T:GFP transgene requires an E-box at position −954 and E-boxes bind basic helix-loop-helix transcription factors like ascl1a, we suspected ascl1a would regulate α1T promoter activity via its −954 E-box. Consistent with this idea, we found that ascl1a transactivated a minimal β-globin promoter harboring three copies of the −954 E-box (CATGTG) but did not transactivate the β-globin promoter with three copies of a mutant E-box (CATGCA) (Fig. 5a). In addition, mutation of this E-box in the context of the −1696α1T promoter also blocked transactivation by ascl1a (Fig. 5b), although transactivation was less dramatic, possibly because of high basal expression caused by endogenous transcriptional activators, which act at sites other than the E-box. We were unsuccessful in demonstrating a direct interaction between ascl1a and the −954 E-box using gel electrophoretic mobility shift assays (our unpublished observations), which may suggest that ascl1a activation of the α1T promoter is indirect or requires the presence of additional DNA-binding and/or ascl1a-interacting proteins.
Figure 5.
Ascl1a regulates the α1T promoter through the −954 E-box. a, Luciferase reporter vectors with either a minimal β-globin promoter alone (light gray bars) or three copies of the −954 E-box (dark gray bars) or the mutant E-box from probe 4–4 (see Fig. 2b) (black bars) were transfected in combination with ascl1a into HEK293T cells. Ascl1a transactivates the reporter when a functional E-box is present but not when the E-box is mutated. b, Full-length α1T constructs harboring either the wild-type (-1696) or mutant E-box promoters were transfected into HEK293T cells with or without ascl1a. Ascl1a induces reporter expression when the E-box is intact, but not when the E-box is mutated. Error bars indicate SEM for three replicates.
Despite the lack of evidence that ascl1a can bind the −954 E-box, the very early induction of ascl1a after retinal injury and its ability to regulate α1T promoter activity suggest it may regulate α1T transcription in vivo. To test this hypothesis, we suppressed ascl1a expression in Müller glia after retinal injury. To do this, we electroporated injured adult retinas with lissamine-labeled antisense MOs that have been demonstrated previously to knockdown ascl1a expression when injected into single-cell zebrafish embryos (Cau and Wilson, 2003). We compared the effects of control or ascl1a MOs on transgene expression in our −1016α1T:GFP transgenic fish. Normally, GFP+ Müller glia are visible at 2 dpi, and by 4 dpi, there are numerous GFP+ Müller glia (Fig. 1, −1016 panel). It was readily apparent that ascl1a knockdown suppressed transgene expression in Müller glia after retinal injury at both 2 and 4 dpi (4 dpi shown in Fig. 6). Specificity was confirmed using a second MO (ascl1a5′UTR), which gave similar results. We counted the number of lissamine+/GFP+ cells in the inner nuclear layer (INL) and ONL by analyzing confocal images and found that 13% of the control MO-treated cells expressed GFP at 2 dpi, whereas only 4% the ascl1a MO-treated cells expressed GFP (three retinas were counted for each group; p = 0.02). At 4 dpi, 22% of the control MO-treated cells expressed GFP, whereas only 2% of the ascl1a and ascl1a5′UTR MO-treated cells expressed GFP, respectively (three retinas were counted for each group; p = 0.03 for the ascl1a MO, p = 0.04 for the ascl1a5′UTR MO). These quantitative data confirm our qualitative observations and suggest ascl1a is required for α1T:GFP transgene expression in vivo.
Figure 6.
Ascl1a is required for transgene expression in vivo. −1016 transgenic fish received retinal injuries and morpholino treatment on day 0. Eyes were harvested on day 4. a, Control morpholino treatment has no effect on transgene expression in Müller glia (arrows), and there are many GFP+ Müller glia (arrowheads). Morpholinos targeting ascl1a (ascl1a and ascl1a5′UTR panels) prevent transgene expression in treated cells (arrows) and cause reduced transgene expression in general, although Müller glia that did not receive ascl1a morpholinos are able to express GFP (arrowheads). GCL, Ganglion cell layer.
Because ascl1a knockdown prevented the α1T transgene from being induced in response to injury, we wondered whether endogenous gene induction was also blocked. For these experiments, we used our −1016α1T:GFP transgenic fish so we could monitor transgene GFP expression by fluorescence microscopy and endogenous α1T expression by in situ hybridization in control and ascl1a MO-treated retinas at 4 dpi. Although the control MO had no effect on transgene expression or endogenous α1T induction (Fig. 7a–c), ascl1a knockdown almost completely abolished transgene and endogenous α1T expression in the morpholino-treated portion of the injured retina (Fig. 7d–f).
