Abstract
Selenium may affect prostate-cancer (PC) risk via its plasma carrier selenoprotein P which shows dramatically reduced expression in PC tumors and cell-lines. The selenoprotein P (SEPP1) Ala234 SNP allele is associated with lower plasma selenoprotein P in men, reducing the concentration/activity of other antioxidant selenoproteins. Selenium status also modifies the effect of mitochondrial superoxide dismutase (SOD2) SNP Ala16Val on PC risk. We investigated the relationship of these SNPs with PC risk. DNA from 2,975 cases and 1,896 age-matched controls from the population-based Prostate Cancer in Sweden (CAPS) study were genotyped using TaqMan® assays. Cases were designated aggressive (APC) or non-aggressive (NPC) at diagnosis by clinical criteria. Association with PC was investigated by logistic regression; gene-gene interaction using a general linear model. Mean plasma selenium measured in 169 controls was relatively-low (76.0±17.2μg/L). SNP genotype-distributions were in Hardy-Weinberg Equilibrium. SOD2-Ala16+ men were at greater PC risk (OR 1.19, 95%CI 1.03-1.37) compared to SOD2-Val16 homozygotes. Men homozygous for SEPP1-Ala234 who were also SOD2-Ala16+ had a higher risk of PC (OR 1.43, 95%CI 1.17-1.76) and APC (OR 1.60, 95%CI 1.22-2.09) than those who were SOD2-Val16 homozygotes (interaction, PC P=0.05, APC P=0.01). This interaction was stronger in ever-smokers: SOD2-Ala16+ men homozygous for SEPP1-Ala234 had an almost doubled risk of PC (OR 1.97, 95%CI 1.33-2.91; interaction P=0.001). In a low selenium population, SOD2-Ala16+ men homozygous for SEPP1-Ala234 are at increased risk of PC/APC especially if ever-smokers, because they are likely to produce more mitochondrial H2O2 that they cannot remove, thereby promoting prostate tumor-cell proliferation and migration.
Keywords: mitochondrial superoxide dismutase, polymorphism, prostate cancer, selenium, selenoprotein P, SNPs
INTRODUCTION
Selenium, an essential nutrient, may reduce the incidence and/or progression of prostate cancer particularly in men with the relatively-low baseline selenium status commonly found in Europe [1-4]. The potential anti-cancer effects of selenium may be exerted through a number of parallel mechanisms some of which involve the selenoproteins [1]. Indeed recent evidence suggests an important role for selenoproteins in cancer [1, 5], specifically in prostate cancer [6].
Selenoprotein P contains at least 40% of the total selenium in plasma [7]. Deletion of the gene for selenoprotein P in mouse models alters the distribution of selenium in body tissues suggesting that selenoprotein P is required for selenium transport [8, 9]. While the human selenoprotein P gene (SEPP1) is abundantly expressed in normal colon mucosa, there is a significant reduction or loss of SEPP1 mRNA expression in colon cancers [10]. Expression of SEPP1 is also dramatically reduced in a subset of human prostate tumors, mouse tumors and in the androgen-dependent (LNCaP) and androgen-independent (PC-3) prostate cancer cell lines [11]. Homozygosity for the Ala234 allele of the SEPP1 Ala234Thr SNP (rs3877899) is associated with a lower concentration of plasma selenoprotein P in men, affecting the concentration and/or activity of other selenoproteins, notably of thioredoxin reductase 1 (TR1) and some of the antioxidant glutathione peroxidases (GPx) [12]. Thus it is conceivable that SEPP1 genotype may affect prostate cancer risk.
Selenium status is linked to the effect of another polymorphism on prostate cancer risk i.e. that of mitochondrial superoxide dismutase (SOD2 or MnSOD) [13], the major detoxifying enzyme in the mitochondrion. This enzyme dismutes superoxide to H2O2, which must itself then be detoxified to water by GPx [13]. A well-characterized polymorphism in SOD2 results in the substitution of alanine (Ala) for valine (Val) at codon 16 (rs4880) and a higher activity of the Ala16 mitochondrial enzyme [14]. Among US men homozygous for Ala16, those whose plasma selenium was in the top quartile had a relative risk (RR) of prostate cancer of 0.3 (95% CI 0.2 to 0.7) and of clinically aggressive prostate cancer of 0.2 (95% CI 0.1 to 0.5) when compared with those whose plasma selenium was in the bottom quartile [13].The dependence on selenium status of this genotype effect on prostate cancer risk may relate to the requirement for adequate GPx to remove the extra H2O2 formed in Ala16 homozygotes [13].
We investigated the relationship of these two putatively-functional polymorphisms to prostate cancer risk in the large CAPS (CAncer Prostate in Sweden) study [15]. We hypothesised that in a low selenium environment, as in Sweden, men who have SNP alleles that both reduce their ability to make functional selenoprotein P (SEPP1 Ala234 homozygotes) and increase their production of H2O2 (SOD2 Ala16) thereby increasing their requirement for GPx, would have a greater risk of prostate cancer than men who do not have these alleles.
SUBJECTS AND METHODS
Participants
The CAPS study is a large-scale, population-based, prostate cancer case-control study, which has been extensively described in previous work [15, 16]. The inclusion criterion for cases was a newly diagnosed, pathologically or cytologically verified adenocarcinoma of the prostate. In total, 3,648 prostate cancer patients were invited to participate in the study and 3,161 (87%) agreed. DNA samples were obtained from a total of 2,915 cases, for which the corresponding clinical data and completed demographic questionnaires were available. Cases were classified as either non-aggressive at diagnosis (tumor stage 1 and 2, Gleason score < 8, Differentiation G1–G2, N0/NX, M0/MX, PSA < 100 μg/L; NPC) or aggressive at diagnosis (tumor stage 3-4, Gleason score ≥ 8, Differentiation G3–G4, N+, M+, PSA ≥ 100 μg/L; APC) [15]. Controls matching the case distribution for age (within 5 year bands) and geographical region were randomly selected from the Swedish Population Registry. A total of 3,153 controls were invited to participate of whom 2,149 agreed. DNA and completed questionnaires were obtained from a total of 1,764 controls, giving a response rate of over 82%. Written informed consent was obtained from all participants and study was approved by the research ethics committees at the Karolinska Institutet and Umeå University Hospital. The University of Surrey research ethics committee approved this genetics study.
