Summary
The transcription factor GATA-1 is required for terminal erythroid maturation and functions as an activator or repressor depending on gene context. Yet its in vivo site selectivity and ability to distinguish between activated versus repressed genes remain incompletely understood. In this study, we performed GATA-1 ChIP-seq in erythroid cells and compared it to GATA-1 induced gene expression changes. Bound and differentially expressed genes contain a greater number of GATA binding motifs, a higher frequency of palindromic GATA sites, and closer occupancy to the transcriptional start site versus non-differentially expressed genes. Moreover, we show that the transcription factor Zbtb7a occupies GATA-1 bound regions of some direct GATA-1 target genes, that the presence of SCL/TAL1 helps distinguish transcriptional activation versus repression, and that Polycomb Repressive Complex 2 (PRC2) is involved in epigenetic silencing of a subset of GATA-1 repressed genes. These data provide insights into GATA-1 mediated gene regulation in vivo.
Keywords: GATA-1, Polycomb, Zbtb7a, erythroid, ChIP-seq
Introduction
Lineage commitment from hematopoietic stem cells involves the activation of specific gene programs and concomitant suppression of multipotential and alternate lineage gene programs (Cantor and Orkin, 2001). These events are regulated in large part by a limited set of lineage-specific master transcription factors (MTFs) that functionally cross-antagonize one another. Studies on MTFs are therefore fundamental to understanding mechanisms of cell fate determination and lineage plasticity.
GATA-1 is a prototypic MTF that is essential for erythroid and megakaryocytic development, and antagonizes neutrophilic differentiation. It was first identified as a protein that binds key cis-regulatory elements within the globin gene loci, but has since been shown to regulate a large number of erythroid-specific genes (Evans and Felsenfeld, 1989; Tsai et al., 1989). Targeted disruption of the GATA-1 gene in mice causes embryonic lethality between embryonic day 10.5 to 11.5 (e10.5-11.5) due to severe anemia from blocked maturation and increased apoptosis of erythroid precursors (Fujiwara et al., 1996; Weiss and Orkin, 1995).
GATA-1 contains two closely spaced zinc fingers. The carboxyl zinc finger binds the DNA consensus sequence (T/A)GATA(A/G) (Evans et al., 1988; Wall et al., 1988). The amino zinc finger binds DNA at certain double GATA sites, and has preference for GATC core motifs (Newton et al., 2001; Trainor et al., 1996). However, simple annotation of GATA consensus sequences, even phylogenetically conserved sites, is a poor predictor of in vivo GATA-1 occupancy as demonstrated by recent chromatin immunoprecipitation (ChIP) studies across extended loci (Bresnick et al., 2005). Thus, additional information must contribute to its in vivo site selectivity.
Studies using a murine GATA-1 null erythroid cell line that stably expresses a GATA-1-estrogen receptor ligand binding domain fusion molecule (G1-ER4 cells) have provided important insights into GATA-1 mediated gene regulation (Gregory et al., 1999). Treatment of these cells with estradiol causes rapid activation of GATA-1, allowing for detailed kinetic study of GATA-1 mediated transcriptional events. cDNA microarray studies using this system show that GATA-1 not only activates a large number of genes, but unexpectedly represses an almost equal number (Welch et al., 2004). How GATA-1 distinguishes between activated and repressed target genes, and how it carries out these opposing transcriptional activities remains incompletely understood.
Further elucidation of the rules that govern GATA-1 in vivo site selectivity and its gene context-dependent activities requires that a large number of bona fide chromatin occupancy sites be identified. In this study, we carried out genome-wide ChIP of GATA-1 in induced mouse erythroleukemia (MEL) cells and compared it to a new comprehensive analysis of GATA-1 induced gene expression changes. Here we report that the number of GATA-1 motifs, the presence of double palindromic sites, and distance from the transcriptional start site (TSS) correlate with GATA-1 occupancy and functional activity. We also identify Zbtb7a as a transcription factor that binds at GATA-1 occupancy sites of a number of direct GATA-1 target genes, and show that Polycomb Repressive Complex 2 (PRC2) is involved in epigenetic silencing of a subset of GATA-1 repressed genes during maturation of primary erythroid precursor cells.