Figure 7.

Ascl1a knockdown prevents induction of endogenous α1T and pax6. Retinas from −1016 transgenic fish were injured and electroporated with lissamine-labeled MOs on day 0 and harvested on day 4. a–f, α1T expression detected by ISH is shown as a dark brown deposit (a, d), native GFP expression is shown in green (b, e), and lissamine-labeled MO is shown in red (b, e). The injury site is marked by an asterisk. a–c, Control MO treatment does not affect endogenous or transgene α1T induction (arrows). d–f, Ascl1a knockdown prevents endogenous and transgene α1T induction. Note the lack of α1T and GFP expression between the asterisk and the arrows, where the retina is treated with MO. Where the retina did not receive MO, α1T and GFP are expressed (arrows). g–l, Pax6 expression detected by ISH is shown as a blue/purple deposit, native GFP expression is shown in green, and MO is shown in red. g–i, Control MO treatment does not affect pax6 induction at 4 dpi. Arrows indicate pax6+/MO+/GFP+ Müller glia. j–l, Ascl1a MO treatment prevents pax6 induction. Arrowheads indicate MO+/pax6− cells. Rare GFP+ cells are sometimes present but were not treated with MO (arrow).
We also examined whether ascl1a knockdown could suppress the increased expression of pax6 that we observed in cycling progenitors located in the INL and ONL at 4 dpi (supplemental Table 1, available at www.jneurosci.org as supplemental material) (Fausett and Goldman, 2006). Because pax6 is constitutively expressed by amacrine cells in the uninjured retina at a position in the INL that is closest to the inner plexiform layer (Fig. 7j), we compared ectopic pax6 expression in the INL and ONL of control and ascl1a MO-treated/injured retinas by ISH (Fig. 7g–i). Interestingly, ascl1a MO-treated retinas exhibited a dramatic reduction in ectopic pax6 expression (Fig. 7j–l) compared with control MO-treated retinas (Fig. 7g–i). The observation that pax6 expression persisted in amacrine cells of the ascl1a MO-treated/injured retina (Fig. 7i) suggests that these cells control pax6 expression in an ascl1a-independent manner and demonstrate the ascl1a MOs are not causing a general decrease in gene expression.
Ascl1a is required to convert Müller glia into cycling population of retinal progenitors
The early induction of ascl1a after retinal injury and its requirement for α1T expression suggests ascl1a induction may be necessary for retina regeneration to occur. One of the hallmarks of retina regeneration is the production of a cycling population of retinal progenitors derived from Müller glia. To determine whether ascl1a induction is necessary for the generation of this cycling progenitor pool, we delivered ascl1a-targeted or control MOs to Müller glia in the injured retina. Because the vast majority of BrdU-labeled cells in the injured retina are derived from Müller glia (Fausett and Goldman, 2006), MO+/BrdU+ cells represent injury-induced retinal progenitors. We therefore injured and simultaneously electroporated −1016:GFP transgenic retinas with lissamine-labeled control or ascl1a MOs and marked dividing cells by housing the fish in BrdU-treated water from 36 to 60 h, when Müller glia are dividing. We harvested the retinas at 10 dpi and quantified the number of lissamine+/BrdU+ cells in the INL by analyzing confocal images of retinal sections (Fig. 8). We found 22% of the control MO-treated cells were labeled with BrdU, whereas two different ascl1a-targeting morpholinos resulted in only 5% (5′UTR MO) and 3.5% (ATG spanning MO) of the cells being labeled with BrdU (three retinas were counted for each group; p < 0.05). Similar results were obtained when we assayed for the earliest proliferating cells by injuring and electroporating the retina on day 0 and housing the fish in BrdU-treated water from 24–48 h and immediately harvesting retinas to assay MO+/BrdU+ cells (supplemental Fig. 5, available at www.jneurosci.org as supplemental material). Although the presence of ascl1a MO treated cells at 10 dpi suggests these MOs do not cause cell death, we examined whether any MO-treated cells were caspase+ at 2 or 4 dpi and did not detect MO+/caspase+ cells, further suggesting ascl1a MOs do not cause cell death (data not shown). Together, these results suggest ascl1a knockdown prevents Müller glia from entering the cell cycle after injury.
