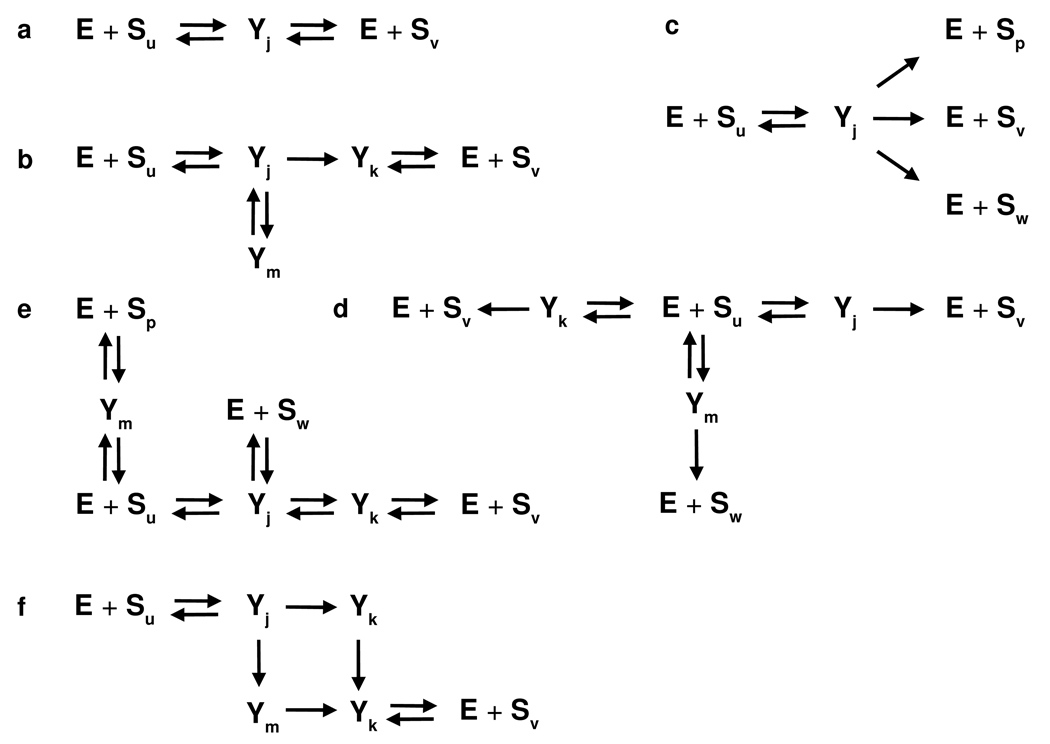Abstract
Post-translational modification of proteins plays a central role in cellular regulation but its study has been hampered by the exponential increase in substrate modification forms (“modforms”) with increasing numbers of sites. We consider here biochemical networks arising from post-translational modification under mass-action kinetics, allowing for multiple substrates, having different types of modification (phosphorylation, methylation, acetylation, etc) on multiple sites, acted upon by multiple forward and reverse enzymes (in total number L), using general enzymatic mechanisms. These assumptions are substantially more general than in previous studies. We show that the steady-state modform concentrations constitute an algebraic variety that can be parameterised by rational functions of the L free enzyme concentrations, with coefficients which are rational functions of the rate constants. The parameterisation allows steady states to be calculated by solving L algebraic equations, a dramatic reduction compared to simulating an exponentially large number of differential equations. This complexity collapse enables analysis in contexts that were previously intractable and leads to biological predictions that we review. Our results lay a foundation for the systems biology of post-translational modification and suggest deeper connections between biochemical networks and algebraic geometry.
Keywords: algebraic geometry, King-Altman, Matrix-Tree theorem, multisite PTM, rational variety, systems biology
1. Introduction
Post-translational modification (PTM) of proteins is a central regulatory mechanism in eukaryotic cells [44]. Although phosphorylation was the first modification to be discovered [12, 24], and remains the best studied, proteins are subject to other types of modification by covalent attachment of molecules to the side chains of amino acid residues. The modifiers include other small molecules, as in methylation, acetylation and sulfation, complex sugars, as in glycosylation, and small protein moieties, as in ubiquitin-like modification [44]. It has become increasingly clear that these modifications work together to orchestrate cellular function [20].
PTMs are dynamically maintained. Forward enzymes, which transfer modifiers from donor molecules to specific residues, are usually competing against reverse enzymes, which hydrolyse modified residues, detaching the modifier and returning it to the pool from which donor molecules are synthesised. In the case of phosphorylation, these enzymes are protein kinases and phosphoprotein phosphatases, respectively. These so-called “futile cycles” of modification and demodification can use energy to keep the concentrations of modified substrates far from equilibrium, testifying to the importance of such processes as regulatory mechanisms.
A given protein may be modified on multiple sites. For instance, the transcription factor and tumour suppressor p53 has 18 serine/threonine sites that can be phosphorylated and 10 lysine sites that can accommodate acetylation, methylation and attachment of ubiquitin, SUMO and NEDD [25]. O-linked modifications like phosphorylation, on residues like serine, threonine or tyrosine, are digital—at most one phosphate group is attached to each residue—but N-linked modifications like methylation, on residues like lysine or arginine, can be more complex [44]. Ubiquitin, in particular, can form linear and branched poly-ubiquitin chains connected by iso-peptide linkages through lysine residues. In general, an individual molecule with n sites of modification may be in one of several global states of modification (“modforms”), whose numbers increase exponentially with n. Cartoon diagrams often pick one of these modforms, usually the maximally modified one, to depict the state of a protein in the cell. However, there is always a population of such molecules present and individual molecules in the population may be in different states. The state of the population is best described as a frequency distribution over these single molecule states. We call this the modform distribution. Not all modforms will necessarily be present at any one time but this begs the question of which modforms are present and of how the relevant forward and reverse enzymes cooperate to shape the modform distribution. The methods of this paper were developed in part to address such questions.
The combinatorial explosion in the number of potential modforms is a challenge for both experiment and theory. Mass spectrometry techniques have only recently begun to provide data on modform distributions [32, 33], prompted, in part, by the growing realisation that different modforms may have distinct biological effects [31, 34, 45]. The theoretical challenge lies in the complexity of any mathematical model, which must accommodate some number, L, of forward and reverse enzymes and some total number, N, of modforms targetted by those enzymes. Additionally, the biochemistry of modification and demodification usually requires intermediate enzyme-substrate complexes associated to each enzyme and its substrates. If there are P such intermediate complexes, then the model will have L+N+P state variables, where N and P grow exponentially with n, while L is relatively much smaller:
.
Because the dynamical equations are non-linear, these models cannot be analytically solved for the temporal trajectories of the variables. Simulation provides the widely used alternative to analytical solution. The trajectories of the system can be calculated by numerical integration once the site-specific rate constants have been given values. These values have usually not been measured and it may be necessary to search through the space of rate constants to determine whether a particular behaviour is robust to the choice of rate constant values [18]. Calculations of this kind rapidly become infeasible with increasing n, which has limited simulation studies to systems with few sites. These difficulties have made it hard to see what, if any, general principles lie behind the widespread use of multisite modification systems in cellular regulation.
In this paper we show that, if attention is restricted to the steady states of a multisite PTM system, then it is not necessary to numerically integrate L + N + P differential equations but only to solve L algebraic equations (Theorem 3). Furthermore, this reduction can be carried out without having to specify the rate constant values in advance. For instance, for the case of two enzymes, the steady states can be found by an analogue of the nullcline analysis that is widely used for two-dimensional systems of differential equations [40]: the steady states correspond to the intersections of two curves in the plane, no matter what the number of sites. This exponential reduction in complexity leads to biological insights in contexts that were previously intractable, as reviewed in §4, and provides a new theoretical foundation for studying multisite PTM systems.
The present paper generalises and conceptually simplifies a method that emerged in previous work for systems with two enzymes and one substrate [27, 41]. It allows for multiple enzymes, multiple types of modification, multiple substrates and complex biochemistry of modification and demodification. These assumptions are substantially more general than any used previously in the literature, [11, 14, 16, 22, 26, 28, 37, 38]. The main restrictions in terms of applicability of our methods are the requirements that enzymes cannot also be substrates and that the recharging mechanism for each modification should keep the concentration of donor molecules constant on the time scale of steady state formation. The latter requirement is widely believed to hold for phosphorylation, in which the donor molecule is ATP, and it has been taken for granted in all mathematical models of phosphorylation, but its validity for other forms of modification appears not to have been widely studied.