Genotyping
DNA was extracted from leukocytes using a Puregene kit (Gentra Systems, Minneapolis MN). All genotyping was performed using TaqMan® assays and the operator was blinded to case/control status. Controls of known genotype for each of the polymorphisms investigated were included in the assay. Non-template controls and duplicate samples were incorporated for quality control purposes. SOD2 Ala16Val (rs4880) PCR primers and dual-labelled, allelic probes were designed and manufactured by Applied Biosystems Inc (CA, USA). Primer and probes were as follows: forward GCTGTGCTTTCTCGTCTTCAG, reverse CTGCCTGGAGCCCAGATAC, Ala16 Probe VIC-CCAAAGCCGGAGCC-TAM, Val16 probe FAM-CCCAAAAGCCGGAGCC-TAM. SEPP1 Ala234Thr (rs3877899) was genotyped using a pre-designed assay (TaqMan® Assay number C_2841533_10) so primer and probe details are not available but the Ala234 allele was detected with VIC and the Thr234 allele with FAM. Each reaction (12.50 μl) contained 2–10 ng of DNA, 1 x PCR master mix (ABgene, Epsom, UK), 1 x TaqMan® assay mix (working concentration of dual labelled probes 100 nM each and PCR primer 900 nM). Reactions were incubated as specified by the manufacturer: 95°C for 15 min, 40 cycles of 95°C for 15 sec and 60°C for 1 min. Fluorescence was measured and genotypes assigned using the ABI prism 7500 and associated software (Applied Biosystems Inc, CA, USA).
Plasma selenium measurement
EDTA plasma samples from 169 controls were stored at -80°C prior to determination of selenium by dynamic reaction cell inductively coupled plasma mass spectrometry (DRC ICP-MS) using an Elan 6100 DRC plus (SCIEX Perkin-Elmer). 78Selenium was measured, employing methane (at 0.5 ml/min) as the DRC gas to remove the argon dimer background [17] and butanol to increase the sensitivity of the signal [18]. Within the plasma Se concentrations used in this study, the within-run coefficient of variation (CV) was 2.1-2.6% while the between-run CV was 3.1-5.6% (n=10). Accuracy was assured by analysis of four internal quality control serum samples (TEQAS, University of Surrey, Guildford) and certified reference materials: Seronorm Serum (Nycomed, Norway) JL4409, mean value (5 determinations) 0.90, SD 0.04 μmolL-1 (certified 0.92, range 0.84 – 1.00) μmolL-1, and NO0371 mean value (5 determinations) 1.76, SD 0.04 (certified 1.72, range 1.61-1.83) μmolL-1. The detection limit was less than 0.01μmolL-1.
Data analysis
All data were analyzed using SPSS v13. Continuous variables were shown to be normally distributed. The Pearson Chi squared test was used to compare observed SNP genotype frequencies with those expected under conditions of Hardy-Weinberg equilibrium. Logistic regression models were used to assess the association between SNP genotypes and prostate cancer risk, with genotypes coded either as number of rare alleles (0, 1, 2) or as minus allele/plus allele (0, 1) as appropriate, with adjustment for the possible confounding factors age and geographical location. Genotype-specific risks were estimated as adjusted odds ratios (OR) with associated 95% confidence intervals (CI) by logistic regression. The interaction between the two SNPs in determining prostate cancer risk was assessed using a general linear model, again adjusting for age and geographical region, with genotypes coded as minus allele/plus allele (0, 1). Data analysis was performed separately in cases with non-aggressive or aggressive disease, and in ever- and never-smokers. A two-tailed P value of less than 0.05 was considered statistically significant.
RESULTS
Demographics
Data on age and smoking status for the cases and controls, and on tumor classification for the cases, are given in supplementary Table 1.
Table 1.
The relationship between SNPs in two genes, SOD2 (Val16Ala) and SePP1 (Ala234Thr) with risk of all prostate cancer, non-aggressive prostate cancer and aggressive prostate cancer in the CAPS study.
| Controls | All prostate cancer | Non-aggressive prostate cancer | Aggressive prostate cancer | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Genotype | n | n | OR (95% CI)* | P | n | OR (95% CI)* | P | n | OR (95% CI)* | P |
| SOD2 Codon 16 | 1636 | 2634 | 1648 | 986 | ||||||
| Val/Val | 423 | 602 | Referent | 382 | Referent | 220 | Referent | |||
| Ala/Val | 789 | 1352 | 1.21 (1.03 to 1.41) | 0.02 | 854 | 1.21 (1.02 to 1.44) | 0.03 | 498 | 1.21 (0.99 to 1.48) | 0.60 |
| Ala/Ala | 424 | 680 | 1.15 (0.97 to 1.37) | 0.11 | 412 | 1.11 (0.91 to 1.36) | 0.29 | 268 | 1.22 (0.98 to 1.53) | 0.78 |
| Ala/Val + Ala/Ala | 1213 | 2032 | 1.19 (1.03 to 1.37) | 0.02 | 1266 | 1.18 (1.00 to 1.38) | 0.05 | 766 | 1.21 (1.01 to 1.46) | 0.04 |
| SEPP1 Codon 234 | 1570 | 2643 | 1653 | 990 | ||||||
| Ala/Ala | 878 | 1522 | Referent | 951 | 571 | Referent | ||||
| Ala/Thr | 595 | 949 | 0.94 (0.82 to 1.07) | 0.37 | 593 | 0.95 (0.82 to 1.11) | 0.54 | 356 | 0.93 (0.79 to 1.10 | 0.40 |
| Thr/Thr | 97 | 172 | 1.09 (0.83 to 1.42) | 0.54 | 109 | 1.13 (0.84 to 1.52) | 0.42 | 63 | 1.03 (0.74 to 1.44) | 0.86 |
| Ala/Thr + Thr/Thr | 692 | 1121 | 0.96 (0.85 to 1.09) | 0.54 | 702 | 0.98 (0.85 to 1.13) | 0.76 | 419 | 0.94 (0.80 to 1.11) | 0.48 |
odds ratio adjusted for age and geographical location, 95% confidence interval
n = Number of subjects
Selenium status
The mean (±SD) plasma selenium was 76.0 ± 17.2 μg/L in 169 CAPS control samples confirming the expected relatively-low selenium status of this group of Swedish men. There was no difference in selenium status between genotypes or by smoking status.