Results
ChIP-Solexa Sequencing of Metabolically Biotin-tagged GATA-1 in MEL cells
Metabolic biotin tagging of recombinant proteins in mammalian cells has recently been developed and applied to ChIP assays (de Boer et al., 2003; Parrott and Barry, 2000; Viens et al., 2004). The exceedingly strong affinity between streptavidin and biotin (Kd ~10−15 M) allows for high stringency washing conditions (including 2% sodium dodecyl sulfate) that are not possible with standard antibody-antigen based methods. This results in comparatively reduced background noise (Viens et al., 2004). Here, we combined metabolic biotin tagging and ChIP-Solexa sequencing (ChIP-seq) to perform genome-wide analysis of GATA-1 occupancy in erythroid cells.
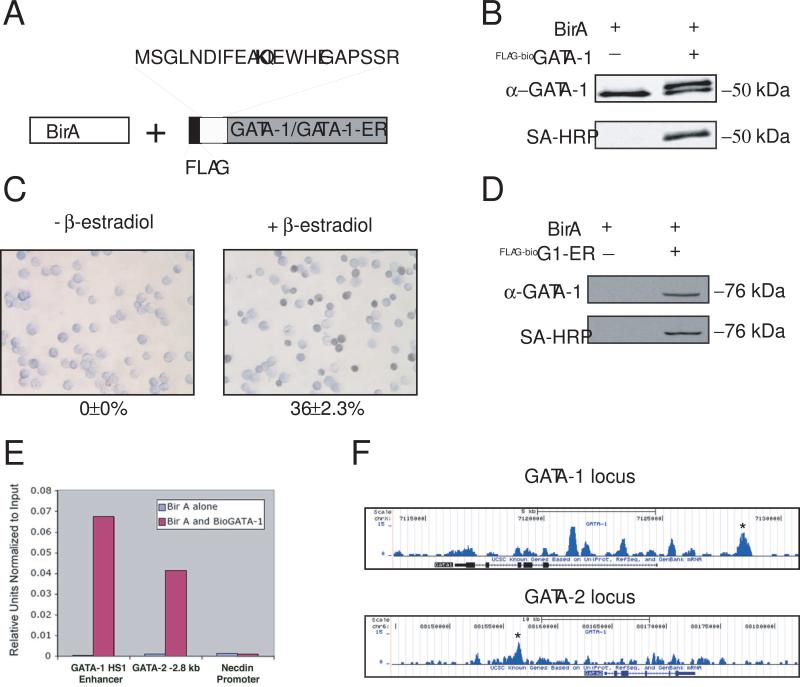
Mouse erythroleukemia (MEL) cell lines were generated that stably express the bacterial biotin ligase birA and recombinant GATA-1 containing a 23 amino acid birA recognition motif fused at its amino terminus (Fig. 1A). A FLAG epitope tag was also included to assist in subsequent co-immunoprecipitation (co-IP) assays. Clones were chosen that express recombinant GATA-1 at levels similar to endogenous GATA-1 (Fig. 1B, upper panel). Western blot analysis using streptavidin-horse radish peroxidase (SA-HRP) demonstrates in vivo biotinylation of the recombinant GATA-1 (Fig. 1B, lower panel). The biotin tagged protein is functionally competent since FLAG-BioGATA-1-ER rescues erythroid maturation of the GATA-1 null G1E cell line after treatment with β-estradiol, as judged by o-dianisodine staining for hemoglobin production (Fig. 1 C and D).
Figure 1.