Figure 8.
Ascl1a is required for proliferation of Müller glia. Morpholinos were electroporated into injured −1016 retinas on day 0 and fish were housed in BrdU-treated water from 36 to 60 h after injury to label dividing cells. The fish recovered until day 10 when eyes were harvested. The control morpholino does not prevent treated cells from labeling with BrdU (arrowheads; top 3 control panels). Morpholinos targeting ascl1a (ascl1a and ascl1a5′UTR) prevent cells from labeling with BrdU (arrows; ascl1a and ascl1a5′UTR panels). Some cells are able to proliferate but were not treated with morpholino (arrowheads; ascl1a and ascl1a5′UTR panels). GCL, Ganglion cell layer.
Discussion
Zebrafish provide an ideal system for exploring the mechanisms underlying nervous system regeneration because of their robust regenerative response to injury and their amenability to experimental manipulation. We generated transgenic zebrafish models for studying regeneration in the zebrafish nervous system, and these models have further facilitated our ability to study the regenerative process (Goldman and Ding, 2000; Goldman et al., 2001; Senut et al., 2004; Fausett and Goldman, 2006). In particular, we used the α1T promoter driving EGFP expression as a reporter of the regenerative response. Transgenic fish harboring wild-type or mutant α1T promoters led us to a critical E-box that is necessary for transgene induction in retinal progenitors after injury. Investigation of the proteins that may activate the α1T promoter via this E-box led us to ascl1a, which proved to be a critical regulator of not only α1T promoter activity but also the regenerative response itself. Ascl1a was recently reported to be induced within 2 dpi, the earliest time point examined (Yurco and Cameron, 2007). Our data show ascl1a is one of the earliest gene inductions in the injured retina (first detectable by 4 hpi), preceding α1T, olig2, pax6, notch, and delta by at least 18 h. Based on their temporal expression pattern, most of these latter genes may contribute to maintaining a population of cycling progenitors, cell migration, and cell type specification. Delta-notch signaling has long been known to regulate cell fate decisions in Drosophila (Bray, 1998), and in zebrafish, notch signaling regulates neuronal cell type specification in the spinal cord (Shin et al., 2007). Pax6 has also been shown to play a role in specifying certain retinal cell types, because its removal at optic cup stages reduces the formation of most retinal neurons (Marquardt et al., 2001). α1T expression may contribute to microtubules that are involved in cell division, cell migration, and cell shape changes.
Ascl1a regulates α1T transgene expression in proliferating Müller glia
We identified a critical E-box (CATGTG) at position −954 within the α1T promoter that is necessary for transgene expression in proliferating Müller glia after retinal injury. Although we demonstrated that this E-box mediates ascl1a-dependent promoter induction, we were unable to show a direct interaction between ascl1a and this E-box in gel electrophoretic mobility shift assays. This may suggest the conditions we used are not appropriate for binding, or that ascl1a has an indirect effect on α1T promoter activity. Although we cannot distinguish between these possibilities, we note that the critical E-box in the α1T promoter deviates from the consensus mouse achaete-scute complex-like 1 (Ascl1) binding site (CAGCTG) (Hu et al., 2004). In addition, α1T activation in the injured retina lags behind ascl1a induction by at least 14 h suggesting ascl1a may induce expression of other basic helix-loop-helix proteins or must cooperate with later induced proteins to bind the −954 E-box of the α1T promoter. Nonetheless, knockdown of ascl1a in the injured retina dramatically suppressed α1T:GFP transgene and endogenous α1T expression. By using two different MOs targeting different ascl1a sequences, we ensure specificity of these results. Therefore, we conclude that ascl1a regulates α1T expression in proliferating Müller glia.
Ascl1a activates quiescent Müller glia to become multipotent progenitors
Müller glia are quiescent cells that respond to retinal injury by generating multipotent retinal progenitors. α1T transgene expression, cell cycle re-entry, and expression of other injury-induced genes are all indications that multipotent progenitors have been formed. What causes these glial cells to become actively dividing retinal progenitors? Although the signaling mechanisms are still unknown, the results presented here suggest ascl1a expression is necessary for converting quiescent Müller glia into actively proliferating progenitors. This conclusion is based on MO-mediated knockdown of ascl1a in adult Müller glia at the time of retinal injury, and the observation that ascl1a is induced within 4 hpi, which is 18 h before the beginning of Müller glia proliferation (Fausett and Goldman, 2006).