SNP genotyping
Through the blinded genotyping of duplicated samples, genotyping error rates were less than 1.5%. Both the SOD2 Ala16Val and SEPP1 Ala234Thr polymorphisms were shown to be in Hardy Weinberg equilibrium and the allele frequencies within the control population were comparable to those in other published data† [12, 13].
SOD2 and SEPP1 genotypes and prostate cancer risk
Individuals with at least one SOD2 Ala16 allele (SOD2 Ala16+) had an almost 20% increased risk of prostate cancer compared to Val16 homozygotes (adjusted OR 1.19; 95% CI 1.03 to1.37; P = 0.02; Table 1). No association between SEPP1 Ala234Thr genotype and prostate cancer risk was observed. The association between the SOD2 polymorphism and either non-aggressive or aggressive disease was of similar magnitude to that with all prostate cancer (non-aggressive plus aggressive) (Table 1). There was no association between SEPP1 genotype and either non-aggressive or aggressive disease.
Interaction between SOD2 and SEPP1 polymorphisms
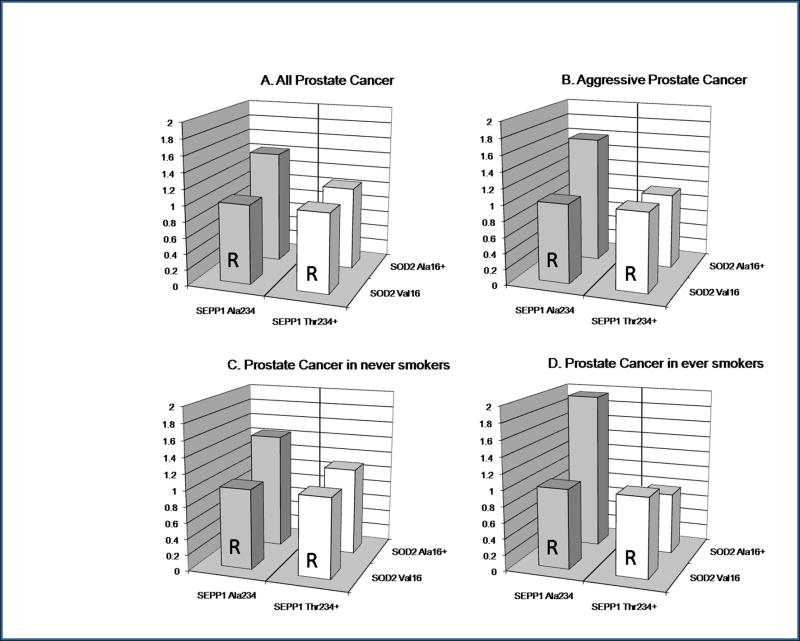
Men homozygous for the SEPP1 Ala234 allele, who were also SOD2 Ala16+, were at 43% greater risk of prostate cancer than SOD2 Val16 homozygotes (adjusted OR 1.43; 95% CI 1.17 to 1.76; P = 0.0005; Table 2, Fig. 1). By contrast, there was no association between SOD2 genotype and cancer risk in SEPP1 Thr234+ men. This interaction between the two SNPs in determining risk of prostate cancer had a borderline statistically significant P value of 0.05.
Table 2.
The interaction between SOD2 Val16Ala and SEPP1 Ala234Thr SNPs and risk of all prostate cancer, non-aggressive prostate cancer and aggressive prostate cancer in the CAPS study.
| Controls | All prostate cancer | Non-aggressive prostate cancer | Aggressive prostate cancer | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Genotype | n | n | OR (95% CI)* | P | n | OR (95% CI)* | P | n | OR (95% CI)* | P |
| SEPP1 Codon 234 Ala/Ala | 816 | 1360 | 854 | 506 | ||||||
| SOD2 Val/Val | 224 | 286 | Referent | 1 | 189 | Referent | 1 | 97 | Referent | 1 |
| SOD2 Ala/Val + Ala/Ala | 592 | 1074 | 1.43 (1.17 to 1.76) | 0.0005 | 665 | 1.36 (1.09 to 1.71) | 0.0073 | 409 | 1.60 (1.22 to 2.09) | 0.0007 |
| SEPP1 Codon 234 Ala/Thr + Thr/Thr | 638 | 1009 | 631 | 378 | ||||||
| SOD2 Val/Val | 161 | 246 | Referent | 1 | 147 | Referent | 1 | 99 | Referent | 1 |
| SOD2 Ala/Val + Ala/Ala | 477 | 763 | 1.06 (0.84 to 1.34) | 0.62 | 484 | 1.14 (0.87 to 1.48) | 0.35 | 279 | 0.95 (0.71 to 1.28) | 0.75 |
| P for interaction | 0.051 | 0.278 | 0.012 | |||||||
odds ratio adjusted for age and geographical location, 95% confidence interval
n = Number of subjects
Figure 1.
The interaction between SOD2 Val16Ala and SEPP1 Ala234Thr SNPs and risk of: A all prostate cancer; B aggressive prostate cancer; C prostate cancer in never-smokers and D prostate cancer in ever-smokers, in the CAPS study. OR = odds ratio, adjusted for age and geographical location. R indicates the referent group (foreground column) for each comparison with the alternative SOD2 genotype group (background column).
In aggressive prostate cancer, the interaction between the SNPs was stronger (P = 0.01) with SEPP1 Ala234 homozygotes who were also SOD2 Ala16+ having a 60% increased risk of aggressive disease compared to SOD2 Val16 homozygotes (adjusted OR 1.60; 95% CI 1.22, 2.09; P = 0.0007; Table 2, Fig. 1), whereas there was no association with SOD2 genotype in the SEPP1 Thr234+ men. Although, the association with SNP genotypes in non-aggressive disease might appear significant, the interaction between the two SNPs was far from statistical significance as demonstrated by the broad overlap between the odds ratio confidence intervals in the two SEPP1 genotype groups (Table 2).
