Abstract
The Escherichia coli Lon protease degrades the E. coli DNA-binding protein HUβ, but not the related protein HUα. Here we show that the Lon protease binds to both HUβ and HUα, but selectively degrades only HUβ in the presence of ATP. Mass spectrometry of HUβ peptide fragments revealed that region K18-G22 is the preferred cleavage site, followed in preference by L36-K37. The preferred cleavage site was further refined to A20-A21 by constructing and testing mutant proteins; Lon degraded HUβ-A20Q and HUβ-A20D more slowly than HUβ. We used optical tweezers to measure the rupture force between HU proteins and Lon; HUα, HUβ, and HUβ-A20D can bind to Lon, and in the presence of ATP, the rupture force between each of these proteins and Lon became weaker. Our results support a mechanism of Lon protease cleavage of HU proteins in at least three stages: binding of Lon with the HU protein (HUβ, HUα, or HUβ-A20D); hydrolysis of ATP by Lon to provide energy to loosen the binding to the HU protein and to allow an induced-fit conformational change; and specific cleavage of only HUβ.
Introduction
The DNA-binding histone-like proteins HUα (HU2) and HUβ (HU1) of Escherichia coli are encoded by two closely related genes, hupA and hupB, located at 90 and 9.8 min of the chromosome, respectively (1,2). The 90-amino-acid HUα and HUβ proteins are 68.9% identical; the functional protein, HU, is isolated mainly as an αβ heterodimer (3,4). HU is highly conserved among prokaryotes, but bacterial species other than E. coli and Salmonella typhimurium synthesize only one type of HU subunit, which forms a homodimer (5,6).
HU plays important pleiotropic roles in DNA replication (7), gene regulation (8,9), translation (10), DNA supercoiling (11,12), and other processes (13). A connection between HU and the ATP-dependent Lon protease is suggested by the role of both Lon and HU in the mucoid phenotype of E. coli (14,15). Lon is most likely involved in controlling gene expression in bacteria and yeast, either by regulating the levels of transcription factors (16,17) or by influencing the structural stability of genomic DNA (18,19). A lon mutation has a pleiotropic phenotype: ultraviolet sensitivity, mucoidy, deficiency for lysogenization by bacteriophage λ and P1, and lower efficiency in the degradation of abnormal proteins (15,20–24). Based on the results of in vivo experiments, it has been suggested that the regulation of HU in E. coli is Lon-dependent and that HU is degraded by Lon or a Lon-dependent protease (25).
Lon protease (EC 3.4.21.53) is a member of the AAA+ superfamily (ATPases associated with diverse cellular activities). This superfamily is characterized by a conserved segment of 220–250 amino acids, referred to as the AAA domain or nucleotide binding domain, which contains several conserved motifs for ATP binding and hydrolysis. Lon and other known members of this superfamily, e.g., FtsH, ClpAP, ClpXP, ClpC, and HslVU are all ATP-dependent proteases (26). Lon and FtsH protease carry both the ATPase and the proteolytic active sites within a single polypeptide chain. In the Clp family, these functions are encoded by separate polypeptide chains. Lon protease functions as a homooligomer, in which each subunit consists of three domains: the amino-terminal domain, possibly involved in substrate recognition and binding (27); the ATPase domain (A domain), containing the ATP-binding motif (28); and the carboxy-terminal proteolytic domain (P domain) (29). Like a molecular chaperone, Lon recognizes a broad range of proteins, both misfolded and properly folded, and mediates their turnover. Through the degradation of various specialized proteins, Lon protease also regulates a number of biological functions (30). These specialized proteins include the λ N protein (31,32), the SulA cell division regulator (33,34), the RcsA positive regulator of capsule synthesis (35), the F factor addiction system protein CcdA (36), and the DNA-binding protein HU (25).
We have previously cloned and investigated the functional domains of the Lon protease from Brevibacillus thermoruber (37,38). Here, we elucidate in detail the substrate recognition of the E. coli Lon protease in the known cleavage of HUβ and the lack of cleavage of HUα (25).
Materials and Methods
Preparation of Lon and HU
Lon protease from Br. thermoruber (Bt-Lon) and E. coli (Ec-Lon) were prepared as described previously (37–39). We sequenced the prepared Ec-Lon and found an E269G mutation. Proteolysis activity was measured in a peptidase assay. E. coli hupA and hupB were cloned from the commercial E. coli strain ECOS 101 (Yeastern Biotech, Taipei, Taiwan, Republic of China). The amino acid sequences of isolated HUα and HUβ were the same as those reported previously (National Center for Biotechnology Information accession numbers BAB38346 and BAB33917). The concentration of the protein in the pooled fractions was determined using the Bradford method (BioRad Laboratories, Hercules, CA).
Identification of the initial cleavage sites
E. coli HUβ (8.4 μg), E. coli HUα (11.3 μg), and Bacillus subtilis HU (Bs-HU) (10.2 μg) were each incubated at 37°C with Ec-Lon (15.2 μg) and at 50°C with Bt-Lon (10 μg) in buffer containing 25 mM Tris-HCl (pH 8.0), 75 mM NaCl, 5 mM MgCl2, 5 mM CaCl2, 5μM ATP, and 10% glycerol in a total volume of 200 μl. After incubation, the reaction was stopped by adding 500 μl 95% acetonitrile/5% H2O/0.1% trifluoroacetic acid. The reaction mixture was centrifuged at 16,060 × g for 10 min. The supernatant was dried, and the residue was dissolved in 5% acetonitrile/95% H2O/0.1% trifluoroacetic acid. The solution was then applied to a Waters C18 column (5 μm particle size, 250 × 4.6 mm), and eluted by reverse-phase high-performance liquid chromatography (1100 series, Agilent, Santa Clara, CA) with a gradient of acetonitrile (5–95% in 30 min). Fractions were collected and lyophilized. Peptides of HU proteins were identified using a nanoESI-Q-TOF mass spectrometer (MicroMass, Cary, NC) (40) with a peptide mass tolerance of 0.25 Da and a tandem mass spectroscopy ion mass tolerance of 0.25 Da with up to one missing cleavage.
Model construction of the homodimeric HU, E. coli HUβ2
The amino acid sequences of E. coli HUα and HUβ were used for BLAST analysis and then aligned with identified homologous proteins using the program CLUSTAL-W (41). The high-resolution structure of HUα (Protein Data Bank code 1MUL) (42), incomplete due to a lacking electron density map, was used to model the main-chain conformation of HUβ. For HUα, the MODELER program of InsightII (Accelrys, San Diego, CA) was used to generate the missing loop (Arg58-Ile72) in the DNA-binding arms (43). This allowed us to obtain a 3-D structure of HUα with a complete internal loop region. The homodimeric proteins were then assembled by superimposing the monomeric model with the crystal structure of HUα. The structure with the lowest violation score and lowest energy was chosen as the candidate. The distribution of the backbone dihedral angles of the model was evaluated using a Ramachandran plot and the program PROCHECK (44). The ProStat module of InsightII was used to analyze the properties of bonds, angles, and torsions. The Profile-3D program was used to check the structure and sequence compatibility (45).