Streptavidin ChIP-Seq of metabolically biotinylated GATA-1 in induced MEL cells. (A) Schematic diagram of metabolic biotin tagging system. birA, E.coli biotin ligase. The birA recognition motif and FLAG epitope tag is shown fused to the amino terminus of GATA-1 or GATA-1ER. The biotin acceptor lysine is indicated in bold. (B) Western blot of nuclear extracts from MEL cell clones expressing either birA alone, or birA and the recombinant GATA-1 (FLAG-BioGATA-1). Upper panel, probed with anti-GATA-1 antibody; Lower panel, same as upper panel after stripping and re-probing with streptavidin-horse radish peroxidase (SA-HRP). (C) O-dianosidine (benzidine) stain of G1E cells expressing FLAG-BioGATA-1-ER without (left) or with (right) treatment with β-estradiol for 48 hrs. Hemoglobinized cells stain dark brown/black. The percentage of positive cells is indicated below the panel (−/+ standard error of the mean (SEM); n=3). (D) Western blot of nuclear extracts from cell shown in “C” probed with anti-GATA-1 antibody (top) and SA-HRP (bottom). (E) Quantitative ChIP assay at two known GATA-1 occupancy sites (GATA-1 HS1 and GATA-2 “−2.8 kb” enhancer) and a negative control site (necdin promoter) using streptavidin-based ChIP from induced MEL cells (DMSO for 24 hrs) expressing birA alone or birA and FLAG-BioGATA-1. (F) Streptavidin ChIP-Seq enrichment profiles for FLAG-BioGATA-1 at the GATA-1 and GATA-2 loci. Enrichment peaks corresponding to GATA-1 HS1 and GATA-2 “−2.8 kb” are indicated with asterisks.
Streptavidin-based ChIP was then performed on MEL cells containing FLAG-BioGATA-1 after dimethyl sulfoxide (DMSO) induced maturation for 24 hours. Occupancy at two well-characterized GATA-1 binding sites, an enhancer element upstream of the GATA-1 gene (GATA-1 HS1) (Vyas et al., 1999b) and a cis-regulatory element of the GATA-2 gene (GATA-2 “−2.8 kb”) (Grass et al., 2003), were initially analyzed using standard quantitative SA-ChIP assays. This revealed marked enrichment and low background signal compared to control assays at the necdin promoter, which does not contain GATA binding motifs, or from cells expressing birA alone (Fig. 1E). Deep sequencing of the SA-ChIP material was then performed using an Illumina Solexa genome analyzer. A total of 6, 036,924 reads were obtained that map to the genome. Enrichment profiles at the GATA-1 and GATA-2 loci are shown in Fig. 1F, with peaks corresponding to the GATA-1 HS1 and GATA-2 “−2.8 kb” sites indicated.
Global GATA-1 Genomic Occupancy
Global analysis of the ChIP-seq dataset identified a total of 4,199 enrichment peaks with a false discovery rate (FDR) <0.01 (Fig S1). Validation studies using standard antibody based ChIP assays in wild type MEL cells and primary e13.5 fetal liver cells were performed on a sample of peaks to confirm the ChIP-seq results. Thirty-one of 32 (97%) and 39 of 43 (91%) enrichment peaks were validated in induced MEL and primary mouse fetal liver cells, respectively (Fig. S2).
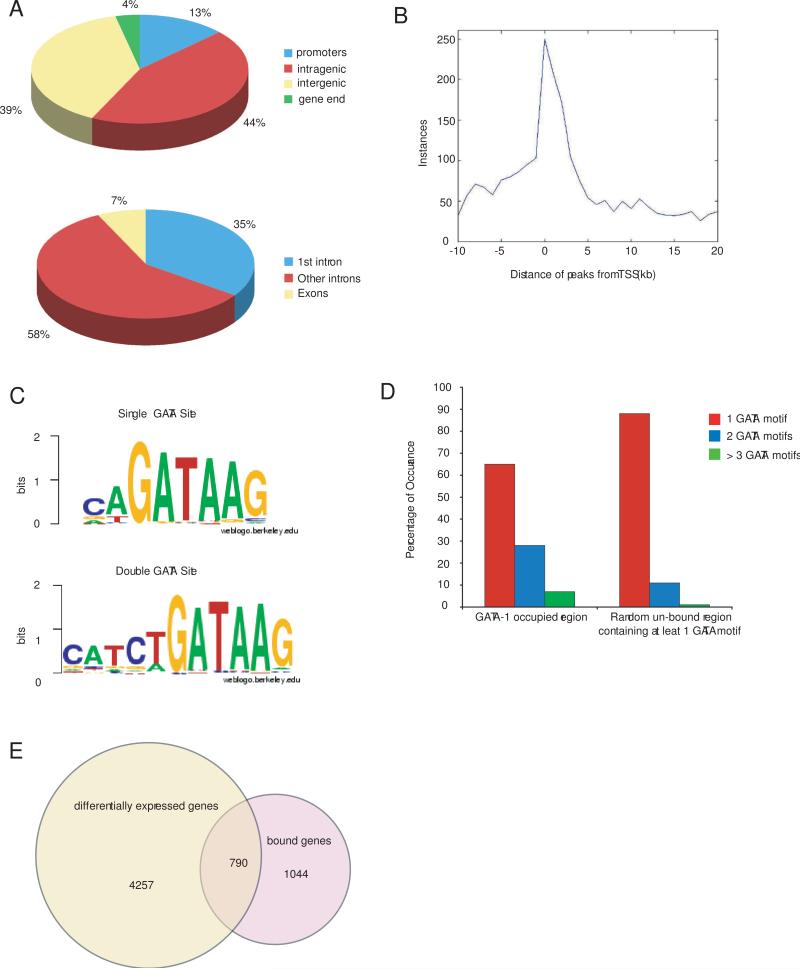