The two most significant examples that currently fall outside the scope of our analysis are kinase cascades and ubiquitin-like modifications. Both violate the first requirement by relying on chains of enzymatic modification. We discuss how our results might be extended to such cases in §4. It is an interesting question whether ubiquitin-like modifications satisfy the second requirement. Peptide modifiers are synthesised by mRNA translation rather than by the cell’s central metabolism, as is the case for small molecule modifiers, and little is known about how effectively this translation process is buffered against varying demand.
Our method of proof is one of hierarchical elimination of variables from the steady-state equations, which, because of mass-action, are polynomials in the variables. The intermediate enzyme-substrate complexes are first eliminated in favour of the substrates and the enzymes (Proposition 1). Using the results of this, the substrates are then eliminated in favour of the enzymes (Proposition 2). Each elimination step is framed as the solution to a system of linear equations (Lemma 2) but is undertaken over an extension field of the real numbers, which carries the nonlinearity that is present in the underlying steady-state equations. This allows us to solve a nonlinear system of equations using essentially linear methods. The extension field also permits the rate constants to be treated symbolically during the elimination. The Matrix-Tree theorem (Theorem 1) plays a key role in identifying the nonlinear coefficients that arise in the elimination steps. This important graph-theoretic result goes back to the 19th century but was first stated in the form we use it in 1948 [43]. It does not seem to have been previously noticed that the Matrix-Tree theorem immediately implies the famous King-Altman method, developed in 1956 [23], for calculating the rate function of an enzyme [5]. We find it intriguing that the method we use to analyse multisite PTM systems is so closely related to a key result of classical biochemistry. §2 reviews the graph theoretic preliminaries, including the Matrix-Tree theorem, before the main results are presented in §3.
Our results are not derivable from any existing mathematical theories of biochemical reaction networks, such as Chemical Reaction Network Theory (CRNT) [9, 15], more recently developed injectivity methods [8, 39] or Monotone Systems Theory [1]. We believe that they are best interpreted in algebraic geometric terms, as discussed at more length in an earlier paper [27]. Under the mass-action kinetics used here, a network of biochemical reactions gives rise to a polynomial dynamical system. Hence, for given rate constant values, the steady states form a real algebraic variety [6]. Despite this, the use of algebraic geometric methods to study biochemical networks has been surprisingly limited, the most interesting exception being the use of toric varieties to reinterpret the Deficiency Zero theorem of CRNT [7, 13]. For multisite PTM systems, we show that the variety of steady-state modform concentrations can be parameterised by L rational functions (Theorem 4). A rational parameterisation provides an explicit description of points on a variety, in contrast to their implicit definition as solutions of polynomial equations. Rationally parameterisable varieties are rare and of considerable interest in their own right. The rationality of multisite PTM systems suggests that algebraic geometry may provide powerful tools for analysing biochemical reaction networks and overcoming molecular complexity. We may hope, thereby, to better see the biological wood for the molecular trees.
2. Preliminaries
2.1. Symbols and polynomials
In this paper we will analyse systems of ordinary differential equations arising from networks of biochemical reactions under mass-action kinetics. The rate constants and dynamical variables are usually treated as real variables in ℝ. In our analysis, we will make use of certain directed graphs whose edges have labels like aX, where a is a rate constant and X is a dynamical variable in steady state. These labels must satisfy a positivity condition, expressed in Lemma 1 below. The rate constants can reasonably be taken to be positive but whether or not a dynamical variable is positive in steady state is more delicate. Even if a system is started with all its variables positive, it may not be persistent within the positive orthant and may reach a steady state on the boundary in which some variables are zero. To avoid having to rule out such situations and thereby limit our analysis, we take a more algebraic approach. We treat the rate constants and the dynamical variables in steady state as uninterpreted symbols in an appropriate extension field of ℝ. We show that the calculations can be carried out over this extension field and, having done them, we then give real values to the symbols and draw conclusions over ℝ. While this avoids the problem and brings added benefits, it incurs some technical cost. The reader will not lose much by assuming that all calculations take place over ℝ and ignoring the problems that arise with loss of positivity in the dynamical variables. The symbolic calculations only appear in §3.3 and §3.4; rate constants and dynamical variables are treated as real variables elsewhere.
For more information about the algebraic methods reviewed here see, for instance, [19]. For any finite set Q = {q1, …, qn}, ℝ[Q] will denote the ring of real polynomials in the elements of Q, considered as algebraically independent symbols. Recall that a polynomial p ∈ ℝ[Q] is a linear combination of monomials, p = ∑αcαqα, where cα ∈ ℝ and each monomial qα is a product of symbols,
ℝ(Q) will denote the field of rational functions in the elements of Q: the smallest field in which the symbols in Q can be added, subtracted, multiplied and divided as if they were (non-zero) numbers. Equivalently, ℝ(Q) is the field of fractions of ℝ[Q]: each element f ∈ ℝ(Q) can be expressed as the ratio of two polynomials, f = p1/p2 where p1, p2 ∈ ℝ[Q]. ℝ[Q] sits inside ℝ(Q) in the obvious way: p = p/1.
For various finite sets Q, we will use elements of ℝ[Q] as labels in graphs, and use ℝ(Q) as a field over which we solve linear equations. For instance, Q may consist of rate constants. To relate calculations over ℝ[Q] or ℝ(Q) back to biology, the symbols ultimately have to be given real values. This is not a problem for the polynomial ring ℝ[Q]. Any assignment, ι : Q → ℝ, extends to a homomorphism of rings ι : ℝ[Q] → ℝ. Hence, any symbolic algebraic expression in ℝ[Q] gives rise to a corresponding expression in ℝ. However, the field of rational functions, ℝ(Q), contains elements like 1/(ι(q1) − q1), which become undefined in ℝ no matter what assignment of real values are made to elements of Q. To infer an expression over ℝ, it is essential to show that nothing blows up in this way.
We make use of S-positivity to do this. A polynomial p ∈ ℝ[Q] is said to be S-positive (“sum positive”) if it is a non-zero sum of positive monomials. That is, p is S-positive if p = ∑αcαqα ≠ 0 and if cα > 0 whenever cα ≠ 0. A rational function in ℝ(Q) is said to be S-positive if it can be expressed as the ratio of two S-positive polynomials. If the elements of Q are given positive real values, which is biochemically realistic for rate constants, then any S-positive rational function in ℝ(Q) will be well defined over ℝ. Note that, if Q = ∅, so that ℝ[Q] = ℝ, then x ∈ ℝ[Q] is S-positive if, and only if, it is positive in the usual sense.
If p = ∑αcαxα ∈ ℝ[Q] then p = 0 means that cα = 0 for all α. If ι : Q → ℝ, then p = 0 in ℝ [Q] implies that ι(p) = 0 ∈ ℝ. The converse, of course, is false: if ι(p) = 0 ∈ ℝ it does not imply that p = 0 ∈ ℝ[Q]. However, if p ≠ 0 then the variety in ℝn corresponding to the set of solutions of p = 0 has dimension strictly less than n. Hence, if ι(p) = 0 for sufficiently many assignments ι, then p = 0. Let ℝ+ denote the positive reals.
Remark 1
If ι(p) = 0 for all ι : Q → ℝ+, then p = 0 ∈ ℝ[Q].
2.2. Graphs and Laplacians
A labelled, directed graph is a triple, (V, E, ℓ), where V is a finite set of nodes, V = {v1, …, vn}, E is a finite set of directed edges, E ⊆ V × V, and ℓ : E → 𝕂 − {0} is a function that associates to each edge a non-zero label in some field 𝕂. Because they take values in a field, labels can be treated algebraically as if they were numbers. Usually, 𝕂 = ℝ(Q) for some set of symbols Q and the labels are elements of ℝ[Q]. In place of a labelling function, the notation will denote an edge from vi to vj with label a. We will sometimes abbreviate this to vi → vj.
If G is a labelled, directed graph, G⋆ will denote the corresponding unlabelled, undirected graph, in which the direction of the edges is forgotten and multiple edges between the same vertices are merged. We use vi ↔ vj to denote an edge in G⋆. Note that this could imply vi → vj or vj → vi or both in G. We say that G is connected if G⋆ is connected: if there is a chain of undirected edges linking any two vertices. Any labelled, directed graph is a disjoint union of connected components. G is strongly connected if there is a directed path between any two distinct vertices.