Circular dichroism spectra of HUβ and HUβ-A20D
E. coli HUβ was incubated at 37°C with Ec-Lon in 25 mM Tris-HCl (pH 8.0), 75 mM NaCl, 5 mM MgCl2, 5 mM CaCl2, and 10% glycerol in a total volume of 25 μl. After incubation, the reaction was stopped by adding 200 μl of the same buffer and freezing at −80°C. The samples were melted before Circular dichroism (CD) measurements. CD spectra were obtained with a J-715 spectropolarimeter (JASCO, Tokyo, Japan). The far-ultraviolet CD spectra were the mean of five accumulations with a 0.1-cm light path. The final concentrations of Ec-Lon, HUβ, HUβ-A20D, and ATP were 3.0 μg ml−1, 23 μg ml−1, 23 μg ml−1, and 0.4 μM, respectively.
Optical tweezers
A schematic diagram of the optical tweezers setup is shown in Fig. S3 in the Supporting Material. A linearly polarized laser beam (λ = 1064 nm, 300 mW, Nd:YVO4 cw laser, LeadLight Technology, Taoyuan, Taiwan, Republic of China) was used for optical trapping. A half-wave plate and a polarizer were used to control the trapping-laser power by rotating the half-wave plate. A beam expander was used to expand and collimate the beam such that the beam diameter (∼1 cm) slightly overfilled the back aperture of the microscope objective. A telescope (telescope 1, consisting of two identical lenses, each with focal length f = 150 mm) was used to change the trapping-beam pathways to manipulate the trapped particle by moving the first lens (in telescope 1). A second laser beam (wavelength 632.8 nm, 5 mW, He-Ne cw laser, Uniphase, Ottawa, Ontario, Canada) provided a tracking beam to track the position of the trapped particle. A spatial filter/beam expander unit was used to expand and collimate the beam to a beam diameter of ∼1 cm. A second telescope (telescope 2) was used to adjust the relative positions of the focal points of the two laser beams in the sample chamber. The two laser beams were combined by a dichroic mirror (DM1, SRR-25.4, Lambda Research Optics, Littleton, MA), which is highly reflective at 632.8 nm and highly transmissive at 1064 nm, and injected into the back aperture of an oil-immersion microscope objective (NA 1.24, 100×, working distance 0.13 mm, Olympus, Melville, NY). The tracking beam, scattered by and diffracted off the trapped particle, was collected by a condenser (NA 0.42, 50×, working distance 20.5 mm, Mitutoyo, Tokyo, Japan) and projected onto a quadrant photodiode (QPD; S7479, Hamamatsu, Hamamatsu City, Japan) to track the motion of the particle in the transverse plane. A notch filter was used to block the trapping beam (1064 nm) from entering the QPD. The electrical signals from the QPD were recoded by a data acquisition system, and wide-field images of the trapped particle were captured by a charge-coupled device camera (WAT-120N, Watec, Vienna, Austria) for optical alignment of the trap and for image observation and analysis.
Immobilized proteins on beads
Polystyrene beads (10 μl) were washed with buffer containing 25 mM Tris-HCl (pH 8.0), 75 mM NaCl, 5 mM MgCl2, 5 mM CaCl2, and 10% glycerol. Large polystyrene beads (12.3 μm in diameter) were incubated with Lon protease at 4°C for 18 h. Small polystyrene beads (2.88 μm in diameter) were incubated at 4°C for 18 h with HUα, HUβ, HUβ-A20D, or bovine serum albumin (BSA). After incubation, the beads were washed three times with 1 ml of the same buffer. The beads were then incubated with 1 ml BSA (10 mg/ml in phosphate-buffered saline) before measurement with optical tweezers. Beads with or without protein coating were coated with 20 nm gold and inspected by scanning electron microscopy (S-2700, Hitachi, Tokyo, Japan) (Fig. S6), using 5000× magnification for large beads and 15,000× for small beads.
Rupture force measurements
Optical tweezers were used to measure the strength of the interaction between HU proteins and Lon protease coated on polystyrene beads (Fig. S5). One bead (2.88 μm in diameter) coated with an HU protein was trapped by the optical tweezers. Its position was tracked by QPD. Another bead (12.3 μm in diameter) coated with the Lon protease and adhered to a coverglass was placed near the trapped bead (Fig. S8). At the beginning of each experiment, the small bead was trapped by optical tweezers at the beam axis, and the displacement of the trapped bead, deduced from the calibrated QPD output signal, was set to 0 μm by fine adjustment of the QPD lateral position. The Lon-protease-coated bead on the coverglass was moved toward the trapped bead semicontinuously, at 37 nm/step/s, by a piezoelectric-transducer (PZT)-driven sample stage until the trapped bead was pushed away by ∼60 nm from the original equilibrium position, as deduced from the precalibrated QPD signal. When the two beads touched, as shown in Fig. S8 A, the stage was stopped for 3 min to allow the enzyme to interact with the proteins. Then, the PZT-driven sample stage was moved semicontinuously (at 37 nm/step/s) away from the HU-protein-coated bead (Fig. S8 B). If the two beads are bound by a protein-protein interaction, the small bead will be gradually pulled away from the trapping center by the larger bead. Since the optical force on the small bead is proportional to its distance from the trapping center (within a linear range on the order of 100 nm), the optical force increased as the displacement of the bead increased. If the trapping force exceeds the enzyme-substrate binding force, the two beads would be pulled apart, and the 2.88-μm bead coated with the HU proteins would bounce back to the trapping center (Fig. S8 C). The maximum displacement of the small bead was measured by QPD and converted into force using Hooke's law. Data of noninteracting cases were not used. Only those cases which showed clear rupture force were taken into account. The force as a function of the displacement of the larger bead (12.3 μm), coated with the Lon protease, is shown in Fig. S9. The three stages described above are illustrated schematically in the force-versus-position plot in Fig. S8 D. In stage B, the optical force increased as the large bead moved away from the small bead, until the force was sufficient to rupture the binding force between the two beads. The maximum force in the plot appears at the boundary between stages B and C, when the two beads are pulled apart (Fig. S8 D). The rupture force that pulled the two beads apart was equal to the maximum force of the force-versus-position plot. The system was computer-controlled (via LAB VIEW) to maintain stability and to allow reproducibility.
Substrate interference assay
Protease activity was measured as described previously (46). The substrate, fluorescein isothiocyanate (FITC)-α-casein (10 μg; Sigma, St. Louis, MO), was incubated with Lon protease (4.6 μg) in 5 mM Tris-HCl (pH 8.0), 1 mM MgCl2, and 1 mM ATP, with ∼2.3 × 10−9 mol of the competing substrate BSA, casein, HUα, HUβ, or HUβA20D in 200 μl. After 1 h incubation at 37°C, 100 μg BSA was added. To precipitate proteins, 100 μl of 10% trichloroacetic acid was added and the solution was placed on ice for 10 min. The solution was then centrifuged, and the pellet was discarded. The pH of the solution was adjusted with 0.2 ml CHES-Na (0.5 M, pH 12.0). Activity was assayed by measuring the fluorescence of the solution (excitation 490 nm, emission 525 nm).
Results
Ec-Lon selectively degrades E. coli HUβ in the presence of ATP
E. coli HU exists mostly as an HUαβ heterodimer, and sometimes as a homodimer, and the composition of the dimer varies during growth (47). Exponentially growing E. coli contains a mixture of HUα2 and HUαβ, but not HUβ2. This indicates that HUβ2 is unstable and might therefore be susceptible to protease processing. To determine whether Lon is the responsible protease, we cloned and expressed HU and Lon from E. coli. After incubation of E. coli HUα or HUβ with Ec-Lon, the samples were denatured and analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). E. coli Lon degraded HUβ in the presence of ATP (Fig. 1 A), but it did not degrade HUα (Fig. 1 B), even after overnight incubation (not shown). This is consistent with a previous in vivo study showing that HUα is not degraded by Lon or Lon-dependent protease (25).