Impact of smoking on the SOD2 and SEPP1 associations with prostate cancer risk
Although smoking status was not available for all subjects, given the known effect of smoking on antioxidant status, we investigated the effect of genotype on prostate cancer risk in the subset of participants (CAPS1 [15]) for whom smoking data were available (Table 3). Neither SOD2 nor SEPP1 genotype taken separately significantly affected the risk of prostate cancer in either never-or ever-smokers. The odds ratio associated with the SOD2 Ala16+ ever-smokers (Table 3) was, however, similar in magnitude to that in the group as a whole (Table 1) although the association did not reach statistical significance.
Table 3.
The relationship between SNPs in two genes, SOD2 (Val16Ala) and SePP1 (Ala234Thr) with risk of prostate cancer, in never-smokers and ever-smokers in the CAPS study.
| Never-Smokers | Ever-Smokers | |||||||
|---|---|---|---|---|---|---|---|---|
| Genotype | Controls (n) | Cases (n) | OR (95% CI)* | P | Controls (n) | Cases (n) | OR (95% CI)* | P |
| SOD2 Codon 16 | 314 | 526 | 487 | 777 | ||||
| Val/Val | 75 | 115 | Referent | 1 | 131 | 178 | Referent | 1 |
| Ala/Val | 160 | 278 | 1.12 (0.79 to 1.59) | 0.53 | 230 | 388 | 1.23 (0.93 to 1.64) | 0.15 |
| Ala/Ala | 79 | 133 | 1.11 (0.74 to 1.67) | 0.61 | 126 | 211 | 1.23 (0.89 to 1.70) | 0.21 |
| Ala/Val + Ala/Ala | 239 | 411 | 1.12 (0.80 to 1.56) | 0.52 | 356 | 599 | 1.23 (0.94 to 1.61) | 0.12 |
| SEPP1 Codon 234 | 260 | 452 | ||||||
| Ala/Ala | 145 | 259 | Referent | 1 | 225 | 417 | Referent | 1 |
| Ala/Thr | 103 | 164 | 0.89 (0.65 to 1.23) | 0.49 | 146 | 234 | 0.89 (0.68 to 1.17) | 0.41 |
| Thr/Thr | 12 | 29 | 1.38 (0.68 to 2.78) | 0.37 | 24 | 30 | 0.79 (0.45 to 1.40) | 0.43 |
| Ala/Thr + Thr/Thr | 115 | 193 | 0.94 (0.69 to 1.28) | 0.71 | 170 | 264 | 0.88 (0.68 to 1.14) | 0.33 |
odds ratio adjusted for age and geographical location, 95% confidence interval
n = Number of subjects
The interaction between SEPP1 and SOD2 SNPs in determining prostate cancer risk was modified by smoking status (Table 4, Fig. 1). Ever-smokers homozygous for SEPP1 Ala234 had a highly-significant two-fold increase in prostate cancer risk if they were also SOD2 Ala16+ (OR 1.97; 95% CI 1.33, 2.91; P = 0.0007). The association between SOD2 genotype and cancer risk in ever-smokers was not observed in SEPP1 Thr234+ men (OR 0.75; 95% CI 0.47, 1.18; P = 0.21). This interaction between SEPP1 and SOD2 SNPs in determining prostate cancer risk in ever-smokers was highly significant (P = 0.0014) contrasting with the lack of interaction found in never-smokers (P = 0.43).
Table 4.
The interaction between SOD2 Val16Ala and SePP1 Ala234Thr SNPs in determining prostate cancer risk in never-smokers and ever-smokers in the CAPS study.
| Never-Smokers | Ever-Smokers | |||||||
|---|---|---|---|---|---|---|---|---|
| Genotype | Controls (n) | Cases (n) | OR (95% CI)* | P | Controls (n) | Cases (n) | OR (95% CI)* | P |
| SEPP1 Codon 234 Ala/Ala | 142 | 252 | 219 | 396 | ||||
| SOD2 Val/Val | 38 | 51 | Referent | 1 | 67 | 73 | Referent | 1 |
| SOD2 Ala/Val + Ala/Ala | 104 | 201 | 1.44 (0.89 to 2.34) | 0.14 | 152 | 323 | 1.97 (1.33 to 2.91) | 0.0007 |
| SEPP1 Codon 234 Ala/Thr + Thr/Thr | 111 | 189 | 167 | 257 | ||||
| SOD2 Val/Val | 27 | 43 | Referent | 1 | 39 | 73 | Referent | 1 |
| SOD2 Ala/Val + Ala/Ala | 84 | 146 | 1.08 (0.62 to 1.87) | 0.80 | 128 | 184 | 0.75 (0.47 to 1.18) | 0.21 |
| P for interaction | 0.43 | 0.0014 | ||||||
odds ratio adjusted for age and geographical location, 95% confidence interval
n = Number of subjects
DISCUSSION
The mean plasma selenium of 76.0 ± 17.2 μg/L in control samples confirms the relatively low selenium intake and status in the Swedish population (cf mean US value of 125 μg/L determined in the Third National Health and Nutrition Examination Survey [19]). This value is less than the 92 μg/L required for maximal plasma GPx activity [20] and considerably less than the plasma concentration required for full expression of SEPP1 [7, 21], demonstrating that the study population has a selenium intake inadequate for optimal selenoprotein synthesis and/or activity. This may be relevant to prostate cancer risk since low selenoprotein production led to higher-grade lesions and aggressive disease in a transgenic mouse model of prostate cancer [6]. We reasoned that in a population with relatively-low selenium status, inter-individual variation in selenium requirement, as determined by selenoprotein genotype, might have a greater effect on the risk of prostate cancer than in a selenium-replete population.