There is a bijective correspondence between matrices and labelled, directed graphs. If A is a n × n matrix over 𝕂, let 𝒢(A) be the associated labelled, directed graph with nodes {1, …, n} and labelled edges , if, and only if, Aij ≠ 0. Note that entry Aij goes fron j to i. If G is a labelled, directed graph on {1, …, n}, let ℳ(G) be the n × n matrix for which ℳ(G)ij = a if, and only if,. Evidently, ℳ(𝒢(A)) = A and 𝒢(M(G)) = G.
If G is a labelled directed graph then its Laplacian matrix, ℒ(G), is given by ℒ(G) = ℳ(G) − diag(1.ℳ(G)). Here, 1 denotes the all 1’s row vector of the appropriate dimension, 1 = (1, …, 1), and diag(v), where v is a row vector, denotes the diagonal matrix with v on the main diagonal
The Laplacian encodes much information about graph structure; see, for instance, [3], but note that the conventions used here are different. Note also that, by construction, 1.ℒ(G) = 0.
Lemma 1
Let G be a labelled, directed graph on n vertices with no self-loops in which each non-zero label is a S-positive element of ℝ[Q]. If G is strongly connected then the rank of ℒ(G) over ℝ(Q) is n − 1.
PROOF
Let u = (u1, …, un) ∈ ℝ(Q)n. Since 1.ℒ(G) = 0, it is sufficient to show that if u.ℒ(G) = 0 then u1 = … = un ∈ ℝ(Q). Express each ui ∈ ℝ(Q) as a fraction ui = fi/gi, where fi, gi ∈ ℝ[Q] and we may suppose that gi ≠ 0. We can now clear the denominators. Let λ = g1g2 … gn ≠ 0 and vi = λui. Then, vi ∈ ℝ[Q] and v.ℒ(G) = 0. We can now operate in the polynomial ring ℝ[Q] in preference to the field of fractions ℝ(Q). The entries in the Laplacian have the following form. If i ≠ j, ℒ(G)ij is either zero or is a S-positive element of ℝ[Q], while the diagonal entries are given by
Hence, for the i-th column of v.ℒ(G) = 0,
| (1) |
So far, this has all been symbolic, in ℝ[Q]. Now suppose that the symbols in Q are given positive real values and, suppressing the corresponding assignment ι : Q → ℝ+ for readability, let us consider the corresponding system of column equations to (1) in ℝ. Since the vi are now real numbers, there is a smallest, vp, for which vi ≥ vp for all i. Let U ⊆ {1, …, n} be the set of those indices i for which vi = vp. U ≠ ∅ since p ∈ U. If m ∈ U, then, in the m-th column equation,
each non-zero term is the product of a nonnegative quantity, vk − vm, since vm is smallest, and a strictly positive quantity ℒ(G)km, since each label in G is S-positive. It follows that vk = vm = vp whenever there is an edge m → k, so that U is closed under outgoing edges. Since G is strongly connected, U = {1, …, n}. Hence, vi = vp ∈ ℝ for all i. Since this holds for any assignment of positive values to symbols in Q we deduce from Remark 1 that v1 = … = vn symbolically in ℝ[Q]. Since λ ≠ 0 we see that u1 = … = un ∈ ℝ(Q), as required.
Some condition on the labels is necessary for the conclusion of Lemma 1. The following labelled, directed graph on {1, 2, 3}, with labels in ℝ, is strongly connected
but its Laplacian has rank 1, not 2:
Remark 2
If M is any n × n matrix, we can construct a labelled directed graph with no self-loops by ignoring the labels on the main diagonal: G = 𝒢(M − diag(Mii)). If, in addition, 1.M = 0, then M is the Laplacian of G:
2.3. The Matrix-Tree Theorem
In what follows we will need to solve linear systems of the form M.z = 0, where M is a n × n matrix over some field 𝕂, z is a column vector of n unknowns and M has rank n − 1. Recall that the adjugate matrix of M, adj(M), is defined by
| (2) |
where M(ji) is the maximal minor given by the determinant of the (n − 1) × (n − 1) matrix obtained from M by removing the jth row and ith column. Note the reversal of indices in (2). The adjugate satisfies the Laplace relations
If rk(M) = n−1, any column vector of adj(M) is a basis for the column null space. Taking the first column, let
| (3) |
so that M.z = 0.
The maximal minors have a particularly striking form when M is a Laplacian matrix. Let G be a labelled, directed graph with no self loops. T is said to be a spanning tree of G if T is a directed subgraph which reaches each node of G such that T⋆ is connected and acyclic. T inherits labels from G. T is said to be rooted at v ∈ G if v is the unique sink in T. That is, v is the only node of T with no edges leaving it, v ↛ w. This implies that any non-root node has exactly one edge leaving it, for otherwise there would either be an additional sink or an undirected cycle. Let Θv(G) denote the set of spanning trees of G rooted at v.
Theorem 1
(The Matrix-Tree Theorem [43, §3.6]) Let G be a directed graph on {1, …, n} with labels in the field 𝕂 and no self-loops. The maximal minors of the Laplacian are given by
It is remarkable that the maximal minor is, up to a sign, just a sum of products of labels, since the determinant itself is a sum with alternating signs. Results like Theorem 1 go back to the 19th century work of Kirchhoff on electrical networks and of Sylvester and others on elimination theory; see [29, Chapter 5] for references. Theorem 1 was first proved by Tutte in 1948 [43]. Combining (3) with Theorem 1, we get the following.
Lemma 2
Let M be a n × n matrix over a field 𝕂 such that 1.M = 0 and rk(M) = n − 1. Let G be the labelled, directed graph constructed as in Remark 2 for which M = ℒ(G). The one dimensional column null space of M is generated by the vector ρ = (ρ1, …, ρn)t, where,
| (4) |
There is a simple condition for x to also be in the null space of M.
Remark 3
Suppose that x, ρ ∈ 𝕂n with ρk ≠ 0 for some 1 ≤ k ≤ n. Then, x = λρ if, and only if, xi = (ρi/ρk)xk for 1 ≤ i ≤ n.
The quantities ρi/ρk will play an important role below; see (8), (20) and (24). In these calculations, the labels will lie in ℝ[Q] and will either be symbols, like q1, or S-positive polynomials, like q1q2+q3q4. Under these conditions, we see from (4) that each ρi is either 0, if there are no spanning trees rooted at vertex i, of, if there are such spanning trees, ρi is (−1)n+1 times a S-positive polynomial. Accordingly, ρi/ρk is a S-positive rational function.
2.4. The King-Altman method
If the biochemical mechanism of an enzyme is known, its rate function is often calculated in the quasi-steady state approximation [5]. King and Altman worked out a graphical method for doing this that is widely used [5, 23]. It seems not to have been previously noticed that this is an immediate application of the Matrix-Tree Theorem. Since we will see the Matrix-Tree Theorem at work in more detail later, we illustrate the application to rate functions with the simple example of reversible Michaelis-Menten kinetics. Here, enzyme E and substrate S reversibly form an enzyme-substrate complex Y, which reversibly yields enzyme and product P (compare Figure 1a):
| (5) |
Figure 1.
Examples of sub-networks. a. Michaelis-Menten style enzyme, with a single enzyme-substrate complex, Yj, and reversible product formation, as in (5). b. Example with two enzyme-substrate complexes, Yj, Yk, leading irreversibly to Sv, along with a dead-end complex Ym. c. Example used in [41], with a single enzyme-substrate complex Yj and multiple products Sp, Sv, Sw. d. Alternative network to c with a different enzyme-substrate complex for each product. e. Example with partially overlapping routes for different products. f. Cyclic network, which may not correspond to known biochemistry but is mathematically allowed.