Figure 1.
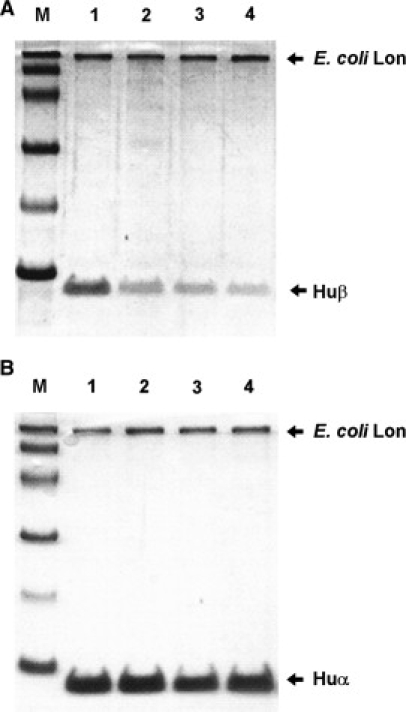
Analysis of the substrates of the E. coli Lon protease by SDS-PAGE. E. coli HUβ (A) and E. coli HUα (B) were incubated with the Lon protease and 5 μM ATP for 0 (lane 1), 1 (lane 2), 2 (lane 3), and 3 h (lane 4). M, size markers (94, 66, 45, 30, 20, and 14 kDa). The reaction volume of the sample was 20 μl. The amount of Lon protease was 1 μg and that of the substrate HUβ was ∼3 μg.
Cleavage sites of the Lon proteases
After incubation of HUβ with Ec-Lon protease and ATP for 30 min, the peptide fragments formed were separated by liquid chromatography and analyzed by nanoESI-Q-TOF mass spectrometry. One main peak at 793.86, 2+, was identified. Mass spectrometry of this peak yielded five major peaks (Fig. S3): 1∗, 584.31; 2∗, 655.34; 3∗, 768.43; 4∗, 881.52; and 5∗, 952.57. Peaks of other peptide fragments were also observed. The mass spectra data were combined into a single-peak list file, which was searched using the program MASCOT. We found 29 peptides that matched; a significant hit with a Mowse score of 1189 indicated that HUβ is a substrate of Ec-Lon. After 30, 10, and 5 min incubation of HUβ with Ec-Lon protease and ATP, 21, 9, and 3 HUβ peptide fragments, respectively, with a Mowse score >25 were formed (Table S1). The peptide fragment AGRALDAIIASVTESL (peptide 9, molecular weight 1585.80) was found in all reactions.
We identified two preferred cleavage sites and regions by aligning the peptide fragments K18-G22 (KAAAG) and L36-K37. The three peptide fragments formed after 5 min incubation of Ec-Lon with HUβ are cleaved at one or both of these sites. We therefore postulated that Ec-Lon preferentially cleaves at the C-terminal end of alanine or leucine, and cleavage occurs within region K18-G22 or region L36-K37 or both, at the beginning of the reaction.
Ec-Lon specificity
We analyzed the specificity of Ec-Lon for HUβ by comparing the amino acid sequences of E. coli HUα, E. coli HUβ, and Bs-HU. HUα and HUβ are 68.9% identical and 80% similar; HUα and Bs-HU are 56.5% identical and 67.4% similar, and HUβ and Bs-HU are 52.2% identical and 75% similar. The sequence alignment of these three proteins (Fig. 2 A) reveals that they differ in amino acid residues 18–22, one of the preferred cleavage sites of Ec-Lon in HUβ: HUβ, KAAAG; HUα, KTQAK; and Bs-HU, KKDAT. HUα and Bs-HU, which are not degraded by Ec-Lon, lack the A-A bond cleaved by Ec-Lon in HUβ.
Figure 2.
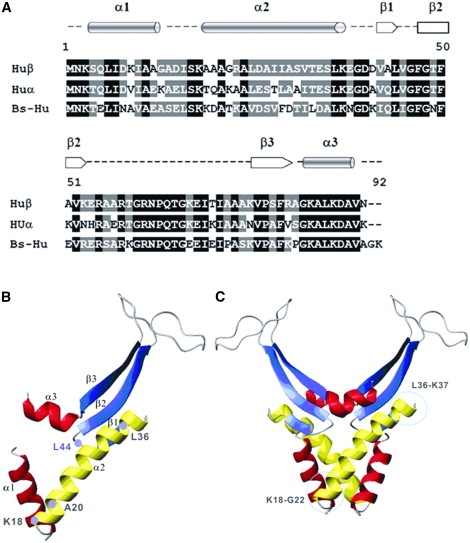
Structures of E. coli HUβ, E. coli HUα, and B. subtilis HU. (A) Alignment of the sequences using Vector NTI Advance 9. Residues identical in all sequences are highlighted in black. Similar residues are highlighted in gray. The secondary structure of the sequence is shown above the primary sequence. (B) The tertiary structure of the Ec-HUβ monomer composed of three α-helices and three β-sheets. Residues K18, A20, L36, and L44, which are part of the preferred cleavage sites, are indicated. The α1-helix and the α3-helix are shown in red, and the α2-helix is shown in yellow. The β-sheets are shown in blue. (C) The quaternary structure of the HUβ homodimer, which can be considered as the association of an α-subdomain and a β-subdomain (42). The DNA-binding domain is located in the β-subdomain. Peptide 9 is located in the α-subdomain.
Although HUα and Bs-HU contain the L36-K37 bond that is cleaved by Ec-Lon in HUβ, Ec-Lon did not cleave either of these proteins, probably because of a difference between them and HUβ somewhere else in the amino acid sequence. We analyzed this hypothesis by creating a homology-based model of HUβ (Fig. 2 B). The structure of HUα (PDB code 1MUL) (42) was used to model the main-chain conformation of HUβ. The HUβ peptide fragment A21GRALDAIIASVTESL36, which is formed by cleavage at both of the preferred cleavage sites, comprises the main part of the α2-helix, with A20-A21 and L36-K37 at the N- and C-terminal ends, respectively, of this helix. The sequence between G13 and K18 is a loop that links the α1- and α2-helices. In the HUα monomer, the tertiary fold is stabilized by intrahelical salt bridges between K22 and E26 (42). In HUβ, however, these amino acid residues differ, and G22 cannot interact with D26. Therefore, the α2-helix of HUβ is weaker than that of HUα. This might explain why Ec-Lon can degrade HUβ, but not HUα.