This proposed role for ascl1a in the regenerating retina is very different from its role in the developing retina, where ascl1 appears to collaborate with other bHLH and homeodomain transcription factors to initiate cell cycle exit and specify retinal neuronal subtype (Akagi et al., 2004; Wang and Harris, 2005; Guillemot, 2007). However, in other parts of the nervous system such as the medial ganglionic eminence of the telencephalon and olfactory epithelium (Cau et al., 1997; Casarosa et al., 1999), ascl1 does contribute to the generation of neural precursors, where it seems to play a role in specifying stem cells to commit to a particular type of transiently proliferating neural progenitor (Torii et al., 1999). We suspect ascl1a expression in zebrafish Müller glia behaves similarly contributing to its transition from a quiescent cell to an actively dividing retinal progenitor. Although the mechanism by which ascl1a accomplishes this is not known, it is possible that the molecular environment and/or ascl1a expression level of Müller glia is such that ascl1a-dependent dedifferentiation and cellular proliferation are favored over cell-cycle exit and retinal cell type differentiation. Ascl1a likely specifies Müller glia to become injury-induced progenitors by collaborating with other transcription factors to initiate the expression of target genes such as α1T and cell cycle regulators (Kassen et al., 2007). Interestingly, the cell cycle regulator cyclin D1 is induced in the injured retina (Kassen et al., 2007) and harbors an E-box (CATGTG) in its promoter that matches the α1T ascl1a-dependent E-box in sequence and position (data not shown).
Might ascl1a also contribute to retinal cell type specification? This is an interesting question, because ascl1a is normally thought to contribute to cell cycle exit and cellular differentiation, and ascl1a expression rises to its maximal level at ∼48 hpi (supplemental Table 1, available at www.jneurosci.org as supplemental material) and persists up to at least 7 dpi (Yurco and Cameron, 2007). However, this question is difficult to address experimentally, because it would require us to knockdown ascl1a expression in the regenerating retina at a time when retinal progenitors are actively dividing (at ∼4 dpi). Because MO-mediated gene knockdown is most effective in the vicinity where it is injected, this would require one to introduce the MO into the injured retina in the same place where the initial needle poke injury occurred, which is very difficult. Therefore, this question may be better addressed by using intense light to injure the retina, which results in a more uniform injury response (Kassen et al., 2007).
The rapid induction of ascl1a in Müller glia after retinal injury suggests it may be regulated by transcription factors already present in Müller glia. One such candidate is stat3, which is expressed in quiescent Müller glia and may be activated by injury-induced extracellular cues (Kassen et al., 2007). Interestingly, FGF can cause Müller glia to re-enter the cell cycle in the absence of injury (Fischer et al., 2002), and ascl1a appears to act downstream of FGF signaling in developing zebrafish pituitary gland (Herzog et al., 2004). Another potential regulator is the wnt signaling pathway. In rats, adding wnt3a to retinal explant cultures increases Müller glia proliferation (Osakada et al., 2007). Wnt signaling is mediated by nuclear localization of β-catenin, which activates genes harboring T cell transcription factor binding sites. Whether wnt3a/β-catenin mediates its effect via ascl1a is not known. Future studies will begin to address the role these signal transduction cascades play in regeneration of the zebrafish retina.
Footnotes
This work was supported by a research grant from the National Eye Institute and by funds from the University of Michigan Endowment for the Basic Sciences (D.G.). B.V.F. was supported in part by a National Institutes of Health Vision Research Training grant. We thank members of the Goldman laboratory and Professors Ben Novitch and Dave Turner for comments and suggestions on this work. We also thank Eric Weinberg, Matthias Hammerschmidt, Bruce Appel, Julian Lewis, Michael Lardelli, and Audrey Seasholtz for providing cDNA clones. We thank Richard Griggs for generating custom electroporation electrodes.