Despite our rationale, we found no effect of SEPP1 genotype per se on the risk of prostate cancer or on non-aggressive or aggressive prostate cancer. A similar null-effect of this genotype was found in colorectal cancer [10]. We did, however, find an effect of genotype in a pathway associated with selenoprotein function: although SOD2 is not a selenoprotein, the product of its activity, H2O2, is a substrate for GPx. The Val16Ala SNP in SOD2 has been shown to alter the secondary structure of the mitochondrial import sequence of the superoxide dismutase protein such that the Ala16 variant is imported more efficiently into the mitochondrial matrix, resulting in higher enzyme activity [22]. Individuals with at least one SOD2 Ala16 allele (Ala16+) therefore generate more active superoxide dismutase (and therefore more H2O2) than those homozygous for the Val16 variant. In our study, SOD2 Ala16+ men were at a 19% increased risk of prostate cancer compared to Val16 homozygotes (Table 1). The mitochondrion contains little or no catalase and so is entirely dependent on the activity of GPx to remove H2O2 [23], though of course H2O2 is sufficiently long-lived to diffuse out of the mitochondrion.
H2O2 promotes prostate cancer cell proliferation and migration [24, 25], and induces matrix metalloproteinases required for tumor invasion [26, 27]. For instance, H2O2 levels rose in cell-lines of the LNCaP series as tumorigenic and metastatic potential increased [24]. Furthermore, addition of ebselen, a GPx mimetic, to the assay completely abolished the chemiluminescence attributable to H2O2 [24]. At low selenium concentration where there is insufficient GPx activity to remove H2O2, the SOD2 16Ala allele would therefore be expected to have a deleterious effect owing to the higher H2O2 production associated with that allele variant. Our population has a mean plasma selenium concentration well below the level required to fully optimise GPx (as plasma GPx) [20] making our observation consistent with predictions.
Furthermore, the mean selenium concentration in our study was below the bottom of the range of plasma selenium (84-131 μg/L) in the study in which Li et al [13] reported an association between SOD2 Ala16Val genotype and prostate cancer risk when subjects were divided according to quartile of selenium status. In that study, high selenium status was advantageous for SOD2 Ala16 homozygotes as the combination of high SOD2 activity in the mitochondrion, together with high selenium/GPx, enabled efficient removal of both reactive oxygen species, superoxide and H2O2. By contrast, Ala16 homozygotes were at increased risk, particularly of aggressive prostate cancer, when their selenium status was in the bottom quartile [13]. While we saw an overall effect of SOD2 Ala16Val genotype in our low-selenium population, Li et al [13] observed no overall effect, perhaps because the range of selenium status in their population encompassed both positive and negative effects of SOD2 genotype on prostate cancer risk leading to a null effect overall. Our findings might help explain why the SOD2 Ala16Val risk allele appears to vary from study to study in a number of cancers and illustrates a gene-nutrient interaction where the effect of genotype on risk reverses with change in nutrient status of the population.
Results from the ATBC study may further illustrate this point. Finnish smokers who were SOD2 Ala16 homozygotes had a 70% increased risk of prostate cancer when compared to Val homozygotes [28]. This is consistent with the findings of Li et al in their lowest selenium quartile where the SOD Ala16 homozygotes had an increased risk of prostate cancer [13] and suggests that men from the ATBC cohort must have had a similar selenium status to the bottom quartile of US men. Although at first sight this might seem surprising since the cohort was recruited after the introduction of selenium-enriched fertilizers in Finland, in fact there are data to show that the ATBC study did indeed include men with very low Se status [29]. When the use of selenized fertilisers was implemented in 1984, mean plasma selenium in Finland was 70 μg/L. However, selenium status did not peak until 1990, reaching a maximum of 120 μg/L (cf mean US value 125 μg/L [19]), before declining to 90 μg/L in 1999 [30]. Thus the mean selenium status of this Finnish cohort, recruited from 1985-88 and followed up until death or 1993, would have been considerably lower than that of the US men and would certainly have been lower during the time this slow-growing cancer was developing, prior to the initiation of the selenized fertilizer program.
We found evidence for a gene-gene interaction between the Ala234Thr polymorphism in the Se transport protein, SEPP1, and the SOD2 Val16Ala polymorphism. Individuals homozygous for SEPP1 Ala234, SOD2 Ala16+ were 43% more likely to have prostate cancer compared to Val16 homozygotes (Table 2); this interaction was stronger in aggressive disease resulting in a 60% increased risk. Prospective cohort studies have shown a stronger association between selenium status and risk of aggressive than localized disease [31-35].
Of relevance to our findings, Méplan et al recently found that male SEPP1 Ala234 homozygotes had lower plasma selenoprotein P than heterozygotes, with an associated trend for reduced activity or protein concentration of GPx1 (cytosolic GPx) and GPx4 (phospholipid GPx) [12]. The authors suggest that the Ala234Thr polymorphism affects the stability of selenoprotein P, possibly through a post-translational modification, affecting protein levels only when selenium intake is suboptimal. Thus genotype-dependent differences in plasma selenoprotein P concentration were only observed under relatively low selenium conditions and disappeared after supplementation with selenium [12]. Our observations in a population of relatively-low selenium status are consistent with those findings.
Because there is strong evidence that selenoprotein P plays a critical role in delivery of hepatic selenium to other tissues [8, 9, 36], we hypothesise that men homozygous for the SEPP1 Ala234 allele have reduced availability of selenium for the production of GPx in the prostate. This would impede the ability of the prostate to remove H2O2 and would therefore increase the risk of prostate cancer [24, 25] (Fig. 2). In our study, this polymorphism only affected prostate cancer risk in conjunction with the SOD2 Ala16 allele, which, by allowing more efficient transport of superoxide dismutase into the mitochondrion, caused increased production of H2O2 that became detrimental in the face of low selenium. The gene-gene interaction between the SEPP1 Ala234Thr and SOD2 Val16Ala polymorphisms was also apparent in current or ex-smokers who had a two-fold increased risk of prostate cancer compared with SOD2 Val16 homozygotes (Table 4). As we have postulated that the mechanism by which these polymorphisms have their combined effect is oxidative-stress related, the greater strength of the interaction in smokers than in the study as a whole, despite smaller numbers, might be explained by an exacerbation of oxidative stress in ever-smokers though it may equally well be related to the lower selenium status seen in smokers [37, 38]. In fact we did not observe lower selenium status among smokers in our study but this may have been due to the small number (n=169) of plasma selenium measurements made.