The main Ec-Lon cleavage site in HUβ
We further tested the main Ec-Lon cleavage site at A20-A21 by constructing HUβ proteins with an A20Q or an A20D mutation. After incubation of HUβ or HUβ-A20Q with Ec-Lon and ATP for 7 h, SDS-PAGE analysis of the products showed that HUβ was completely degraded and that the same concentration of HUβ-A20Q was degraded only to a small extent (Fig. S4). The minor bands that appeared are only Lon protease autodegradation products (48). These results indicated that the A20Q mutation reduced the degradation rate of HUβ by Ec-Lon. We also analyzed the reaction of Ec-Lon and HUβ, as well as that of Ec-Lon and HUβ-A20D, using SDS-PAGE (Fig. 3) and CD spectrometry (Fig. 4). SDS-PAGE indicated that HUβ-A20D is resistant to degradation by Ec-Lon (Fig. 3). The CD spectrum of Ec-Lon incubated with HUβ revealed two negative peaks at 205 and 222 nm and was similar to the spectrum of HUβ at the same concentration. After incubation of Ec-Lon, HUβ, and ATP for 6 h, the spectrum showed that HUβ was degraded by Ec-Lon, and a random coil signature was evident (Fig. 4 A). In contrast, after incubation of Ec-Lon, HUβ-A20D, and ATP for 6 h, the spectrum indicated an α-helix as a major secondary structure (Fig. 4 B), i.e., Lon protease degraded HUβ-A20D more slowly than it degraded HUβ. These results indicate that residue A20 has a powerful impact on the degradation of HUβ by Lon protease. Recently, synthetic peptides were used to evaluate the interaction of amino acid residues in peptide substrate with the proteolytic site of Ec-Lon (49). The degradation rate of the substrate peptide with glutamic acid was significantly slower compared to the rate with serine under the same substrate concentration, which implies that negative charge is not favored at the P1′ site of the Ec-Lon substrate. Our data support this point of view.
Figure 3.
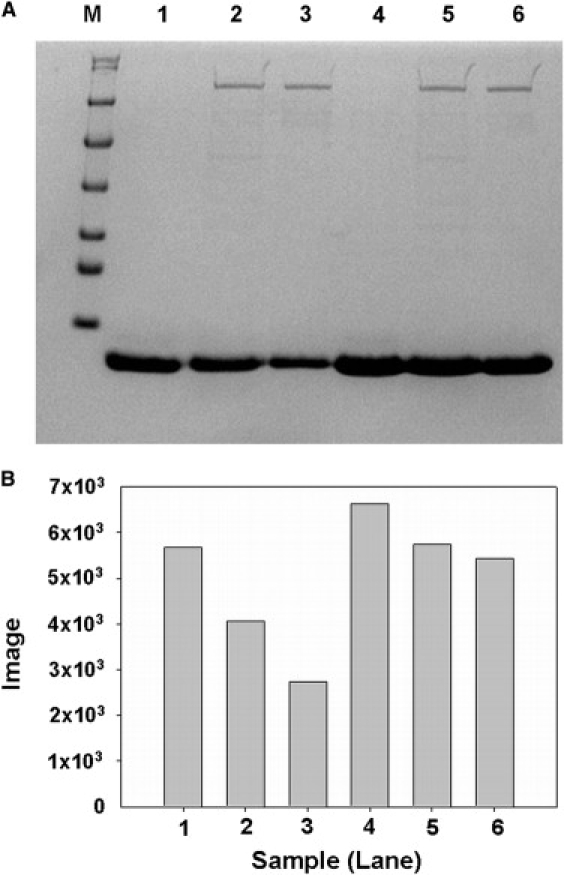
Importance of residue 20 in the cleavage of HUβ by Lon. (A) HUβ-A20D (0.08 mg ml−1) and HUβ (0.08 mg ml−1) were each incubated with Ec-Lon (0.1 mg ml−1) with or without ATP (5 μM) for 6 h. Samples were then denatured by heating and analyzed by SDS-PAGE. Amounts loaded were 3 μg HUβ, 3 μg HUβ-A20D, and 1 μg Ec-Lon. M, Size markers (116, 66, 45, 35, 25, 18.4, and 14.4 kDa); lane 1, HUβ; lane 2, Ec-Lon and HUβ without ATP; lane 3, Ec-Lon and HUβ with ATP; lane 4, HUβ-A20D; lane 5, Ec-Lon and HUβ-A20D without ATP; and lane 6, Ec-Lon and HUβ-A20D with ATP. (B) The bands of HUβ and HUβ-A20D were analyzed by IQuant software (Molecular Dynamics).
Figure 4.
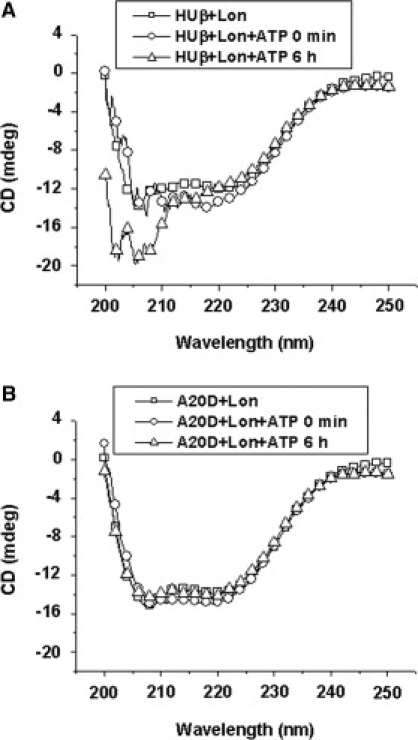
(A) Degradation of HUβ by Lon protease. (B) Lack of degradation of mutant HUβ-A20D by Lon protease. The change in the CD spectrum of HUβ incubated with Lon and ATP for 6 h indicates the formation of a random coil, i.e., degradation.
Force measurements
We determined the rupture force of HU-Lon protein-protein binding using HU-coated polystyrene beads and optical tweezers. The force was calculated from the displacement of the small bead when the force-versus-position curve peaked and the rupture force broke the binding (see Fig. S8 D). The average rupture forces in the presence and absence of ATP were determined (Fig. 5). Without ATP, the average rupture forces of Ec-Lon with HUα, HUβ, and HUβ-A20D were 24 ± 6, 28 ± 5, and 31 ± 4 pN, respectively, i.e., the binding forces between Ec-Lon and each of the three different HU proteins are similar. In the presence of ATP, the average rupture forces of Ec-Lon with HUα, HUβ, and A20D-HUβ were 18 ± 2, 11 ± 3, and 15 ± 2 pN, respectively, i.e., all of the rupture forces decreased in the presence of ATP. The average and the standard deviation were obtained by repeating the experiments with ∼10 pairs of samples for each case (see Table S3 for details).
Figure 5.
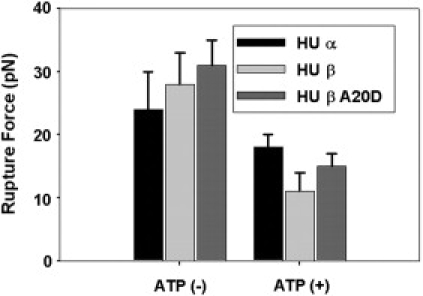
Results of rupture-force measurements. The average rupture forces of HUα, HUβ, and A20D-HUβ without ATP were 24 ± 6, 28 ± 5, and 31 ± 4 pN, respectively. The average rupture forces of HUα, HUβ, and A20D-HUβ became smaller in the presence of ATP and they were 18 ± 2, 11 ± 3, and 15 ± 2 pN, respectively.
Substrate interference assay
We further studied Lon protease in a substrate interference assay using FITC-α-casein in competition with equimolar concentrations of unlabeled HUα, HUβ, A20D-HUβ, casein, or BSA (Fig. 6). The fluorescence of cleaved FITC-α-casein was measured at 525 nm. Cleavage of the other, unlabeled substrate decreased the amount of FITC-α-casein cleaved, and therefore lowered the fluorescence. All proteins except BSA competed similarly with FITC-α-casein as substrates of Ec-Lon. In keeping with these results, it has been reported previously that native BSA is nondegradable by Lon protease (50), and casein and HUβ are both substrates of Lon protease. HUα, however, is not cleaved by Lon protease in the presence of ATP, yet still competed with FITC-α-casein. In addition, similar interference effects of A20D-HUβ and HUβ may indicate that position 20 is not involved in the substrate binding of Lon protease.