References
- Akagi T, Inoue T, Miyoshi G, Bessho Y, Takahashi M, Lee JE, Guillemot F, Kageyama R. Requirement of multiple basic helix-loop-helix genes for retinal neuronal subtype specification. J Biol Chem. 2004;279:28492–28498. doi: 10.1074/jbc.M400871200. [DOI] [PubMed] [Google Scholar]
- Barthel LK, Raymond PA. In situ hybridization studies of retinal neurons. Methods Enzymol. 2000;316:579–590. doi: 10.1016/s0076-6879(00)16751-5. [DOI] [PubMed] [Google Scholar]
- Bernardos RL, Barthel LK, Meyers JR, Raymond PA. Late-stage neuronal progenitors in the retina are radial Müller glia that function as retinal stem cells. J Neurosci. 2007;27:7028–7040. doi: 10.1523/JNEUROSCI.1624-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray S. Notch signalling in Drosophila: three ways to use a pathway. Semin Cell Develop Biol. 1998;9:591–597. doi: 10.1006/scdb.1998.0262. [DOI] [PubMed] [Google Scholar]
- Cameron DA, Gentile KL, Middleton FA, Yurco P. Gene expression profiles of intact and regenerating zebrafish retina. Mol Vis. 2005;11:775–791. [PubMed] [Google Scholar]
- Casarosa S, Fode C, Guillemot F. Mash1 regulates neurogenesis in the ventral telencephalon. Development. 1999;126:525–534. doi: 10.1242/dev.126.3.525. [DOI] [PubMed] [Google Scholar]
- Cau E, Wilson SW. Ash1a and neurogenin1 function downstream of floating head to regulate epiphysial neurogenesis. Development. 2003;130:2455–2466. doi: 10.1242/dev.00452. [DOI] [PubMed] [Google Scholar]
- Cau E, Gradwohl G, Fode C, Guillemot F. Mash1 activates a cascade of bHLH regulators in olfactory neuron progenitors. Development. 1997;124:1611–1621. doi: 10.1242/dev.124.8.1611. [DOI] [PubMed] [Google Scholar]
- Fausett BV, Goldman D. A role for alpha1 tubulin-expressing Müller glia in regeneration of the injured zebrafish retina. J Neurosci. 2006;26:6303–6313. doi: 10.1523/JNEUROSCI.0332-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fimbel SM, Montgomery JE, Burket CT, Hyde DR. Regeneration of inner retinal neurons after intravitreal injection of ouabain in zebrafish. J Neurosci. 2007;27:1712–1724. doi: 10.1523/JNEUROSCI.5317-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer AJ, Reh TA. Müller glia are a potential source of neural regeneration in the postnatal chicken retina. Nat Neurosci. 2001;4:247–252. doi: 10.1038/85090. [DOI] [PubMed] [Google Scholar]
- Fischer AJ, McGuire CR, Dierks BD, Reh TA. Insulin and fibroblast growth factor 2 activate a neurogenic program in Müller glia of the chicken retina. J Neurosci. 2002;22:9387–9398. doi: 10.1523/JNEUROSCI.22-21-09387.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman D, Ding J. Different regulatory elements are necessary for alpha1 tubulin induction during CNS development and regeneration. NeuroReport. 2000;11:3859–3863. doi: 10.1097/00001756-200011270-00051. [DOI] [PubMed] [Google Scholar]
- Goldman D, Hankin M, Li Z, Dai X, Ding J. Transgenic zebrafish for studying nervous system development and regeneration. Transgenic Res. 2001;10:21–33. doi: 10.1023/a:1008998832552. [DOI] [PubMed] [Google Scholar]
- Guillemot F. Spatial and temporal specification of neural fates by transcription factor codes. Development. 2007;134:3771–3780. doi: 10.1242/dev.006379. [DOI] [PubMed] [Google Scholar]
- Herzog W, Sonntag C, Walderish B, Odenthal J, Maischein HM, Hammerschmidt M. Genetic analysis of adenohypophysis formation in zebrafish. Mol Endocrinol. 2004;18:1185–1195. doi: 10.1210/me.2003-0376. [DOI] [PubMed] [Google Scholar]