The rate constants, a, b, c, d are all taken to be positive. The quasi-steady state approximation assumes that Y reaches steady state while substrate is being converted to product. Under mass-action kinetics, the differential equations for E and Y are
| (6) |
Note that E is at steady-state, if, and only if, Y is at steady state (compare Lemma 3). Hence, (5) is in quasi-steady state, if, and only, if, (6) is in steady state. At steady-state, the right hand side of (6) is a system of linear equations in E and Y. We can ignore the uninteresting case when S = P = 0 and assume that the coefficients of (6) are positive, thereby avoiding symbolic calculations. Let M be the corresponding matrix over ℝ for the basis {E, Y}. We see from (6) that 1.M = 0. Since rk(M) = 1 by inspection, we can use Lemma 2 to find solutions of M.z = 0. The labelled, directed graph formed from M according to Remark 2, is
| (7) |
This has a single spanning tree rooted at E, , and a single spanning tree rooted at Y, . By Lemma 2, a basis for the column null space of M is
By Remark 3, M.z = 0 if, and only, if
| (8) |
We see that, at steady state, the enzyme-substrate complex is the free enzyme times a linear combination of substrate and product. The coefficients of the linear form are reciprocals of the forward and reverse Michaelis-Menten constants, Kf = (b + c)/a, and Kr = (b + c)/d [5] (compare Proposition 1). The rate function can now be determined using the conservation law for the enzyme, zE + zY = Etot, giving, as in [5],
This simple calculation may provide some orientation for the more involved treatment that now follows.
3. Results
3.1. The system equations
We begin by setting up a general model for a PTM system. There are three kinds of chemical species in the system: enzymes, substrates and intermediate enzyme-substrate complexes. Let Enz, denote a non-empty, finite set of enzymes, Sub, a non-empty, finite set of substrate modforms and Int, a non-empty, finite set of enzyme-substrate complexes. A non-empty, finite set of sub-networks, Net, is defined in terms of these:
Each sub-network, T ∈ Net, consists of an associated enzyme, e(T) ∈ Enz, a non-empty subset of modforms, σ(T) ⊆ Sub, a non-empty subset of enzyme-substrate complexes, γ(T) ⊆ Int, and a reaction network, N(T), defined below. The sub-networks encode the biochemical details of how enzymes convert modforms.
This formulation allows for multiple forward and reverse enzymes, which may catalyse different types of modification and demodification. Multiple substrates are also permitted; the distinction between them will emerge in the calculation, as part of Condition 2. The combinatorics of multisite modification are not directly represented: the modforms of all substrates are simply listed 1, …, N. As discussed in the Introduction, N and P may be exponentially larger than L.
The enzyme-substrate subsets must be disjoint between distinct sub-networks: if i ≠ j then γ(Ti) ∩ γ(Tj) = ∅. However, distinct sub-networks may share both modforms and enzymes. We assume, without loss of generality, that each substrate is in some σ(T) and each enzyme-substrate complex in some γ(T)
Given Y ∈ Int, t(Y) ∈ Net denotes the (unique) sub-network containing Y.
For E = e(T), any Su ∈ σ(T) and any Yv, Yi, Yj ∈ γ(T), the sub-network N(T) is made up of any reactions of the following three kinds,
| (9) |
We further assume Condition 1 in §3.3 and Condition 2 in §3.4. These are strong connectivity conditions on certain graphs that allow Lemma 1 to be used. They will be stated after introducing additional concepts below.
The reactions (9) imply that enzyme is conserved—it is either free or bound in some enzyme-substrate complex—while substrate can flow between different modforms. While not much is known about the biochemical details of how enzymes modify multisite substrates, the sub-network assumptions allow considerable flexibility. They can accommodate, for instance, overlapping site preferences, arbitrary orders of modification and demodification, distributivity or processivity [11] and intricate hierarchical dependencies between enzymes [10, 35]. Some simple examples are shown in Figure 1. The main assumption behind (9) is that the donor molecules that provide the modifier are kept at constant concentration, on the time scale of the PTM dynamics, by mechanisms that are not explicitly modelled. As mentioned in the Introduction, this assumption has always been made for phosphorylation but needs further investigation for other modifications. It means that the donor molecules do not have to be treated as dynamical variables; their affects can be absorbed into the rate constants. This permits both forward and reverse reactions to be bimolecular with only enzyme and substrate. (In fact, we are implicitly making a similar assumption for the reverse reactions by ignoring the water molecules needed for hydrolysis.) Enzymes would normally be expected to give rise to tree networks, as in Figure 1a–e, but cyclic networks like Figure 1f are mathematically allowed.
The data above give rise to a polynomial dynamical system defined by mass-action kinetics on the set of chemical species, Sub ∪ Enz ∪ Int. For 1 ≤ u ≤ N; for 1 ≤ v ≤ P, T = t(Yv), E = e(t(Yv)); and for 1 ≤ w ≤ L,
| (10) |
| (11) |
| (12) |
Terms like in equation (11) are indexed over edges in the undirected graph N⋆ (T). If one or other corresponding directed edge is not present in N(T), the associated label should be treated as if it were zero. In other words,
Indexing over N⋆(T) is purely a notational convenience, which allows for a more compact syntax.
3.2. Conservation laws
In this section, the rate constants and dynamical variables in (10)–(12) are treated as real variables. The structure of these equations can be better seen in terms of fT, the net flux of enzyme out of sub-network T. By definition,
so that equation (12) can be rewritten as
| (13) |
Moreover, if (11) is added up for all Yv ∈ T, we get a sum of two terms. The first term counts each binomial twice with opposite sign and hence vanishes. The second term is just −fT. Hence,
| (14) |
Finally, the enzyme flux from T is equal to the flux of all substrate modforms from T. Summed over all sub-networks, this gives the total substrate flux. More formally, if equation (10) is added up for all substrates, then, rearranging the order of summation, and noting that each E + S ↔ Y is unique since the Y are not shared, we see that
| (15) |
From (13) and (14) we see that
The term being differentiated is evidently the total amount of enzyme Ew in the system, which we conclude to be the same at all times. Similarly, from (14) and (15) we see that
The term being differentiated is the total amount of substrate in the system, which must also be the same at all times. Let Stot be the total amount of substrate and Ew,tot the total amount of enzyme Ew. We see that, at all times, the following L + 1 conservation laws hold.
| (16) |
| (17) |
Of course, these conservation laws are evident from the form of the allowed reactions in (9) but the derivation above checks the correctness of the system equations and illuminates the role of fT. Reinforcing that, the following immediate consequence of (14) is a key result.
Lemma 3
In any steady state, for any sub-network T, fT = 0.
It is instructive to see this another way, which does not depend on the details of how (14) is proved. In any steady state, the enzyme flux out of T cannot be negative. In other words, there cannot be positive flux of enzyme into T. If there were, this flux cannot escape through any Yv ∈ T, since the enzyme-substrate complexes are not shared between sub-networks. Hence, it would accumulate somewhere, violating the steady state assumption. Accordingly, fT ≥ 0. But then from (13), dEw/dt is a sum of nonnegative terms, so that if dEw/dt = 0, then fT = 0 for each e(T) = Ew.
3.3. Generalised Michaelis-Menten constants
Lemma 3 says that, at steady state, not merely is dEw/dt = 0, but each individual fT = 0. This decouples the system at steady state and allows us to treat each sub-network in isolation.
In this section and the next we will work symbolically. Let Con denote the set of all rate constants for all sub-networks T ∈ Net and all reactions (9) in N(T),
Let T be any sub-network and E = e(T). In any steady state of the system, it follows from (11) and Lemma 3 that for all Yv ∈ γ(T) the following equations are satisfied
| (18) |
| (19) |
These form a system of linear equations in the variables E and Yv ∈ γ(T), with coefficients in ℝ[Con ∪ Sub]. Assume, without loss of generality, that γ(T) = {Y1, …, Yp−1} where p − 1 ≤ P. Let us use the notation Yp = E temporarily, for the purposes of this argument. Let MT denote the p × p matrix over the field ℝ(Con ∪ Sub) corresponding to (18) and (19). Evidently, MT.Yt = 0 where Y is the row vector, Y = (Y1, …, Yp). Furthermore, it follows from (14) that 1.MT = 0.