Figure 6.
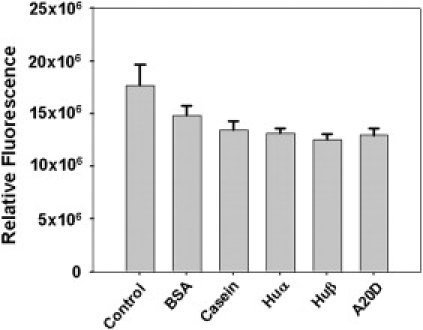
Competition of FITC-α-casein with BSA, unlabeled casein, HUα, HUβ, and HUβ-A20D as substrates of the Lon protease. In the control sample, there was no competing protein. The activity was assayed by measuring the fluorescence of released FITC (excitation, 490 nm; emission, 525 nm).
Discussion
A protease in E. coli and other bacteria must select its substrates from ∼4300 different proteins. Cleavage of the wrong proteins could be fatal, and recognition of the correct substrates is therefore critical. In eukaryotes, the 26S proteasome recognizes and degrades proteins modified by the attachment of ubiquitin. In bacteria, the Clp and Lon proteases interact directly with their protein targets. Even though a recent report indicated that the Lon protease degrades tmRNA-tagged proteins (51), accumulated evidence suggests that the sensor and substrate-discrimination domain, also named the α-domain, of Clp and Lon proteases interacts with polypeptide regions with a hydrophobic patch (52,53). For example, the C-terminus of the Lon substrate SulA is sufficient for recognition, but not for degradation (54). Recognition of a degradation tag can be the sole determinant of targeted proteolysis. The interaction between protease and substrate can be considered as the initial step. Once the substrate is recognized, it will be transferred to the proteolytic active site. The degradation progresses, and the fragments are released.
Lon protease has been shown to cut native proteins including RcsA (55), ribosomal S2 protein (56), and SulA (34,57). Earlier results indicated that Lon or a Lon-dependent protease degrades HU (25), and our results here showed that Ec-Lon degrades Ec-HUβ, but not Ec-HUα. In addition, the heterodimeric HUαβ is stable in the presence of Ec-Lon and ATP (see Fig. S1). Our in vitro findings are in line with those of previous in vivo studies (25).
The Ec-Lon cleavage sites of various peptides and proteins have been reported. Ec-Lon cleaves SulA, which is synthesized only as part of the SOS response to DNA damage (58,59). Cleavage occurs mainly at sites where L and S are in the P1 and P1′ positions; the other cleavage sites have various residues in the P1 position, such as A, V, M, T, S, L, F, Q, and G (57). Ec-Lon cleaves the ribosomal S2 protein at 45 sites, whose P1 and P3 positions are dominantly occupied by hydrophobic residues (56). In our present and past studies, both Ec-Lon and Bt-Lon cleaved at sites with A or L in the P1 position and in most cases with an uncharged amino acid, but also with a positively charged amino acid (e.g., K) in the P1′ position.
Our alignments of the HUβ sequence with the peptide fragments revealed the major Ec-Lon cleavage sites in HUβ. One major cleavage site is located at K18-G22, specifically at A20-A21 within an AAA cluster. In an earlier study of fluorogenic peptide substrates of Ec-Lon (60), the best substrates were those with an A-A bond (glutaryl- or succinyl-A-A-F-methoxynaphthylamine); substitution of A-A with G-G generated a very poor substrate. Ec-Lon was unable, however, to degrade glutaryl-A-A-A-methoxynaphthylamine. Another favored cleavage site of Ec-Lon in HUβ is L36-K37. Our results showed that the cleavage within K18-G22 and at L36-K37 occurs early in the degradation of HUβ by Ec-Lon (Table S1). Since fragments 12, 14, 18, 21, 22, and 23 have an intact K18-G22 and fragments 17, 19, 20, and 22 have an intact L36-K37 (Table S1), there might be more than one initial cleavage site in HUβ. Since all these fragments were found after 10 min of incubation, it is reasonable to postulate that one subunit of the dimer is degraded first, which leads to more cleavage sites being exposed on the other subunit. In such a scenario, large fragments would be present even after longer incubation.
The sequence of HUβ differs from that of HUα and Bs-HU particularly in the K18-G22 region, i.e., the initial Ec-Lon cleavage site in HUβ (Table S1). HUα and Bs-HU are not degraded by Ec-Lon and the mutants HUβ-A20Q and HUβ-A20D are degraded at a lower rate than HUβ. We therefore propose that region K18-G22 is involved in the Ec-Lon recognition of HUβ. The other favored Ec-Lon cleavage site in HUβ is L36-K37. Based on the results of MS analysis, L36-K37 became more accessible for cleavage after the first cleavage at K18-G22 near the N-terminus of HUβ. The cleavage mechanism of the Lon protease may be similar to that of ClpP (61). The active sites in the Lon protease that catalyze peptide-bond cleavage are sequestered in a hollow interior chamber formed by the subunits. The loop near K18-G22 of HUβ enters this chamber through axial channels. Since there is more stereohindrance near L36-K37, K18-G22 is the major cleavage site.
Taken together, we propose a mechanism for the degradation of a HUβ homodimer. The model of the homodimer (Fig. 2) can be considered as the association of two subdomains. In the cleavage mechanism, Ec-Lon recognizes K18-G22 at the end of the α2-helix of one HUβ subunit and cleaves in this region, thereby exposing the α2-helix so that L36-K37 can be cleaved. The loss of the structural integrity of HUβ would then enable cleavage at the hydrophobic core of HUβ, so that the fragment with cleavage site F47-G48 can be accessed. The degradation of the first subunit of the HUβ homodimer leads to the cleavage of more sites in the second subunit.
The heterodimer HUαβ, on the other hand, is not degraded by Ec-Lon in the presence of ATP, even though K18-G22 is present in the HUβ subunit. This lack of cleavage could be explained by the greater thermodynamic stability of the heterodimeric form over that of the homodimeric forms (42). HUαβ forms a spiral structure, residues L6, I7, I10, A11, A21, and L25 of HUβ interact with each other to generate a V-shape between the α1-helix and the α2-helix, and the amino acids in the turn between these helices could interact with another protein (62). It might therefore be difficult for Ec-Lon to unfold the heterodimeric HUαβ, and hence degradation by Lon is arrested.
It appears that only a few research findings are available to date concerning the substrate recognition of Lon protease (54,63). Our attempts to study the substrate recognition using surface plasmon resonance and isothermal titration calorimetry failed, because the interaction between the Lon protease and HUβ were too weak. Recent developments with protein-coated polystyrene beads and optical tweezers (64) enabled us to measure this interaction. Our results indicated that both HUα and HUβ can interact with the Lon protease. It is very unlikely that the rupture force we measured was the result of a single molecular-pair interaction. The force measured will thus depend on the number of molecules present in the contact area of the two beads, and this can fluctuate significantly from sample to sample. This is evident from the relatively large error bars in the results (Fig. 5) and from the statistics shown in Table S3. Despite this deficiency, the comparison of the relative average values of the rupture force under different conditions is still quite informative. It reveals not only that HUα, HUβ, and HUβ-A20D can interact with the Lon protease, but also that the interactions become weaker in the presence of ATP. There is an assertion that approximately equal rupture force in all three cases (i.e., for the binding of Ec-Lon with HUα, HUβ, or HUβ-A20D) implies random and unknown interaction sites. However, our current data indicate that in the presence of ATP, the rupture forces in all three cases were significantly reduced compared to the corresponding one without ATP. Therefore, it is more likely that such interaction is site-specific.