- Hieber V, Dai X, Foreman M, Goldman D. Induction of alpha1-tubulin gene expression during development and regeneration of the fish central nervous system. J Neurobiol. 1998;37:429–440. doi: 10.1002/(sici)1097-4695(19981115)37:3<429::aid-neu8>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- Hitchcock PF, Macdonald RE, VanDeRyt JT, Wilson SW. Antibodies against Pax6 immunostain amacrine and ganglion cells and neuronal progenitors, but not rod precursors, in the normal and regenerating retina of the goldfish. J Neurobiol. 1996;29:399–413. doi: 10.1002/(SICI)1097-4695(199603)29:3<399::AID-NEU10>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Hu Y, Wang T, Stormo GD, Gordon JI. RNA interference of the achaete-scute homolog 1 in mouse prostate neuroendocrine cells reveals its gene targets and DNA binding sites. Proc Natl Acad Sci USA. 2004;101:5559–5564. doi: 10.1073/pnas.0306988101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasoni CL, Reh TA. Temporal and spatial pattern of MASH 1 expression in the developing rat retina demonstrates progenitor cell heterogeneity. J Comp Neurol. 1996;369:319–327. doi: 10.1002/(SICI)1096-9861(19960527)369:2<319::AID-CNE11>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Kassen SC, Ramanan V, Montgomery JE, Burket C, Liu CG, Vihtelic TS, Hyde DR. Time course analysis of gene expression during light-induced photoreceptor cell death and regeneration in albino zebrafish. Develop Neurobiol. 2007;67:1009–1031. doi: 10.1002/dneu.20362. [DOI] [PubMed] [Google Scholar]
- Marquardt T, Ashery-Padan R, Andrejewski N, Scardigli R, Guillemot F, Gruss P. Pax6 is required for multipotent state of retinal progenitor cells. Cell. 2001;105:43–55. doi: 10.1016/s0092-8674(01)00295-1. [DOI] [PubMed] [Google Scholar]
- Mensinger AF, Powers MK. Visual function in regenerating teleost retina following surgical lesion. Vis Neurosci. 2007;24:1–9. doi: 10.1017/S0952523807070265. [DOI] [PubMed] [Google Scholar]
- Nordeen SK. Luciferase reporter gene vectors for analysis of promoters and enhancers. BioTechniques. 1998;6:454–458. [PubMed] [Google Scholar]
- Ooto S, Akagi T, Kageyama R, Akita J, Mandai M, Honda Y, Takahashi M. Potential for neural regeneration after neurotoxic injury in the adult mammalian retina. Proc Natl Acad Sci USA. 2004;101:13654–13659. doi: 10.1073/pnas.0402129101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osakada F, Ooto S, Akagi T, Mandai M, Akaike A, Takahashi M. Wnt signaling promotes regeneration in the retina of adult mammals. J Neurosci. 2007;27:4210–4219. doi: 10.1523/JNEUROSCI.4193-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senut M, Gulati-Leekha A, Goldman D. An element in the α1-tubulin promoter is necessary for retinal expression during optic nerve regeneration but not after eye injury in the adult zebrafish. J Neurosci. 2004;24:7663–7673. doi: 10.1523/JNEUROSCI.2281-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, Poling J, Park H, Appel B. Notch signaling regulates neural precursor allocation and binary neuronal fate decisions in zebrafish. Development. 2007;134:1911–1920. doi: 10.1242/dev.001602. [DOI] [PubMed] [Google Scholar]
- Torii M, Matsuzaki F, Osumi N, Kaibuchi K, Nakamura S, Casarosa S, Guillemot F, Nakafuku M. Transcription factors Mash-1 and Prox-1 delineate early steps in differentiation of neural stem cells in the developing central nervous system. Development. 1999;126:443–456. doi: 10.1242/dev.126.3.443. [DOI] [PubMed] [Google Scholar]
- Vihtelic TS, Hyde DR. Light-induced rod and cone cell death and regeneration in the adult albino zebrafish (Danio rerio) retina. J Neurobiol. 2000;44:289–307. doi: 10.1002/1097-4695(20000905)44:3<289::aid-neu1>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- Wang JC-C, Harris WA. The role of combinational coding by homeodomain and bHLH transcription factors in retinal cell fate specification. Dev Biol. 2005;285:101–115. doi: 10.1016/j.ydbio.2005.05.041. [DOI] [PubMed] [Google Scholar]
- Yun K, Fischman S, Johnson J, Hrabe de Angelis M, Weinmaster G, Rubenstein JL. Modulation of the notch signaling by Mash1 and Dlx1/2 regulates sequential specification and differentiation of progenitor cell types in the subcortical telencephalon. Development. 2002;129:5029–5040. doi: 10.1242/dev.129.21.5029. [DOI] [PubMed] [Google Scholar]
- Yurco P, Cameron DA. Cellular correlates of proneural and notch-delta gene expression in the regenerating zebrafish retina. Vis Neurosci. 2007;24:437–443. doi: 10.1017/S0952523807070496. [DOI] [PubMed] [Google Scholar]