We first examined the location of GATA-1 enrichment sites relative to annotated gene structures based on the UCSC Genome Browser Database (Karolchik et al., 2008). Five hundred and forty nine (13%) of the peaks occur within gene promoters (defined here as within 10 kb 5’ to the TSS), 1853 (44%) are within genes, 146 (4%) are located within 3 kb 3’ to the end of the gene, and 1651 (39%) are intergenic (Fig. 2A). Of the intragenic sites, 1717 (93%) are within introns (645 in the first intron) and 136 (7%) are within exons. Peaks within introns tend to be nearer the TSS, whereas exonic peaks typically occur at the beginning or end of the coding region (Fig. S3). Of all the peaks located between −10 kb of the TSS to +3 kb from the 3’ end of the gene, marked enrichment for binding was observed closest to the TSS (Fig. 2B). A total of 1,834 genes were identified in which the GATA-1 enrichment peak fell within a region encompassing −10 kb of the TSS to +3 kb from the 3’of gene end (hereafter defined as “bound genes”) (Table S1).
Figure 2.
Bioinformatic analysis of GATA-1 global occupancy. (A) Top, pie diagram showing distribution of GATA-1 enrichment peaks located in the promoter (−10 kb to the TSS), intragenic sites, 3’end of gene (3 kb 3’ to the gene end), or intergenic sites (outside of these defined regions). Bottom, pie diagram showing distribution of intronic versus exonic GATA-1 enrichment peaks among intragenic sites. (B) Frequency of GATA-1 enrichment peaks according to their position relative to the TSS. (C) Single (top) and double (bottom) GATA binding motif preferences based on occupancy at the 200 highest promoter region peaks using THEME. (D) Graph showing the percentage of occurrences of 1, 2, or > 3 GATA binding motifs in GATA-1 occupied regions versus 10 sets of random non-bound regions that contain at least one GATA binding motif. (E) Ven diagram showing the number of differentially expressed genes after induction of G1-ER4 cells, the number of genes bound by GATA-1, and the overlap between the two.
In Vivo GATA-1 Binding Sequence Preference
We examined our dataset using the motif search algorithm THEME (Macisaac et al., 2006) to assess whether the canonical GATA-1 binding sequence (T/A)GATA(A/G) (Evans et al., 1988; Martin et al., 1989; Wall et al., 1988) predicts global in vivo occupancy, or whether variants and/or extended motifs contribute to site selectivity. When considering the sequences corresponding to the 200 highest peaks in promoter regions (those with the greatest number of sequence reads), c(T/A)GATAAG was the best predictor of GATA-1 occupancy (Fig. 2C). When we extended our analysis, the same motif remained a good predictor regardless of the position of the peak relative to the gene, or to the peak height. Thus, GATA-1's global in vivo site selectivity reflects the canonical binding sequence, but has additional preferences including cytosine at the −2 position and adenine and guanine at the +1 and +2 positions, respectively, relative to the core “GATA” based on this computational analysis.