Our results indicate that Ec-Lon recognizes and interacts with both HUα and HUβ, but only degrades HUβ in the presence of ATP. Ec-Lon also binds HUβ-A20D, which has a negative charge at position 20, but does not cleave the mutated protein well. HUα, which has glutamine at this position, is also not cleaved. Alanine 20 is therefore not involved in the binding to Ec-Lon, but is involved in the recognition for cleavage by Ec-Lon. In a comparative study, the substrate competition assay also revealed that HUα, HUβ, and HUβ-A20D show similar competitive ability with α-casein (Fig. 6). In other words, Lon protease cannot distinguish HUα from HUβ by interaction with these two proteins. It is obvious that appropriate substrates of Lon protease should be recognized and should have special cleavage sites. BSA is not recognized by Lon protease and therefore cannot be further degraded by Lon protease (Fig. S11). HUα can be recognized by Lon protease but does not have special cleavage sites.
The selection of appropriate substrates by proteases is a critical step in regulatory degradation. Once the substrate is recognized, it is transferred to the proteolytic active site. For the Clp and FtsH proteases, short sequence motifs serve as recognition tags (61,65–67). In this study, the recognition tags have not been found. Here, we must emphasize the difference between initial recognition tags and the initial cleavage site. The region K18-G22 is the initial cleavage site of HUβ by Ec-Lon, but is not involved in substrate binding of Ec-Lon. The Lon protease interacts with certain specific recognition elements within HU proteins. It has been proposed that ATP hydrolysis is necessary to unfold and/or translocate the substrate before its degradation, thereby weakening the binding force between enzyme and substrate and facilitating substrate translocation (68–70). We showed here that the rupture forces do weaken in the presence of ATP. Our results indicate that Lon protease cleaves HUβ in three stages. In the first, less specific step, Lon binds the HU protein (HUβ, HUα, or HUβ-A20D). Lon then hydrolyzes ATP, providing energy to loosen the binding, conformationally changes the substrate, and translocates the substrate to the active site if the sequence of the N-terminus of the substrate allows. Finally, the translocated substrate, HUβ, but not HUα or HUβ-A20D, is cleaved. Substrates with high affinity will replace unfavorable, bound substrates. With such a mechanism, the Lon protease has at least two checkpoints to ensure that appropriate substrates are degraded.
Supporting Material
Additional text, a table, and 12 figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(09)01566-5.
Supporting Material
Acknowledgments
We thank Dr. Po-Huang Liang (Institute of Biological Chemistry, Academia Sinica, Taipei, Taiwan, Republic of China) for providing us with the E. coli Lon protease clone.
This research is supported by the National Science Council, Taipei, Taiwan, Republic of China (grants NSC 95-2311-B-001-033, NSC 96-2311-B-001-010, NSC 96-2752-E010-001-PAE, NSC 96-2627-B-001-003, and NSC 96-2627-M-010-004), and by the “Aim for the Top University Plan”, a grant from the Ministry of Education of the Republic of China.
Contributor Information
Arthur Chiou, Email: aechiou@ym.edu.tw.
Shih-Hsiung Wu, Email: shwu@gate.sinica.edu.tw.
References
- 1.Kano Y., Wada M., Imamoto F. Genetic characterization of the gene hupA encoding the HU-2 protein of Escherichia coli. Gene. 1988;69:331–335. doi: 10.1016/0378-1119(88)90443-x. [DOI] [PubMed] [Google Scholar]
- 2.Storts D.R., Markovitz A. Construction and characterization of mutations in hupB, the gene encoding HU-β (HU-1) in Escherichia coli K-12. J. Bacteriol. 1988;170:1541–1547. doi: 10.1128/jb.170.4.1541-1547.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Laine B., Kmiecik D., Cohen-Solal M. Complete amino-acid sequences of DNA-binding proteins HU-1 and HU-2 from Escherichia coli. Eur. J. Biochem. 1980;103:447–461. doi: 10.1111/j.1432-1033.1980.tb05968.x. [DOI] [PubMed] [Google Scholar]
- 4.Rouvière-Yaniv J., Kjeldgaard N.O. Native Escherichia coli HU protein is a heterotypic dimer. FEBS Lett. 1979;106:297–300. doi: 10.1016/0014-5793(79)80518-9. [DOI] [PubMed] [Google Scholar]
- 5.Drlica K., Rouviere-Yaniv J. Histonelike proteins of bacteria. Microbiol. Rev. 1987;51:301–319. doi: 10.1128/mr.51.3.301-319.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Micka B., Groch N., Marahiel M.A. Molecular cloning, nucleotide sequence, and characterization of the Bacillus subtilis gene encoding the DNA-binding protein HBsu. J. Bacteriol. 1991;173:3191–3198. doi: 10.1128/jb.173.10.3191-3198.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dixon N.E., Kornberg A. Protein HU in the enzymatic replication of the chromosomal origin of Escherichia coli. Proc. Natl. Acad. Sci. USA. 1984;81:424–428. doi: 10.1073/pnas.81.2.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flashner Y., Gralla J.D. DNA dynamic flexibility and protein recognition: differential stimulation by bacterial histone-like protein HU. Cell. 1988;54:713–721. doi: 10.1016/s0092-8674(88)80016-3. [DOI] [PubMed] [Google Scholar]
- 9.Lewis D.E., Geanacopoulos M., Adhya S. Role of HU and DNA supercoiling in transcription repression: specialized nucleoprotein repression complex at gal promoters in Escherichia coli. Mol. Microbiol. 1999;31:451–461. doi: 10.1046/j.1365-2958.1999.01186.x. [DOI] [PubMed] [Google Scholar]
- 10.Balandina A., Claret L., Rouviere-Yaniv J. The Escherichia coli histone-like protein HU regulates rpoS translation. Mol. Microbiol. 2001;39:1069–1079. doi: 10.1046/j.1365-2958.2001.02305.x. [DOI] [PubMed] [Google Scholar]
- 11.Rouvière-Yaniv J., Yaniv M., Germond J.E. E. coli DNA binding protein HU forms nucleosomelike structure with circular double-stranded DNA. Cell. 1979;17:265–274. doi: 10.1016/0092-8674(79)90152-1. [DOI] [PubMed] [Google Scholar]
- 12.Broyles S.S., Pettijohn D.E. Interaction of the Escherichia coli HU protein with DNA. Evidence for formation of nucleosome-like structures with altered DNA helical pitch. J. Mol. Biol. 1986;187:47–60. doi: 10.1016/0022-2836(86)90405-5. [DOI] [PubMed] [Google Scholar]
- 13.Lavoie B.D., Shaw G.S., Chaconas G. Anatomy of a flexer-DNA complex inside a higher-order transposition intermediate. Cell. 1996;85:761–771. doi: 10.1016/s0092-8674(00)81241-6. [DOI] [PubMed] [Google Scholar]
- 14.Painbéni E., Mouray E., Rouvière-Yaniv J. An imbalance of HU synthesis induces mucoidy in Escherichia coli. J. Mol. Biol. 1993;234:1021–1037. doi: 10.1006/jmbi.1993.1656. [DOI] [PubMed] [Google Scholar]