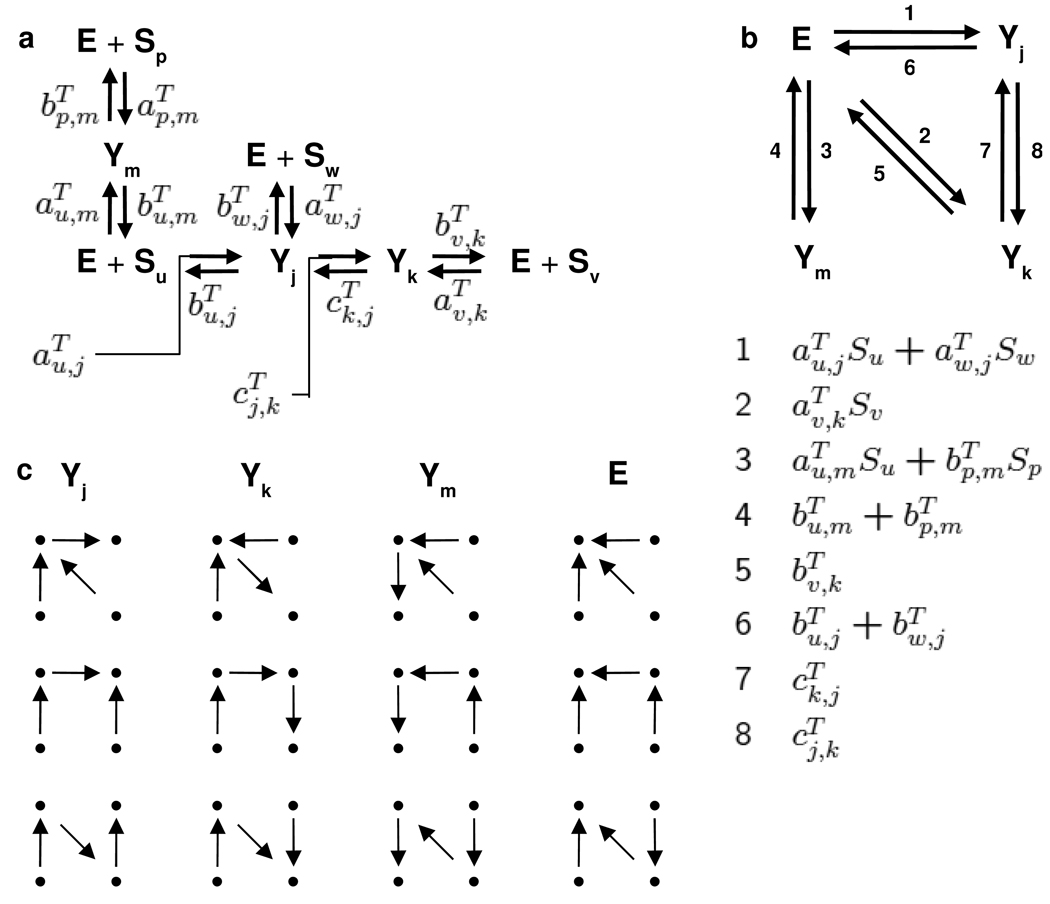
Let GT be the labelled, directed graph formed from MT as in Remark 2, so that MT = ℒ(GT). Figure 2b shows this graph for the sub-network in Figure 1e. GT is identical to N(T) for all edges between Yi for 1 ≤ i < p. However, each edge in N(T) corresponds to the edge Yp → Yv in GT with added to its label. Similarly, each edge in N(T) corresponds to the edge Yv → Yp in GT with added to its label. The labels in GT are all S-positive elements of ℝ[Con ∪ Sub]. We can now state the first additional condition.
Figure 2.
Steady state calculation for the sub-network in Figure 1e. a. The network in Figure 1e with labels on the reactions according to (9). b. The modified graph, GT, on the vertices Yj, Yk, Ym and E with the new labels in ℝ[Con ∪ Sub], as listed in the table below. The edges outgoing from E, numbered 1, 2 and 3, have labels which are linear and homogeneous in the modforms, while all other edges have labels which are rate constants. c. The spanning trees rooted at each vertex of GT, from which the maximal minors in (4) are calculated.
Condition 1
For any sub-network T, GT is strongly connected.
All the examples in Figure 1 satisfy this condition, which seems biochemically reasonable.
By Lemma 1, MT has rank p − 1. Accordingly, a basis vector for the column null space is given by (4). The labels on the edges of GT are rate constants, , except for the edges outgoing from Yp, whose labels are homogeneous linear combinations of modforms (Figure 2b). It follows that each spanning tree rooted at Yp has a label product in ℝ[Con], while any spanning tree rooted at Yi, for i ≠ p, has a label product that is homogeneous linear in the modforms with coefficients in ℝ[Con]. Hence, by (4), ρp is a S-positive element of ℝ[Con], while ρi, for i ≠ p, is a homogeneous linear combination of modforms whose non-zero coefficients are S-positive elements of ℝ[Con]. Let be such that, for 1 ≤ i < p,
| (20) |
The are, by definition, generalised Michaelis-Menten constants (compare the discussion in §2.4). Note that, as defined here, these are reciprocals of the usual Michaelis-Menten constants [5]. We prefer this convention because it allows these constants to be 0 when necessary, in preference to having to be ∞. By construction, the Michaelis-Menten constants are either 0 or are S-positive elements of ℝ(Con). In particular, they are well defined for any positive values of the rate constants in Con. The Michaelis-Menten constants for the example in Figure 1e are shown in Table 1.
Table 1.
Generalised Michaelis-Menten constants for the example in Figure 1e. The entries in the first three rows are the defined in Proposition 1 for x = j, k, m and y = u, v, w, p. The last row gives the denominator term D appearing in the entries for Yj and Yk. Note the S-positivity of all entries.
| Su | Sv | Sw | Sp | ||||
|---|---|---|---|---|---|---|---|
| Yj | 0 | ||||||
| Yk | 0 | ||||||
| Ym | 0 | 0 | |||||
| D | |||||||
Using Remark 3, we have proved the following generalisation of (8).
Proposition 1
For any sub-network T ∈ Net, the sub-network equation (18) and equation (19) are satisfied, if, and only if,
| (21) |
for 1 ≤ i < p.
3.4. Linearising the modforms
In steady state, the enzyme-substrate complexes satisfy (21). Substituting these into the expressions for the modforms given by (10) we obtain
| (22) |
These expressions are linear in the Su with coefficients which are S-positive polynomials in ℝ(Con)[Enz]. Note the critical need at this point for Yi in (21) to be linear in the Su. Let MS be the corresponding N × N matrix over ℝ(Con ∪ Enz) for the basis S1, …, SN. By Lemma 3, fT = 0 in steady state for all sub-networks T ∈ Net. Hence, by (15), 1.MS = 0. Let GS be the labelled, directed graph obtained from MS as in Remark 2, so that MS = ℒ(GS). To understand the structure of GS, it is convenient to extend the definition of the Michaelis-Menten constants so that for all Su ∉ σ(T). We can then replace Su ∈ σ(T) in (20) with Su ∈ Sub. With this convention, after collecting the coefficients of Sv in (22), we see that the label on the edge Sv → Su may be written
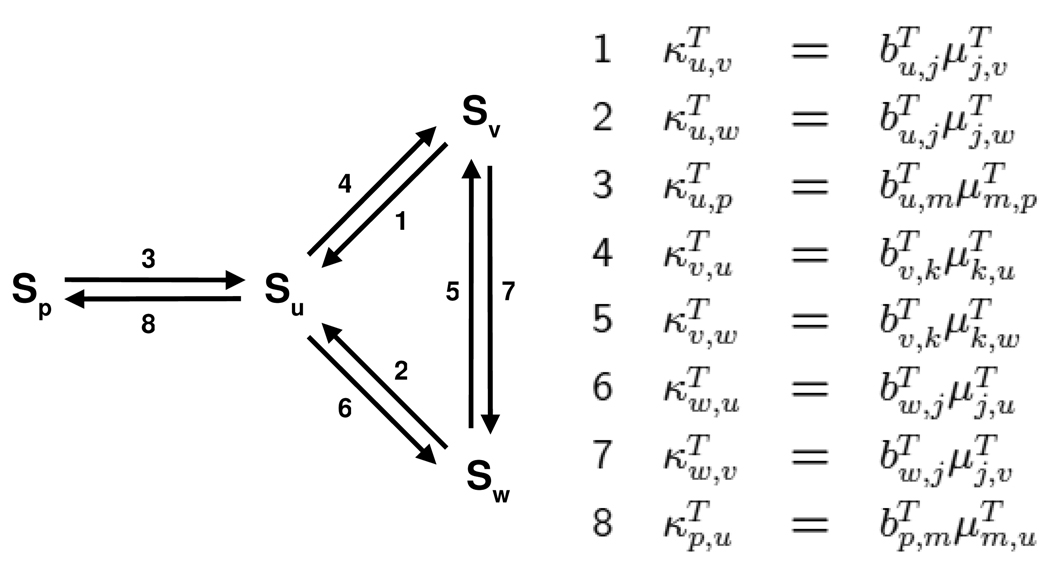
whenever that expression is non-zero. Terms like are familiar to biochemists as catalytic efficiencies. For each T ∈ Net and any distinct pair Su, Sv ∈ Sub, define the (generalised) catalytic efficiency, , by
Note that if Sv ∉ σ(T). If not 0, is a S-positive element of ℝ(Con). With this notation, we can rewrite the label on the edge Sv → Su as
| (23) |
whenever that expression is non-zero. Figure 3 shows GS for the example in Figure 1e.