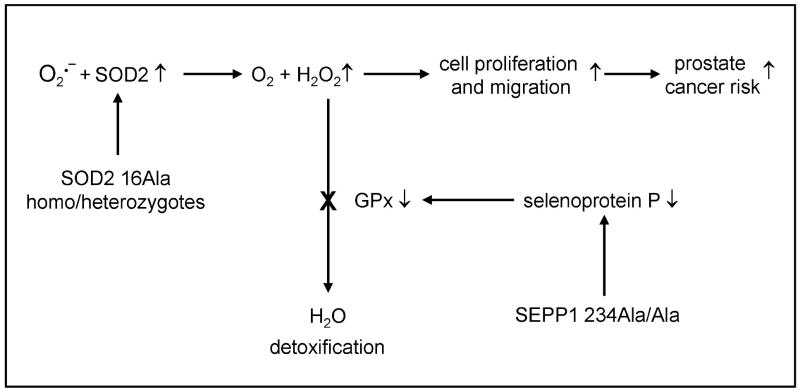
Figure 2.
Postulated mitochondrial mechanism for the interaction between SEPP1 and SOD2 polymorphisms and the risk of prostate cancer. Possession of a SOD2 Ala16 allele promotes higher import of mitochondrial superoxide dismutase to the mitochondrion resulting in higher activity and greater production of H2O2 than with the SOD2 Val16 variant. The availability of selenium for the production of GPx in the prostate, already low in the Swedish population, is further reduced in men homozygous for the SEPP1 Ala 234 allele, reducing the ability of the prostate to protect against excess hydrogen peroxide (promoter of cell proliferation and migration[24, 25]) and cancer.
Our study has limitations in that HapMap‡ shows considerable linkage disequilibrium (LD) at both these gene loci so we cannot be sure that these polymorphisms are the only functional SNPs affecting risk of prostate cancer in the SOD2 and SEPP1 genes. However, given the published studies implying functional consequences of the amino-acid changes, it would seem plausible that they have some function in this context. Both of these SNPs were included in the recent genome-wide screen of a subset of our case-control study (498 aggressive cases and 494 controls) and results are consistent with the present data on a larger sample [39]. Other recent genome-wide SNP screens in prostate cancer have not implicated either of these genes [40-42], but the UK study did find an associated SNP (rs9364554) in the SLC22A3 gene [40] on chromosome 6, just over 700kb telomeric of SOD2. Analysis of HapMap CEPH data reveals no LD between this SNP and SOD2 Val16Ala, suggesting that the SOD2 SNP is independently associated with prostate cancer risk and is not acting as a marker for SLC22A3 association.
If confirmed, the evidence presented here would allow identification of individuals who would particularly benefit from selenium supplementation to prevent prostate cancer. Among the Swedish men in our study, 41% had the high risk genotype combination (homozygosity for SEPP1 Ala 234 and possession of an SOD2 Ala16 allele). Similar allele frequencies have been identified in other populations of European descent † [12, 13]. We have observed the combined effect of the SOD2 Val16Ala and SEPP1 Ala234Thr SNPs under conditions of limited selenium availability. Such an effect can probably not be observed in areas of higher selenium status such as North America where plentiful selenium supply can probably obviate the disadvantage of homozygosity for SEPP1 234 Ala except for those with the lowest intakes [13]. Optimising the selenium intake of individuals homozygous for SEPP1 Ala234 will improve their ability to supply selenium via selenoprotein P for GPx synthesis and H2O2 removal [12]. Confirmation of our findings in other populations with low selenium intake should be a high priority. So far, few genetic tests have had the potential to ameliorate risk by identifying the need for a simple, inexpensive, nutritional supplement.
Acknowledgments
The authors are grateful to Dr Katarina Bälter for her work on the CAPS study, to Professor Jan-Erik Johansson and his colleagues at the Swedish Prostate Cancer Registry, and to Bram Bekaert for helpful discussions.
Funding For the CAPS study, Swedish Cancer Society (Cancerfonden) and Swedish Academy of Sciences; for the current investigation, National Institutes of Health (R03CA110893-2); the UK Prostate Cancer Charitable Trust; Lallemand Animal Nutrition (postgraduate studentship to MLC).
Abbreviations
- PC
prostate cancer
- APC
aggressive prostate cancer at diagnosis
- NPC
non-aggressive prostate cancer at diagnosis
- SNP
single nucleotide polymorphism
- SEPP1
selenoprotein P gene
- SOD2
mitochondrial superoxide dismutase gene
- CAPS
CAncer of the Prostate in Sweden study
Footnotes
NCBI SNP database www.ncbi.nlm.nih.gov/SNP/
References
- 1.Rayman MP. Selenium in cancer prevention: a review of the evidence and mechanism of action. Proc Nutr Soc. 2005;64:527–42. doi: 10.1079/pns2005467. [DOI] [PubMed] [Google Scholar]
- 2.Duffield-Lillico AJ, Dalkin BL, Reid ME, Turnbull BW, Slate EH, Jacobs ET, et al. Selenium supplementation, baseline plasma selenium status and incidence of prostate cancer: an analysis of the complete treatment period of the Nutritional Prevention of Cancer Trial. BJU Int. 2003;91:608–12. doi: 10.1046/j.1464-410x.2003.04167.x. [DOI] [PubMed] [Google Scholar]
- 3.Duffield-Lillico AJ, Reid ME, Turnbull BW, Combs GF, Jr, Slate EH, Fischbach LA, et al. Baseline characteristics and the effect of selenium supplementation on cancer incidence in a randomized clinical trial: a summary report of the Nutritional Prevention of Cancer Trial. Cancer Epidemiol Biomarkers Prev. 2002;11:630–9. [PubMed] [Google Scholar]
- 4.Clark LC, Combs GF, Jr, Turnbull BW, Slate EH, Chalker DK, Chow J, et al. Effects of selenium supplementation for cancer prevention in patients with carcinoma of the skin. A randomized controlled trial. Nutritional Prevention of Cancer Study Group. Jama. 1996;276:1957–63. [PubMed] [Google Scholar]