- 15.Markovitz A. Regulatory mechanisms for synthesis of capsular polysaccharide in mucoid mutants of Escherichia coli K12. Proc. Natl. Acad. Sci. USA. 1964;51:239–246. doi: 10.1073/pnas.51.2.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gill R.E., Karlok M., Benton D. Myxococcus xanthus encodes an ATP-dependent protease which is required for developmental gene transcription and intercellular signaling. J. Bacteriol. 1993;175:4538–4544. doi: 10.1128/jb.175.14.4538-4544.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmidt R., Decatur A.L., Losick R. Bacillus subtilis Lon protease prevents inappropriate transcription of genes under the control of the sporulation transcription factor σ G. J. Bacteriol. 1994;176:6528–6537. doi: 10.1128/jb.176.21.6528-6537.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suzuki C.K., Suda K., Schatz G. Requirement for the yeast gene LON in intramitochondrial proteolysis and maintenance of respiration. Science. 1994;264:891. doi: 10.1126/science.8178144. Erratum for Science. 1994. 264:273–276. [DOI] [PubMed] [Google Scholar]
- 19.Van Dyck L., Pearce D.A., Sherman F. PIM1 encodes a mitochondrial ATP-dependent protease that is required for mitochondrial function in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 1994;269:238–242. [PubMed] [Google Scholar]
- 20.Maurizi M.R., Trisler P., Gottesman S. Insertional mutagenesis of the lon gene in Escherichia coli: lon is dispensable. J. Bacteriol. 1985;164:1124–1135. doi: 10.1128/jb.164.3.1124-1135.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gottesman S., Halpern E., Trisler P. Role of sulA and sulB in filamentation by lon mutants of Escherichia coli K-12. J. Bacteriol. 1981;148:265–273. doi: 10.1128/jb.148.1.265-273.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Apte B.N., Rhodes H., Zipser D. Mutation blocking the specific degradation of reinitiation polypeptides in E. coli. Nature. 1975;257:329–331. doi: 10.1038/257329a0. [DOI] [PubMed] [Google Scholar]
- 23.Bukhari A.I., Zipser D. Mutants of Escherichia coli with a defect in the degradation of nonsense fragments. Nat. New Biol. 1973;243:238–241. doi: 10.1038/newbio243238a0. [DOI] [PubMed] [Google Scholar]
- 24.Tsilibaris V., Maenhaut-Michel G., Van Melderen L. Biological roles of the Lon ATP-dependent protease. Res. Microbiol. 2006;157:701–713. doi: 10.1016/j.resmic.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 25.Bonnefoy E., Almeida A., Rouviere-Yaniv J. Lon-dependent regulation of the DNA binding protein HU in Escherichia coli. Proc. Natl. Acad. Sci. USA. 1989;86:7691–7695. doi: 10.1073/pnas.86.20.7691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dougan D.A., Mogk A., Bukau B. AAA+ proteins and substrate recognition, it all depends on their partner in crime. FEBS Lett. 2002;529:6–10. doi: 10.1016/s0014-5793(02)03179-4. [DOI] [PubMed] [Google Scholar]
- 27.Ebel W., Skinner M.M., Trempy J.E. A conserved domain in Escherichia coli Lon protease is involved in substrate discriminator activity. J. Bacteriol. 1999;181:2236–2243. doi: 10.1128/jb.181.7.2236-2243.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fischer H., Glockshuber R. A point mutation within the ATP-binding site inactivates both catalytic functions of the ATP-dependent protease La (Lon) from Escherichia coli. FEBS Lett. 1994;356:101–103. doi: 10.1016/0014-5793(94)01244-x. [DOI] [PubMed] [Google Scholar]
- 29.Amerik AYu, Antonov V.K., Shimbarevich E.V. Site-directed mutagenesis of La protease. A catalytically active serine residue. FEBS Lett. 1991;287:211–214. doi: 10.1016/0014-5793(91)80053-6. [DOI] [PubMed] [Google Scholar]
- 30.Gottesman S. Proteases and their targets in Escherichia coli. Annu. Rev. Genet. 1996;30:465–506. doi: 10.1146/annurev.genet.30.1.465. [DOI] [PubMed] [Google Scholar]
- 31.Maurizi M.R. Degradation in vitro of bacteriophage λN protein by Lon protease from Escherichia coli. J. Biol. Chem. 1987;262:2696–2703. [PubMed] [Google Scholar]
- 32.Gottesman S., Gottesman M., Pearson M.L. Protein degradation in E. coli: the lon mutation and bacteriophage λN and cII protein stability. Cell. 1981;24:225–233. doi: 10.1016/0092-8674(81)90518-3. [DOI] [PubMed] [Google Scholar]
- 33.Mizusawa S., Gottesman S. Protein degradation in Escherichia coli: the lon gene controls the stability of sulA protein. Proc. Natl. Acad. Sci. USA. 1983;80:358–362. doi: 10.1073/pnas.80.2.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sonezaki S., Ishii Y., Kato Y. Overproduction and purification of SulA fusion protein in Escherichia coli and its degradation by Lon protease in vitro. Appl. Microbiol. Biotechnol. 1995;43:304–309. doi: 10.1007/BF00172829. [DOI] [PubMed] [Google Scholar]
- 35.Torres-Cabassa A.S., Gottesman S. Capsule synthesis in Escherichia coli K-12 is regulated by proteolysis. J. Bacteriol. 1987;169:981–989. doi: 10.1128/jb.169.3.981-989.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van Melderen L., Thi M.H., Maurizi M.R. ATP-dependent degradation of CcdA by Lon protease. Effects of secondary structure and heterologous subunit interactions. J. Biol. Chem. 1996;271:27730–27738. doi: 10.1074/jbc.271.44.27730. [DOI] [PubMed] [Google Scholar]
- 37.Lee A.Y., Tsay S.S., Wu S.H. Identification of a gene encoding Lon protease from Brevibacillus thermoruber WR-249 and biochemical characterization of its thermostable recombinant enzyme. Eur. J. Biochem. 2004;271:834–844. doi: 10.1111/j.1432-1033.2004.03988.x. [DOI] [PubMed] [Google Scholar]
- 38.Lee A.Y., Hsu C.H., Wu S.H. Functional domains of Brevibacillus thermoruber lon protease for oligomerization and DNA binding: role of N-terminal and sensor and substrate discrimination domains. J. Biol. Chem. 2004;279:34903–34912. doi: 10.1074/jbc.M403562200. [DOI] [PubMed] [Google Scholar]
- 39.Goldberg A.L., Moerschell R.P., Maurizi M.R. ATP-dependent protease La (lon) from Escherichia coli. Methods Enzymol. 1994;244:350–375. doi: 10.1016/0076-6879(94)44027-1. [DOI] [PubMed] [Google Scholar]
- 40.Lee C.L., Hsiao H.H., Khoo K.H. Strategic shotgun proteomics approach for efficient construction of an expression map of targeted protein families in hepatoma cell lines. Proteomics. 2003;3:2472–2486. doi: 10.1002/pmic.200300586. [DOI] [PubMed] [Google Scholar]
- 41.Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramstein J., Hervouet N., Castaing B. Evidence of a thermal unfolding dimeric intermediate for the Escherichia coli histone-like HU proteins: thermodynamics and structure. J. Mol. Biol. 2003;331:101–121. doi: 10.1016/s0022-2836(03)00725-3. [DOI] [PubMed] [Google Scholar]