Figure 3.
Modform graph, GS, for the example in Figure 1e with the labels shown as generalised catalytic efficiencies, , in the accompanying table. The are given in Figure 2a while the generalised Michaelis-Menten constants, are given in Table 1.
The catalytic efficiencies provide a more concise set of labels for GS than the original rate constants. Let Cat denote the set of all non-zero generalised catalytic efficiencies:
Note that ℝ(Cat) is a subfield of ℝ(Con) and that any S-positive element of ℝ(Cat) is also S-positive as an element of ℝ(Con). The labels on GS are S-positive polynomials in ℝ[Cat ∪ Enz], which are homogeneous and linear in the Ew. We may regard MS as a N × N matrix over ℝ(Cat ∪ Enz).
Since the modforms may include distinct substrates, we cannot assume that GS is connected. We can now state the second condition for the systems considered here.
Condition 2
The connected components of GS are strongly connected.
This condition is biochemically reasonable. For a given substrate, each modification is usually balanced by another enzyme carrying out a de-modification. This implies strong connectivity of the corresponding component of GS. Distinct substrates will give rise to distinct components. Although Figure 1e has only a single enzyme, the reversibility of the sub-network is sufficient in this case to give a single component which is strongly connected, as in Figure 3.
Condition 2 implies that MS is block diagonal, with each block corresponding to one of the connected components. Each block can be treated separately. To avoid further complicating the exposition, we assume from now on that MS is a single block and that GS is strongly connected. We point out what needs to be done for the general case but leave it to the reader to write down the details. Since, by Lemma 1, rk(MS) = N − 1, we can apply Lemma 2 to obtain a basis vector for the column null space of MS. Since GS is strongly connected, it has at least one spanning tree rooted at each vertex. It follows from (23) that, in the notation of (4), ρi is a S-positive element of ℝ[Cat ∪ Enz] which is homogeneous in the Ew and of degree N − 1, since there are that many edges in any spanning tree. Hence, ρu/ρ1 is a rational function in ℝ(Cat ∪ Enz), which we may write as a rational function of the enzymes,
| (24) |
whose non-zero coefficients are S-positive elements of ℝ[Cat]. Here, we have, without loss of generality, chosen S1 as a reference modform. With multiple components, a reference modform will be needed for each component. Since the ρu are each homogeneous of degree N − 1, it follows that, for λ ∈ ℝ,
| (25) |
so that the ru are, in fact, inhomogeneous functions of any L − 1 of the variables. For instance, if EL ≠ 0, then
Using Lemma 2 and Remark 3, we deduce the following analogue of Proposition 1.
Proposition 2
The modform equations (22) are satisfied if, and only if,
| (26) |
Examples of the rational functions ru for the case of a single kinase, single phosphatase and single substrate with 2 sites are given in [27].
3.5. The main results
We can now put everything together and return from symbols to real variables. Let us consider any multisite PTM system satisfying the assumptions in §3.1 along with Condition 1 and Condition 2. We assume that the rate constants have positive real values. In this case, the maximal minor ρ1 appearing in (24) is a real polynomial of degree N − 1 in the enzymes and ρ1 = 0 defines a hypersurface in ℝL. As long as the vector of enzyme values lies off this hypersurface, the rational functions in (24) are well defined. If GS has many components, a similar proviso must be made for each of them, in respect of the maximal minor for the corresponding reference modform. Note that, since ρ1 is S-positive in ℝ[Enz], any vector in the positive orthant will satisfy ρ1 ≠ 0. Hence, under biochemically realistic conditions, the ru are always well defined.
Theorem 2
In any steady state of the system for which the enzyme values satisfy ρ1 ≠ 0, equation (26) and equation (21) hold. Conversely, if the enzymes have values in ℝ for which ρ1 ≠ 0, S1 has values in ℝ, the Su are defined by (26) and the Yv are defined by (21), then these quantities form a steady state of the system.
PROOF
We have shown the first part above. Now suppose that Ei ∈ ℝ, such that ρ1(E1, …, EL) ≠ 0, and S1 ∈ ℝ. Define Su by (26), which we may do since ρ1 ≠ 0, and Yv by (21), as specified. For any T ∈ Net, it follows from Proposition 1 that the corresponding sub-network equation (18) and equation (19) are satisfied. Since (18) is just (11) at steady state, we see that dYv/dt = 0 for all Yv ∈ T. Furthermore, the expression on the left hand side of (19) is, by definition, the net flux of enzyme out of T, fT . Hence, fT = 0. Since this holds for all T ∈ Net, we see from equation (13) that dEw/dt = 0. Since (26) had to be satisfied to define the Su, it follows from Proposition 2 that (22) is satisfied. Since this is just (10) at steady state after substitution of (21), which has also been satisfied, we see that dSu/dt = 0. It follows that the system is at steady state.
If the system has specified total amounts of substrate and enzymes, it satisfies the conservation laws (16) and (17). These provide L + 1 equations for the L enzymes and the substrate. For each sub-network T ∈ Net, let ϕT ∈ ℝ[Enz] denote the S-positive polynomial
It follows from Proposition 1, that, whenever ρ1 ≠ 0,
Let Δ ∈ ℝ[Enz] denote the S-positive polynomial,
| (27) |
Rewriting (17) to solve for S1 in terms of Stot, we see that, whenever Δ ≠ 0,
| (28) |
We can then rewrite (16) to get
| (29) |
These L equations are well-defined whenever Δ ≠ 0. With multiple components, Stot can be apportioned among the components and each reference modform will have a corresponding equation to (28).
Let Φ : ℝL × ℝ → ℝL be defined by the left-hand side of (29) so that, for 1 ≤ w ≤ L,
Theorem 3
The steady states of a multisite PTM system are given by the solutions of a system of L equations for the L free enzyme concentrations. More precisely, if A, E ∈ ℝL, S ∈ ℝ, Δρ1 ≠ 0 and Φ(E, S) = A, then there is a steady state of the system for which E gives the free enzyme concentrations, S = Stot and Aw = Ew,tot. Conversely, any steady state of the system having these totals, for which Δρ1 ≠ 0, arises in this way.
PROOF
We have shown above that any steady state of the system having the specified totals, for which Δρ1 ≠ 0, satisfies Φ(E, S) = A. Now suppose that E ∈ ℝL, S ∈ ℝ satisfy Δρ1 ∈ 0 and that Φ(E, S) = A. Define so that
which we may do since Δ ≠ 0. Now use Theorem 2 to define a steady state of the system, , which we may do since ρ1 ≠ 0. We need only check that the total amount of substrate, , and enzymes, , satisfy and . The total amount of substrate is
Since (26) and (21) are satisfied through the use of Theorem 2, this gives
as required. Similarly, the total amount of enzyme Ew is
as required.
Since Δρ1 ∈ ℝ[Enz] is S-positive, it never vanishes for positive free enzyme concentrations, which corresponds to the biochemically realistic case.
3.6. Algebraic geometry of the steady state
As suggested in the Introduction, Theorem 2 should be seen as an assertion that an appropriate algebraic variety is rationally parameterisable. This implies that points on the variety can be explicitly constructed as rational functions of some auxiliary parameters, in contrast to the implicit definition of points as solutions of polynomial equations [6]. For instance, x2 + y2 = 1 provides an implicit definition of the unit circle, while the expression of x and y as
| (30) |
provides an explicit rational parameterisation. In general, a rational parameterisation may be undefined at points where the denominators of the rational functions vanish (which is not the case for (30)) and not all points on the variety may be represented (such as the point (0, 1) in (30)).