- 5.Irons R, Carlson BA, Hatfield DL, Davis CD. Both selenoproteins and low molecular weight selenocompounds reduce colon cancer risk in mice with genetically impaired selenoprotein expression. J Nutr. 2006;136:1311–7. doi: 10.1093/jn/136.5.1311. [DOI] [PubMed] [Google Scholar]
- 6.Diwadkar-Navsariwala V, Prins GS, Swanson SM, Birch LA, Ray VH, Hedayat S, et al. Selenoprotein deficiency accelerates prostate carcinogenesis in a transgenic model. Proc Natl Acad Sci U S A. 2006;103:8179–84. doi: 10.1073/pnas.0508218103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moschos MP. Selenoprotein P. Cell Mol Life Sci. 2000;57:1836–45. doi: 10.1007/PL00000665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hill KE, Zhou J, McMahan WJ, Motley AK, Atkins JF, Gesteland RF, et al. Deletion of selenoprotein P alters distribution of selenium in the mouse. J Biol Chem. 2003;278:13640–6. doi: 10.1074/jbc.M300755200. [DOI] [PubMed] [Google Scholar]
- 9.Schomburg L, Schweizer U, Holtmann B, Flohe L, Sendtner M, Kohrle J. Gene disruption discloses role of selenoprotein P in selenium delivery to target tissues. Biochem J. 2003;370:397–402. doi: 10.1042/BJ20021853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Al-Taie OH, Uceyler N, Eubner U, Jakob F, Mork H, Scheurlen M, et al. Expression profiling and genetic alterations of the selenoproteins GI-GPx and SePP in colorectal carcinogenesis. Nutr Cancer. 2004;48:6–14. doi: 10.1207/s15327914nc4801_2. [DOI] [PubMed] [Google Scholar]
- 11.Calvo A, Xiao N, Kang J, Best CJ, Leiva I, Emmert-Buck MR, et al. Alterations in gene expression profiles during prostate cancer progression: functional correlations to tumorigenicity and down-regulation of selenoprotein-P in mouse and human tumors. Cancer Res. 2002;62:5325–35. [PubMed] [Google Scholar]
- 12.Meplan C, Crosley LK, Nicol F, Beckett GJ, Howie AF, Hill KE, et al. Genetic polymorphisms in the human selenoprotein P gene determine the response of selenoprotein markers to selenium supplementation in a gender-specific manner (the SELGEN study) Faseb J. 2007;21:3063–74. doi: 10.1096/fj.07-8166com. [DOI] [PubMed] [Google Scholar]
- 13.Li H, Kantoff PW, Giovannucci E, Leitzmann MF, Gaziano JM, Stampfer MJ, et al. Manganese superoxide dismutase polymorphism, prediagnostic antioxidant status, and risk of clinical significant prostate cancer. Cancer Res. 2005;65:2498–504. doi: 10.1158/0008-5472.CAN-04-3535. [DOI] [PubMed] [Google Scholar]
- 14.Sutton A, Imbert A, Igoudjil A, Descatoire V, Cazanave S, Pessayre D, et al. The manganese superoxide dismutase Ala16Val dimorphism modulates both mitochondrial import and mRNA stability. Pharmacogenet Genomics. 2005;15:311–9. doi: 10.1097/01213011-200505000-00006. [DOI] [PubMed] [Google Scholar]
- 15.Lindmark F, Zheng SL, Wiklund F, Bensen J, Balter KA, Chang B, et al. H6D polymorphism in macrophage-inhibitory cytokine-1 gene associated with prostate cancer. J Natl Cancer Inst. 2004;96:1248–54. doi: 10.1093/jnci/djh227. [DOI] [PubMed] [Google Scholar]
- 16.Olsson M, Lindstrom S, Haggkvist B, Adami HO, Balter K, Stattin P, et al. The UGT2B17 gene deletion is not associated with prostate cancer risk. Prostate. 2008;68:571–5. doi: 10.1002/pros.20700. [DOI] [PubMed] [Google Scholar]
- 17.Sloth JJ, Larsen EH. The application of inductively coupled plasma dynamic reaction cell mass spectrometry for measurement of selenium isotopes, isotope ratios and chromatographic detection of selenoamino acids. Journal of Analytical Atomic Spectrometry. 2000;15:669–672. [Google Scholar]
- 18.Sieniawska CE, Mensikov R, Delves HT. Determination of total selenium in serum, whole blood and erythrocytes by ICP-MS. Journal of Analytical Atomic Spectrometry. 1999;14:109–112. [Google Scholar]
- 19.Niskar AS, Paschal DC, Kieszak SM, Flegal KM, Bowman B, Gunter EW, et al. Serum selenium levels in the US population: Third National Health and Nutrition Examination Survey, 1988-1994. Biol Trace Elem Res. 2003;91:1–10. doi: 10.1385/BTER:91:1:1. [DOI] [PubMed] [Google Scholar]
- 20.Duffield AJ, Thomson CD, Hill KE, Williams S. An estimation of selenium requirements for New Zealanders. Am J Clin Nutr. 1999;70:896–903. doi: 10.1093/ajcn/70.5.896. [DOI] [PubMed] [Google Scholar]
- 21.Xia Y, Hill KE, Byrne DW, Xu J, Burk RF. Effectiveness of selenium supplements in a low-selenium area of China. Am J Clin Nutr. 2005;81:829–34. doi: 10.1093/ajcn/81.4.829. [DOI] [PubMed] [Google Scholar]
- 22.Sutton A, Khoury H, Prip-Buus C, Cepanec C, Pessayre D, Degoul F. The Ala16Val genetic dimorphism modulates the import of human manganese superoxide dismutase into rat liver mitochondria. Pharmacogenetics. 2003;13:145–57. doi: 10.1097/01.fpc.0000054067.64000.8f. [DOI] [PubMed] [Google Scholar]
- 23.Chance B, Sies H, Boveris A. Hydroperoxide metabolism in mammalian organs. Physiol Rev. 1979;59:527–605. doi: 10.1152/physrev.1979.59.3.527. [DOI] [PubMed] [Google Scholar]