Subsequently, we used this motif to test if the frequency of finding multiple GATA-1 motifs within peaks is greater than one would expect based on random non-bound DNA sequences. Considering only bound regions and random non-bound sequences with a least one GATA-1 motif, we found that the number of GATA-1 motifs was significantly higher in bound peaks compared to 10 sets of random non-bound DNA sequences by Mann-Whitney U test using Benjamini-Hochberg correction for multiple hypothesis testing (p=6.6E-64) (Fig. 1D). Thus, the presence of multiple GATA binding motifs is predictive of in vivo GATA-1 occupancy.
Chromatin Occupancy at Sites Containing Double GATA-1 Binding Motifs
We next examined the correlation between GATA-1 occupancy and the occurrence of double GATA sites. In humans, two naturally occurring mutations in the amino zinc finger, GATA-1R216Q and GATA-1R216W, selectively disrupt binding to double GATA sites and lead to X-linked β-thalassemia, congenital erythropoietic porphyria, and/or macrothrombocytopenia, (Phillips et al., 2006; Tubman, 2005; Yu et al., 2002). Using THEME, we tested different hypotheses concerning the sequence specificity of the double GATA-1 binding site since a number of variations have been reported (Pedone et al., 1997; Trainor et al., 2000; Trainor et al., 1996). We found that the previously characterized mGATApal motif (Trainor et al., 2000), described by the palindromic consensus sequence catctGATAAG (Fig. 2C), is the best descriptor of the double GATA-1 binding sites overall, and occurs in 1680 (40%) out of the 4199 GATA-1 ChIP-seq enrichment peaks. There are 972 bound genes with mGATApal motifs and these are enriched for heme biosynthesis (p=0.012), glycoprotein biosynthetic process (p=0.011), and Ras protein signal transduction (p=0.011) (Table S2). There is significant overlap (Fisher's exact test p=0.0032) between genes with mGATApal binding sites and genes that are bound and up regulated (see “Direct GATA-1 Target Genes”). Bound regions with double GATA-1 binding sites have a higher number of GATA motifs overall (p= 7.8E-80) and a higher peak height (p= 1.6E-12) compared to all bound regions. Thus, the presence of double GATA binding sites is a more general finding than perhaps previously thought, and marks genes containing positive GATA-1 transcriptional activity.
Direct GATA-1 Target Genes
A comprehensive analysis of GATA-1 induced gene expression changes was performed using the G1-ER4 cell system and current generation Affymetrix Moe430v2 cDNA microarray GeneChips™. This new analysis increases the number of probe sets analyzed by about 4-fold, and nearly doubles the number of genes interrogated compared to the prior study by Welch et al. (Welch et al., 2004). A total of 5047 genes (9401 probe IDs) were differentially expressed over the 30 hr induction time course (see Materials and Methods). Of these, 790 genes (16%) contained GATA-1 enrichment peaks within a region extending from −10 kb upstream of the TSS to 3 kb downstream of the gene end (“bound genes”), likely representing functional direct GATA-1 target genes (Fig. 2E and Table S3). Four hundred and fifty four (57%) were up regulated, 325 (41%) were down regulated, and 11 (1.4%) had probe sets that were both up regulated and down regulated. The 4257 genes that changed expression but were not bound likely represent indirect gene expression changes. However, we cannot exclude regulation via long-range interactions (binding outside of our defined window), false negative signals in our ChIP-seq dataset, or gene expression differences between MEL and G1-ER4 cells.
One thousand and forty four genes had GATA-1 peaks within −10 kb to + 3 kb of the gene, but were not differentially expressed (Fig. 2E, Table S4). It is possible that these genes respond at levels below our cut-off values, change expression under conditions other than those tested, and/or represent true non-functional GATA-1 occupancy sites.
Features of Bound and Differentially Expressed Genes
We used K-means clustering to group the bound and differentially expressed genes based on the similarity of their expression profiles. We identified three distinct clusters: one for activated genes and two for repressed genes (Fig. S4). One of the repressed gene clusters contains genes that are down regulated relatively quickly, but rebound after 20 hours of cell induction (“immediately down regulated”). The other repressed gene cluster has a somewhat delayed change, but then steadily decreases in expression following 15 hours (“delayed down regulated”). The up regulated genes are enriched for hemoglobin biosynthesis and Ras/GTPase signaling pathways (Table S5). Many genes belonging to the delayed down regulated genes have ATP binding and adenyl ribonucleotide binding function, and/or are cell cycle related genes. The immediately down-regulated genes are enriched for genes in the ‘intracellular signaling cascade’ gene ontology (GO) category.