Recall from §3.1 that there are L + N + P dynamical variables in our system. Let V ⊆ ℝL+N+P denote the steady-state variety, corresponding to the simultaneous solutions of the system equation (10)–equation (12). V is a real algebraic variety. Let π: ℝL+N+P → ℝN denote the projection on to the space of modforms. π(V) may lack some points required for it to be an algebraic variety but it may be completed to one, if required, [6].
The set π(V) has additional structure, not present in V . Equation (26) gives the modforms only up to a constant. If λ ∈ ℝ, then it is easy to see from the system equation (10)–equation (12) that multiplying each modform and each enzyme-substrate complex by λ, changes all the rates by λ. In particular, if the system is at steady state, then it is still at steady state after such a change. Hence, π(V) is a projective set: given x ∈ π(V) the line through the origin and x also lies in π(V). We can therefore consider π(V) as a subset, π(V)ℙ, of the real projective space ℝℙN−1. The following is a corollary of Theorem 2
Theorem 4
π(V)ℙ has a rational parameterisation. Specifically, the rational functions ru in (24) define a surjective mapping ℝL → π(V)ℙ, which is well-defined away from the hypersurface ρ1 = 0.
In view of (25), the dimension of π(V)ℙ, once completed to a variety, is at most L − 1. For instance, in the case considered in [27, 41], with a single kinase and a single phosphatase, π(V)ℙ is a rational curve.
4. Discussion
Our results show that the exponentially large number of state variables, L + N + P, of a multisite PTM system is determined at steady state by a relatively small “core” of L variables. We have provided in Propositions 1 and 2 a linear algebraic algorithm for calculating all steady-state variables in terms of the core. Tools like Mathematica can readily carry out linear algebra over symbolic fields like ℝ(Q) and an example of such a program is available as the Supplementary Information to [27].
Previous steady-state analyses of multisite PTMs have largely focussed on phosphorylation. They have either used approximations, such as Michaelis-Menten or linear kinetics [14, 26, 28, 37], which ignore sequestration effects when enzymes have multiple substrates [4], or made simplifying biological assumptions, such as small numbers of sites [27] or sequential modification [16], which limits their applicability. The results of the present paper provide a foundation for developing models that are closer to biological reality while also extending the scope of analysis to allow for multiple enzymes, multiple types of modification and multiple substrates. The permitted biochemical mechanisms are also considerably enlarged. Our method of proof reveals the significance of the Matrix-Tree theorem, which seems to play a key role in several forms of algebraic elimination in biochemical networks [7, 23].
The present paper develops the methodology. Applications of these results are found in previous papers which have focussed on phosphorylation with a single kinase, single phosphatase and single substrate [27, 41]. The capability to treat the number, n, of sites as a variable has allowed us to show that, for appropriate rate constants, the number of stable phospho-form distributions can be as many as ⌊(n + 2/2⌋, where ⌊x⌋ denotes the greatest integer not greater than x [41]. In particular, the number of stable phospho-form distributions increases with n. While some of these distributions are focussed on a few phospho-forms, others are more diffuse. Since different phospho-forms may have distinct biological effects [31, 34, 45], the phospho-proteome could be capable of substantial information processing. Indeed, it has been suggested that the remarkable variety of PTMs found on histone proteins in chromatin form a “code” for transcriptional regulation [21, 42]. The analysis in [41] provides the first example of a PTM mechanism which is capable of encoding arbitrary amounts of information and gives an estimate of its information capacity.
A second application, for systems with two sites, has shown that the steady-state geometry can distinguish between different reaction networks [27]. The geometry is detected algebraically through the use of “invariants”, or polynomial functions of the steady-state phospho-form concentrations which depend only on the rate constants [17]. Invariants take the same value no matter what amounts of enzymes and substrates are present or what steady state is formed. To exploit such results we have been developing, in collaboration with Hanno Steen’s group at Children’s Hospital in Boston, mass-spectrometric methods for accurately quantifying phospho-form distributions. We are using this to map out, for the first time, the steady-state geometry of a kinase, phosphatase, substrate system: the MAP kinase Erk, which is doubly phosphorylated by the MAP kinase kinase Mek and dephosphorylated by the dual-specificity phosphatase MKP3. Our experimental studies have already shown that these proteins engage in a more complex set of reactions than is commonly described in the literature and we are developing the method of invariants as a tool to work out the missing pieces.
Our results suggest several directions for future investigation. Which biochemical reaction networks have “cores”, or small subsets of variables in terms of which all others can be calculated at steady state? If a core exists, is it unique? How can cores be identified and how can the functional relationship with non-core variables be algorithmically determined? Do these functional relationships give rational parameterisations of the steady-state variety? A particularly interesting generalisation would be to allow substrates to also be enzymes, thereby accommodating kinase cascades. The difficulty here is that those substrates that are also enzymes can, presumably, no longer be eliminated and must hence be in the “core”, while, at the same time, as substrates, these variables may have non-trivial algebraic dependencies with other core variables. In the case treated here, the variables in the core are independent: their values can be arbitrarily assigned (Theorem 2). In more general cases there may be additional algebraic constraints on the core variables. We believe that the language and methods of algebraic geometry [6, 27] will be particularly useful for unravelling such issues.
Looking further ahead, it is a tantalising question as to whether the elimination procedures developed here can be extended from the steady state to the dynamics, perhaps, initially, to the local vicinity of the steady state. Since steady-state stability is determined by the eigenvalues of the Jacobian, it does not seem implausible that differential algebraic methods could encompass both the steady state itself as well as its local vicinity. More globally, multisite PTM systems have other attractors, such as limit cycles, of which the cyanobacterial circadian oscillator is a particularly significant example [30, 36]. Hilbert’s sixteenth problem asks about the number of limit cycles of two-dimensional polynomial dynamical systems. It remains unsolved, although some lower bounds are known [2]. Several lines of evidence, including the work presented here, suggest that the polynomial dynamics arising from biochemical reaction networks has very good properties at steady state. Could the same be true for other attractors like limit cycles?
Biologists are continually striving to elicit general principles from experimental data. Mathematical methods have been of less help in this respect than in accounting for the results of individual experiments. The present paper provides the tools to reason about post-translational modification systems without having to fix in advance many of the individual details, such as the number of enzymes or the combinatorics of modification. Being able to rise above the molecular complexity while retaining biological and biochemical realism provides a complementary capability to that of simulation. If this capability can be extended to a broad range of cellular processes, we will have a powerful tool with which to articulate the principles of cellular information processing.