- 43.Sali A., Blundell T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 44.Morris A.L., MacArthur M.W., Thornton J.M. Stereochemical quality of protein structure coordinates. Proteins. 1992;12:345–364. doi: 10.1002/prot.340120407. [DOI] [PubMed] [Google Scholar]
- 45.Bowie J.U., Lüthy R., Eisenberg D. A method to identify protein sequences that fold into a known three-dimensional structure. Science. 1991;253:164–170. doi: 10.1126/science.1853201. [DOI] [PubMed] [Google Scholar]
- 46.Twining S.S. Fluorescein isothiocyanate-labeled casein assay for proteolytic enzymes. Anal. Biochem. 1984;143:30–34. doi: 10.1016/0003-2697(84)90553-0. [DOI] [PubMed] [Google Scholar]
- 47.Claret L., Rouviere-Yaniv J. Variation in HU composition during growth of Escherichia coli: the heterodimer is required for long term survival. J. Mol. Biol. 1997;273:93–104. doi: 10.1006/jmbi.1997.1310. [DOI] [PubMed] [Google Scholar]
- 48.Vasilyeva O.V., Kolygo K.B., Ovchinnikova T.V. Domain structure and ATP-induced conformational changes in Escherichia coli protease Lon revealed by limited proteolysis and autolysis. FEBS Lett. 2002;526:66–70. doi: 10.1016/s0014-5793(02)03117-4. [DOI] [PubMed] [Google Scholar]
- 49.Patterson-Ward J., Tedesco J., Lee I. Utilization of synthetic peptides to evaluate the importance of substrate interaction at the proteolytic site of Escherichia coli Lon protease. Biochim. Biophys. Acta. 2009;1794:1355–1363. doi: 10.1016/j.bbapap.2009.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Waxman L., Goldberg A.L. Protease La from Escherichia coli hydrolyzes ATP and proteins in a linked fashion. Proc. Natl. Acad. Sci. USA. 1982;79:4883–4887. doi: 10.1073/pnas.79.16.4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Choy J.S., Aung L.L., Karzai A.W. Lon protease degrades tmRNA-tagged proteins. J. Bacteriol. 2007;189:6564–6571. doi: 10.1128/JB.00860-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith C.K., Baker T.A., Sauer R.T. Lon and Clp family proteases and chaperones share homologous substrate-recognition domains. Proc. Natl. Acad. Sci. USA. 1999;96:6678–6682. doi: 10.1073/pnas.96.12.6678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gonciarz-Swiatek M., Wawrzynow A., Zylicz M. Recognition, targeting, and hydrolysis of the λO replication protein by the ClpP/ClpX protease. J. Biol. Chem. 1999;274:13999–14005. doi: 10.1074/jbc.274.20.13999. [DOI] [PubMed] [Google Scholar]
- 54.Higashitani A., Ishii Y., Koriuchi K. Functional dissection of a cell-division inhibitor, SulA, of Escherichia coli and its negative regulation by Lon. Mol. Gen. Genet. 1997;254:351–357. doi: 10.1007/s004380050426. [DOI] [PubMed] [Google Scholar]
- 55.Stout V., Torres-Cabassa A., Gottesman S. RcsA, an unstable positive regulator of capsular polysaccharide synthesis. J. Bacteriol. 1991;173:1738–1747. doi: 10.1128/jb.173.5.1738-1747.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nishii W., Suzuki T., Takahashi K. Cleavage mechanism of ATP-dependent Lon protease toward ribosomal S2 protein. FEBS Lett. 2005;579:6846–6850. doi: 10.1016/j.febslet.2005.11.026. [DOI] [PubMed] [Google Scholar]
- 57.Nishii W., Maruyama T., Takahashi K. The unique sites in SulA protein preferentially cleaved by ATP-dependent Lon protease from Escherichia coli. Eur. J. Biochem. 2002;269:451–457. doi: 10.1046/j.0014-2956.2001.02668.x. [DOI] [PubMed] [Google Scholar]
- 58.Huisman O., D'Ari R. An inducible DNA replication-cell division coupling mechanism in E. coli. Nature. 1981;290:797–799. doi: 10.1038/290797a0. [DOI] [PubMed] [Google Scholar]
- 59.Mizusawa S., Court D., Gottesman S. Transcription of the sulA gene and repression by LexA. J. Mol. Biol. 1983;171:337–343. doi: 10.1016/0022-2836(83)90097-9. [DOI] [PubMed] [Google Scholar]
- 60.Waxman L., Goldberg A.L. Protease La, the lon gene product, cleaves specific fluorogenic peptides in an ATP-dependent reaction. J. Biol. Chem. 1985;260:12022–12028. [PubMed] [Google Scholar]
- 61.Baker T.A., Sauer R.T. ATP-dependent proteases of bacteria: recognition logic and operating principles. Trends Biochem. Sci. 2006;31:647–653. doi: 10.1016/j.tibs.2006.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guo F., Adhya S. Spiral structure of Escherichia coli HUαβ provides foundation for DNA supercoiling. Proc. Natl. Acad. Sci. USA. 2007;104:4309–4314. doi: 10.1073/pnas.0611686104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ondrovicová G., Liu T., Suzuki C.K. Cleavage site selection within a folded substrate by the ATP-dependent lon protease. J. Biol. Chem. 2005;280:25103–25110. doi: 10.1074/jbc.M502796200. [DOI] [PubMed] [Google Scholar]
- 64.Salomo M., Keyser U.F., Struhalla M., Kremer F. Optical tweezers to study single protein A/immunoglobulin G interactions at varying conditions. Eur. Biophys. J. 2008;37:927–934. doi: 10.1007/s00249-008-0310-3. [DOI] [PubMed] [Google Scholar]
- 65.Gottesman S., Roche E., Sauer R.T. The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system. Genes Dev. 1998;12:1338–1347. doi: 10.1101/gad.12.9.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Flynn J.M., Levchenko I., Baker T.A. Overlapping recognition determinants within the ssrA degradation tag allow modulation of proteolysis. Proc. Natl. Acad. Sci. USA. 2001;98:10584–10589. doi: 10.1073/pnas.191375298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ito K., Akiyama Y. Cellular functions, mechanism of action, and regulation of FtsH protease. Annu. Rev. Microbiol. 2005;59:211–231. doi: 10.1146/annurev.micro.59.030804.121316. [DOI] [PubMed] [Google Scholar]
- 68.Lee I., Berdis A.J., Suzuki C.K. Recent developments in the mechanistic enzymology of the ATP-dependent Lon protease from Escherichia coli: highlights from kinetic studies. Mol. Biosyst. 2006;2:477–483. doi: 10.1039/b609936j. [DOI] [PubMed] [Google Scholar]
- 69.Lee I., Suzuki C.K. Functional mechanics of the ATP-dependent Lon protease- lessons from endogenous protein and synthetic peptide substrates. Biochim. Biophys. Acta. 2008;1784:727–735. doi: 10.1016/j.bbapap.2008.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Licht S., Lee I. Resolving individual steps in the operation of ATP-dependent proteolytic molecular machines: from conformational changes to substrate translocation and processivity. Biochemistry. 2008;47:3595–3605. doi: 10.1021/bi800025g. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