In order to further understand the differences between the genes in the three clusters and the genes that are bound but not differentially expressed, we compared these groups for four peak characteristics (Fig. S5): First, bound and differentially expressed genes have a better match to the GATA-1 motif (Mann-Whitney U test p<0.02). Second, bound regions of up regulated and delayed down regulated genes are significantly closer to the TSS (median 4,330.5 and 5,723.5 bp, respectively) than those that are not differentially expressed (median 12,536 bp) (Mann-Whitney U test p=5.5E-04 and p=0.0038, respectively). Third, peak heights are higher in up regulated genes (median value = 21) compared to bound and non-differentially expressed genes (median value = 19) (Mann-Whitney U test p= 0.0011). Lastly, the number of GATA-1 motifs is higher in bound and differentially expressed genes compared to bound and non-differentially expressed genes (Mann-Whitney U test p= 4.0E-04).
Transcription Factor Binding Motif Enrichment at GATA-1 Occupancy Sites
In order to identify other proteins that might contribute to GATA-1 target gene regulation, we used THEME to compare transcription factor binding motifs at GATA-1 occupancy sites of activated versus repressed genes. Motifs with an adjusted p-value < 0.005 (corrected for multiple hypothesis testing) are reported in Table 1. Among the set of enriched motifs at activated genes were binding sites for the transcription factors NF-E2 (p= 5.7E-04), SCL/TAL1 (p=1.3E-03), and Zbtb7a (p=4.1E-03). NF-E2 and SCL/TAL1 have previously been implicated in globin gene expression and hematopoiesis (Andrews et al., 1993; Porcher et al., 1996), and SCL/TAL1 has recently been shown to selectively occupy a number of activated versus repressed GATA-1 target genes in G1-ER4 cells (Tripic et al., 2008).
Table 1.
Enriched transcription factor binding motifs at GATA-1 bound regions of activated compared to repressed genes using THEME (Macisaac et al., 2006). The following symbols represent nucleotide ambiguity: R=A/G, Y=C/T, S=C/G, W=A/T, K=G/T, M=A/C, and “.” = A/C/G/T. Upper case letters indicate nucleotide assignments with high information content. The 3rd column represents the cross-validation error of each hypothesis. Lower cross-validation (cv) scores indicate better motifs. The 4th column reports the Log-Likelihood Ratio (LLR) match threshold, which ranges from 0-1. Higher values indicate closer match to the refined hypothesis. The 5th column reports the adjusted p-value, which is obtained by comparing the mean five-fold cv error to a distribution of cv errors obtained by running THEME with the same hypothesis on randomly selected sequences. P-values are adjusted for multiple hypothesis testing.
| Transcription Factor |
Refined Hypothesis |
Mean 5-fold cv error |
LLR match threshold |
Adjusted p-value |
|---|---|---|---|---|
| Activated Genes | ||||
| SCL/Tal-1 | ...rmCAGaTGky | 0.39 | 0.47 | 1.3E-03 |
| NFE2L1 | GTCAt... | 0.39 | 0.73 | 5.7E-04 |
| SOX-9 | .aaCAATrg | 0.39 | 0.60 | 2.0E-03 |
| ZIC2 | kgggtggtc | 0.42 | 0.69 | 6.8E-04 |
| Zbtb7a | r..rmCCCm | 0.42 | 0.69 | 4.1E-03 |
| Repressed Genes | ||||
| STAT | TGAGmkCTCA | 0.37 | 0.58 | 1.7E-05 |
| NF-Y | cTGATTGGytas. | 0.39 | 0.54 | 3.4E-03 |
| PPAR-gamma | ..w..AA.tCccArgwc | 0.40 | 0.43 | 5.4E-05 |
| CUX1 | cCAaTAAtcrat | 0.40 | 0.36 | 3.5E-03 |
| Meis-1a | TgACAGkTtwaygr | 0.41 | 0.48 | 6.6E-06 |
| PPAR-gamma | .gg.cAarGgtcw | 0.42 | 0.63 | 3.5E-03 |
| Msx-1 | c.gtaw.Tg | 0.42 | 0.71 | 3.5E-04 |
| Pax-5 | rr.swga.w.ct.rRGCGkracsry.s. | 0.43 | 0.30 | 8.1E-04 |
| HSF | tTCyagaas.TTC | 0.44 | 0.47 | 4.1E-03 |
Zbtb7a (also known as LRF, Pokemon or FBI-1) is a member of the POZ/BTB and Krüppel (POK) family of transcription factors and is involved in B- and T-cell lineage fate determination. It has only recently been recognized as a factor required for erythroid development (Maeda et al., 2007). However, its erythroid target genes and association with GATA-1 transcriptional activities has not been previously described.