- 24.Lim SD, Sun C, Lambeth JD, Marshall F, Amin M, Chung L, et al. Increased Nox1 and hydrogen peroxide in prostate cancer. Prostate. 2005;62:200–7. doi: 10.1002/pros.20137. [DOI] [PubMed] [Google Scholar]
- 25.Polytarchou C, Hatziapostolou M, Papadimitriou E. Hydrogen peroxide stimulates proliferation and migration of human prostate cancer cells through activation of activator protein-1 and up-regulation of the heparin affin regulatory peptide gene. J Biol Chem. 2005;280:40428–35. doi: 10.1074/jbc.M505120200. [DOI] [PubMed] [Google Scholar]
- 26.Zhang HJ, Zhao W, Venkataraman S, Robbins ME, Buettner GR, Kregel KC, et al. Activation of matrix metalloproteinase-2 by overexpression of manganese superoxide dismutase in human breast cancer MCF-7 cells involves reactive oxygen species. J Biol Chem. 2002;277:20919–26. doi: 10.1074/jbc.M109801200. [DOI] [PubMed] [Google Scholar]
- 27.Kinnula VL, Crapo JD. Superoxide dismutases in malignant cells and human tumors. Free Radic Biol Med. 2004;36:718–44. doi: 10.1016/j.freeradbiomed.2003.12.010. [DOI] [PubMed] [Google Scholar]
- 28.Woodson K, Tangrea JA, Lehman TA, Modali R, Taylor KM, Snyder K, et al. Manganese superoxide dismutase (MnSOD) polymorphism, alpha-tocopherol supplementation and prostate cancer risk in the alpha-tocopherol, beta-carotene cancer prevention study (Finland) Cancer Causes Control. 2003;14:513–8. doi: 10.1023/a:1024840823328. [DOI] [PubMed] [Google Scholar]
- 29.Hartman TJ, Taylor PR, Alfthan G, Fagerstrom R, Virtamo J, Mark SD, et al. Toenail selenium concentration and lung cancer in male smokers (Finland) Cancer Causes Control. 2002;13:923–8. doi: 10.1023/a:1021912117067. [DOI] [PubMed] [Google Scholar]
- 30.Eurola M, Hietaniemi V. Publications of Agricultural Research Centre of Finland. Vol. 24 Jokioinen: Agricultural Research centre of Finland; 2000. Report of the Selenium Monitoring Programme 1997-1999. [Google Scholar]
- 31.Yoshizawa K, Willett WC, Morris SJ, Stampfer MJ, Spiegelman D, Rimm EB, et al. Study of prediagnostic selenium level in toenails and the risk of advanced prostate cancer. J Natl Cancer Inst. 1998;90:1219–24. doi: 10.1093/jnci/90.16.1219. [DOI] [PubMed] [Google Scholar]
- 32.Helzlsouer KJ, Huang HY, Alberg AJ, Hoffman S, Burke A, Norkus EP, et al. Association between alpha-tocopherol, gamma-tocopherol, selenium, and subsequent prostate cancer. J Natl Cancer Inst. 2000;92:2018–23. doi: 10.1093/jnci/92.24.2018. [DOI] [PubMed] [Google Scholar]
- 33.Nomura AM, Lee J, Stemmermann GN, Combs GF., Jr Serum selenium and subsequent risk of prostate cancer. Cancer Epidemiol Biomarkers Prev. 2000;9:883–7. [PubMed] [Google Scholar]
- 34.van den Brandt PA, Zeegers MP, Bode P, Goldbohm RA. Toenail selenium levels and the subsequent risk of prostate cancer: a prospective cohort study. Cancer Epidemiol Biomarkers Prev. 2003;12:866–71. [PubMed] [Google Scholar]
- 35.Li H, Stampfer MJ, Giovannucci EL, Morris JS, Willett WC, Gaziano JM, et al. A prospective study of plasma selenium levels and prostate cancer risk. J Natl Cancer Inst. 2004;96:696–703. doi: 10.1093/jnci/djh125. [DOI] [PubMed] [Google Scholar]
- 36.Renko K, Werner M, Renner-Muller I, Cooper TG, Yeung CH, Hollenbach B, et al. Hepatic selenoprotein P (SePP) expression restores selenium transport and prevents infertility and motor-incoordination in Sepp-knockout mice. Biochem J. 2008;409:741–9. doi: 10.1042/BJ20071172. [DOI] [PubMed] [Google Scholar]
- 37.Bates CJ, Thane CW, Prentice A, Delves HT. Selenium status and its correlates in a British national diet and nutrition survey: people aged 65 years and over. J Trace Elem Med Biol. 2002;16:1–8. doi: 10.1016/s0946-672x(02)80002-5. [DOI] [PubMed] [Google Scholar]
- 38.Northrop-Clewes CA, Thurnham DI. Monitoring micronutrients in cigarette smokers. Clin Chim Acta. 2007;377:14–38. doi: 10.1016/j.cca.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 39.Duggan D, Zheng SL, Knowlton M, Benitez D, Dimitrov L, Wiklund F, et al. Two genome-wide association studies of aggressive prostate cancer implicate putative prostate tumor suppressor gene DAB2IP. J Natl Cancer Inst. 2007;99:1836–44. doi: 10.1093/jnci/djm250. [DOI] [PubMed] [Google Scholar]
- 40.Eeles RA, Kote-Jarai Z, Giles GG, Olama AA, Guy M, Jugurnauth SK, et al. Multiple newly identified loci associated with prostate cancer susceptibility. Nat Genet. 2008 doi: 10.1038/ng.90. [DOI] [PubMed] [Google Scholar]
- 41.Gudmundsson J, Sulem P, Rafnar T, Bergthorsson JT, Manolescu A, Gudbjartsson D, et al. Common sequence variants on 2p15 and Xp11.22 confer susceptibility to prostate cancer. Nat Genet. 2008 doi: 10.1038/ng.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thomas G, Jacobs KB, Yeager M, Kraft P, Wacholder S, Orr N, et al. Multiple loci identified in a genome-wide association study of prostate cancer. Nat Genet. 2008 doi: 10.1038/ng.91. [DOI] [PubMed] [Google Scholar]