Acknowledgements
The research undertaken here was supported in part by NIH under grant R01-GM081578. We thank Bernd Sturmfels for pointing us to the Matrix-Tree theorem, Alicia Dickenstein for many stimulating scientific discussions and the Statistical and Applied Mathematical Sciences Institute (SAMSI) for supporting an extended visit by JG to the Program on Algebraic Methods in Systems Biology and Statistics, during which this paper was drafted.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Angeli D, Ferrell JE, Sontag ED. Detection of multistability, bifurcations, and hysteresis in a large class of biological positive-feedback systems. Proc. Natl. Acad. Sci. USA. 2004;101:1822–1827. doi: 10.1073/pnas.0308265100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Christopher CJ, Lloyd NG. Polynomial systems: a lower bound for the Hilbert numbers. Proc. Roy. Soc. Lond. A. 1995;450:219–224. [Google Scholar]
- 3.Chung FRK. Spectral Graph Theory. No. 92 in Regional Conference Series in Mathematics. American Mathematical Society. 1997 [Google Scholar]
- 4.Ciliberto A, Capuani F, Tyson JJ. Modeling networks of coupled enzymatic reactions using the total quasi-steady state approximation. PLoS Comp. Biol. 2007;3:e45. doi: 10.1371/journal.pcbi.0030045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cornish-Bowden A. Fundamentals of Enzyme Kinetics. 2nd Edition. London, UK: Portland Press; 1995. [Google Scholar]
- 6.Cox D, Little J, O’Shea D. Ideals, Varieties and Algorithms. 2nd Edition. Springer; 1997. [Google Scholar]
- 7.Craciun G, Dickenstein A, Shiu A, Sturmfels B. Toric dynamical systems. j. Symb. Comp., to appear. 2008 [Google Scholar]
- 8.Craciun G, Tang Y, Feinberg M. Understanding bistability in complex enzyme-driven reaction networks. Proc. Natl. Acad. Sci. USA. 2006;103:8697–8602. doi: 10.1073/pnas.0602767103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feinberg M. Lectures on Chemical Reaction Networks, lecture notes, Mathematics Research Center, University of Wisconsin, 1979. 1979 www.che.eng.ohio-state.edu/~feinberg/research/ [Google Scholar]
- 10.Ferrarese A, Marin O, Bustos VH, Venerando A, Antonelli M, Allende JE, Pinna LA. Chemical dissection of the APC Repeat 3 multistep phosphorylation by the concerted action of protein kinases CK1 and GSK3. Biochemistry. 2007;46:11902–11910. doi: 10.1021/bi701674z. [DOI] [PubMed] [Google Scholar]
- 11.Ferrell JE, Bhatt RR. Mechanistic studies of the dual phosphorylation of mitogen-activated protein kinase. J. Biol. Chem. 1997;272:19008–19016. doi: 10.1074/jbc.272.30.19008. [DOI] [PubMed] [Google Scholar]
- 12.Fischer EH. Protein phosphorylation and cellular regulation, II. In: Ringertz N, editor. Nobel Lectures, Physiology or Medicine 1991–1995. Singapore: World Scientific; 1997. [Google Scholar]
- 13.Gatermann K, Huber B. A family of sparse polynomial systems arising in chemical reaction systems. J. Symbolic Computation. 2002;33:273–305. [Google Scholar]
- 14.Goldbeter A, Koshland DE. An amplified sensitivity arising from covalent modification in biological systems. Proc. Natl. Acad. Sci. USA. 1981;78(11):6840–6844. doi: 10.1073/pnas.78.11.6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gunawardena J. 2003. Chemical Reaction Network Theory for in-silico biologists, Lecture notes, Harvard Univ, 2003. vcp.med.harvard.edu/papers/crnt.pdf. [Google Scholar]
- 16.Gunawardena J. Multisite protein phosphorylation makes a good threshold but can be a poor switch. Proc. Natl. Acad. Sci. USA. 2005;102:14617–14622. doi: 10.1073/pnas.0507322102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gunawardena J. Distributivity and processivity in multisite phosphorylation can be distinguished through steady-state invariants. Biophys. J. 2007;93:3828–3834. doi: 10.1529/biophysj.107.110866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gunawardena J. Models in systems biology: the parameter problem and the meanings of robustness. In: Lodhi H, Muggleton S, editors. Elements of Computational Systems Biology. Wiley Book Series on Bioinformatics. John Wiley and Sons, Inc.; 2010. [Google Scholar]
- 19.Herstein I. Topics in Algebra. Wiley; 1975. [Google Scholar]
- 20.Hunter T. The age of crosstalk: phosphorylation, ubiquitination and beyond. Mol. Cell. 2007;28:730–738. doi: 10.1016/j.molcel.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 21.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 22.Kim SY, Ferrell JE. Substrate competition as a source of ultrasensitivity in the inactivation of Wee1. Cell. 2007;128:1133–1145. doi: 10.1016/j.cell.2007.01.039. [DOI] [PubMed] [Google Scholar]
- 23.King EL, Altman C. A schematic method of deriving the rate laws for enzyme-catalyzed reactions. J. Phys. Chem. 1956;60:1375–1378. [Google Scholar]
- 24.Krebs EG. Protein phosphorylation and cellular regulation, I. In: Ringertz N, editor. Nobel Lectures, Physiology or Medicine 1991–1995. Singapore: World Scientific; 1997. [Google Scholar]
- 25.Kruse J-P, Gu W. SnapShot: p53 posttranslational modifications. Cell. 2008;133:930. doi: 10.1016/j.cell.2008.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lisman JE. A mechanism for memory storage insensitive to molecular turnover: a bistable autophosphorylating kinase. Proc. Natl. Acad. Sci. USA. 1985;82:3055–3057. doi: 10.1073/pnas.82.9.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manrai A, Gunawardena J. The geometry of multisite phosphorylation. Biophys. J. 2008;95:5533–5543. doi: 10.1529/biophysj.108.140632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Markevich NI, Hoek JB, Kholodenko BN. Signalling switches and bistability arising from multisite phosphorylation in protein kinase cascades. J. Cell Biol. 2004;164:353–359. doi: 10.1083/jcb.200308060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moon JW. Counting Labelled Trees. No. 1 in Canadian Mathematical Monographs. Canadian Mathematical Congress. 1970 [Google Scholar]
- 30.Nakajima M, Imai K, Ito H, Nishiwaki T, Murayama Y, Iwasaki H, Oyama T, Kondo T. Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science. 2005;308:414–415. doi: 10.1126/science.1108451. [DOI] [PubMed] [Google Scholar]
- 31.Park K-S, Mohapatra DP, Misonou H, Trimmer JS. Graded regulation of the Kv2.1 potassium channel by variable phosphorylation. Science. 2006;313:976–979. doi: 10.1126/science.1124254. [DOI] [PubMed] [Google Scholar]
- 32.Pesavento JJ, Bullock CR, LeDuc RD, Mizzen CA, Kelleher NL. Combinatorial modification of human histone H4 quantitated by two-dimensional liquid chromatography coupled with top down mass spectrometry. J. Biol. Chem. 2008;283:14927–14937. doi: 10.1074/jbc.M709796200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Phanstiel D, Brumbaugh J, Berggren WT, Conrad K, Feng X, Levenstein ME, McAllister GC, Thomson JA, Coon JJ. Mass spectrometry identifies and quantifies 74 unique histone H4 isoforms in differentiating human embryonic stem cells. Proc. Natl. Acad. Sci. USA. 2008;105:4093–4098. doi: 10.1073/pnas.0710515105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pufall MA, Lee GM, Nelson ML, Kang H-S, Velyvis A, Kay LE, McIntosh LP, Graves BJ. Variable control of Ets-1 DNA binding by multiple phosphates in an unstructured region. Science. 2005;309:142–145. doi: 10.1126/science.1111915. [DOI] [PubMed] [Google Scholar]
- 35.Roach PJ. Multisite and hierarchal protein phosphorylation. Journal of Biological Chemistry. 1991;266:14139–14142. [PubMed] [Google Scholar]
- 36.Rust MJ, Markson JS, Lane WS, Fisher DS, O’Shea E. Ordered phosphorylation governs oscillation of a three-protein circadian clock. Science. 2007;318:809–812. doi: 10.1126/science.1148596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Salazar C, Höfer T. Versatile regulation of multisite protein phosphorylation by the order of phosphate processing and protein-protein interactions. FEBS J. 2007;274:1046–1060. doi: 10.1111/j.1742-4658.2007.05653.x. [DOI] [PubMed] [Google Scholar]
- 38.Shacter-Noiman E, Chock PB, Stadtman ER. Protein phosphorylation as a regulatory device. Philos. Trans. R. Soc. Lond. B. 1983;302:157–166. doi: 10.1098/rstb.1983.0049. [DOI] [PubMed] [Google Scholar]
- 39.Soulé C. Graphic requirements for multistationarity. ComPlexUs. 2003;1:123–133. [Google Scholar]
- 40.Strogatz SH. Nonlinear Dynamics and Chaos: With Applications to Physics, Biology, Chemistry and Engineering. Perseus Books. 2001 [Google Scholar]
- 41.Thomson M, Gunawardena J. Unlimited multistability in multisite phosphorylation systems. Nature, to appear. 2009 doi: 10.1038/nature08102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Turner B. Cellular memory and the histone code. Nature. 2009;460:274–277. [Google Scholar]
- 43.Tutte WT. The dissection of equilateral triangles into equilateral triangles. Proc. Camb. Phil. Soc. 1948;44:463–482. [Google Scholar]
- 44.Walsh CT. Roberts and Company. Colorado: Englewood; 2006. Posttranslational Modification of Proteins. [Google Scholar]
- 45.Wu RC, Qin J, Yi P, Wong J, Tsai SY, Tsai MJ, O’Malley BW. Selective phosphorylations of the SRC-3/AIB1 coactivator integrate genomic responses to multiple cellular signaling pathways. Mol. Cell. 2004;15:937–949. doi: 10.1016/j.molcel.2004.08.019. [DOI] [PubMed] [Google Scholar]