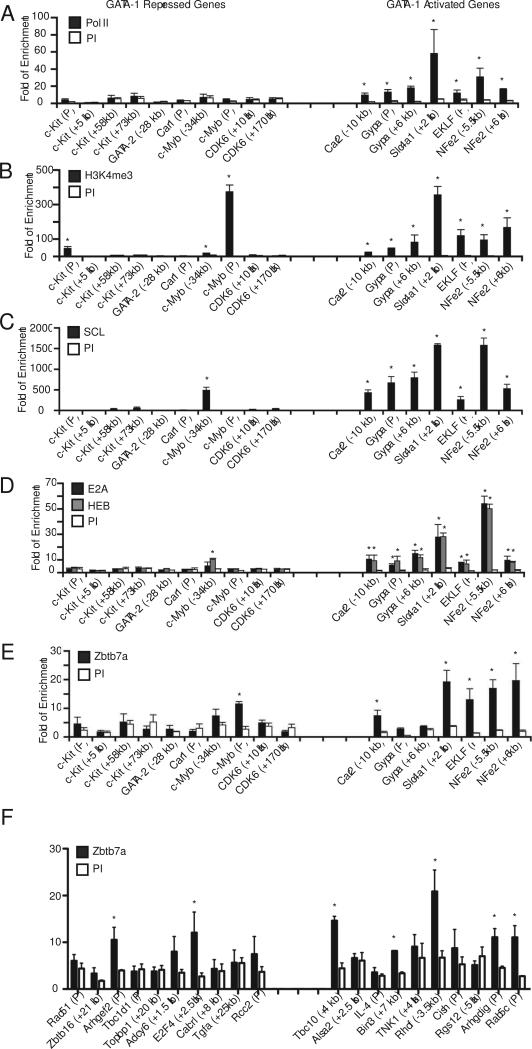
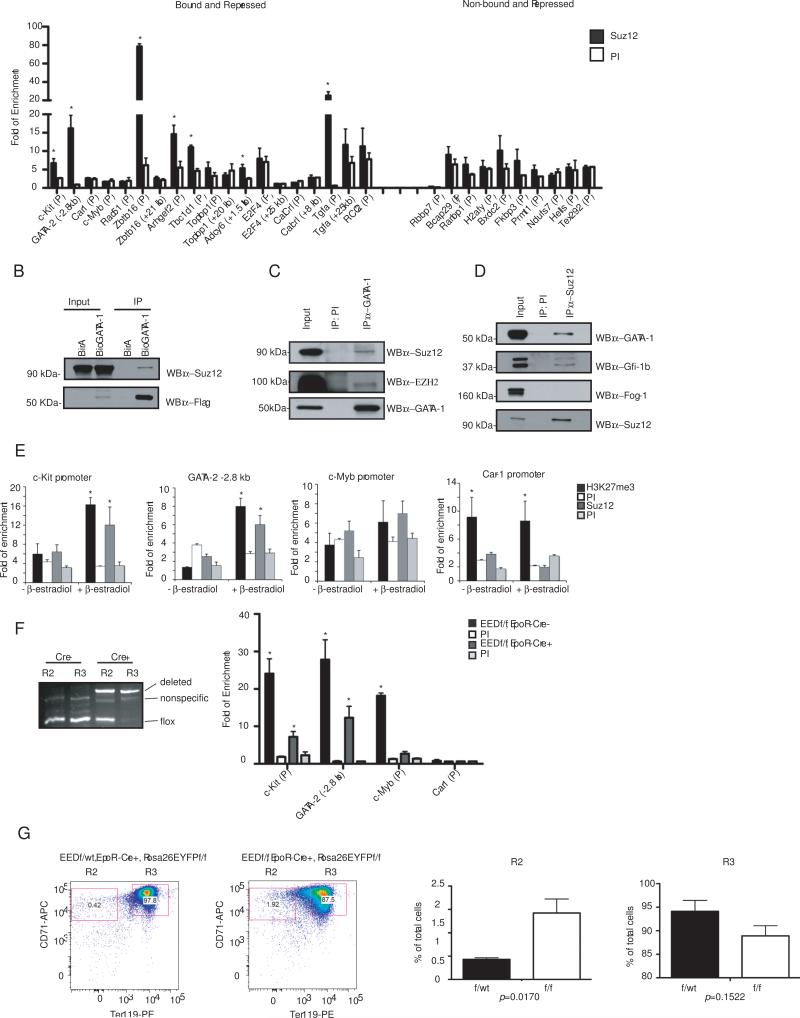
In order to test the relevance of these findings in a physiologic setting, all GATA-1 occupancy sites associated with five bound and up regulated genes (Car2, Gypa, Slc4a1, Klf1, and NFE2-p45) and five bound and down regulated genes (c-kit, GATA-2, Car1, c-myb and CDK6) were initially examined for selected transcription factor occupancy and histone modification in FACS-purified CD71+/low, Ter119+ cells from e13.5-14.5 murine fetal liver. This “R3-4” population represents late stage nucleated erythroid progenitors undergoing terminal maturation (Zhang et al., 2003). Expression changes of the selected genes in this population compared to more primitive CD71+Ter119− (“R2”) cells are similar to that seen during G1ER-4 and MEL cell induction (Fig. S6). As expected, Pol II and the activating histone mark H3K4me3 were present at GATA-1 occupancy sites of all the activated genes (Fig. 3A, B). H3K4me3 was also present at c-myb and c-kit. However, these sites also contain the repressive mark H3K27me3 (Fig. 4D), indicating that they are “bivalently” marked in this cell population. Consistent with the results of Tripic et al. (Tripic et al., 2008), we found occupancy of SCL/TAL1 and its heterodimeric partners HEB and E2A at all of the activated genes, but not at any of the repressed genes except the c-myb upstream region (Fig 3. C,D). Examination of Zbtb7a showed enrichment at GATA-1 occupancy sites of 4 of the 5 activated genes (5 of 7 associated GATA-1 occupancy sites), as well as at the c-myb promoter (Fig. 3E). Examination of 20 additional genes showed enrichment at 5 of 10 activated, but also 2 of 10 repressed genes, indicating that Zbtb7a can occupy both GATA-1 activated and repressed genes (Fig. 3F).
Figure 3.
ChIP analysis for occupancy of candidate activating factors and histone modifications at GATA-1 bound regions of five activated and repressed genes in sorted CD71+/lowTer119+ primary e13.5-14.5 fetal liver cells. (A) Pol II, (B) H3K4me3, (C) SCL/TAL1, (D) E2A/HEB, and (E, F) Zbtb7a. PI, pre-immune or random pooled species-matched IgG. Results are shown as fold enrichment compared to a region located 2 kb 5’ to GATA-1 HS1, which is devoid of GATA-1 binding sites. The mean of 3 independent ChIP assays is shown −/+ SEM. Enrichment differences between specific and control antibodies > 2-fold and with p-values < 0.05 (Student's t-test) are indicated with an asterisk. All GATA-1 occupancy sites detected in the ChIP-seq dataset from induced MEL cells were included. The promoter regions of c-kit and c-myb were also included, even though GATA-1 occupancy was not detected in this region. The c-myb occupancy site was located beyond the −10 kb to TSS window, but was included here given the role of c-myb in erythroid development.
Figure 4.
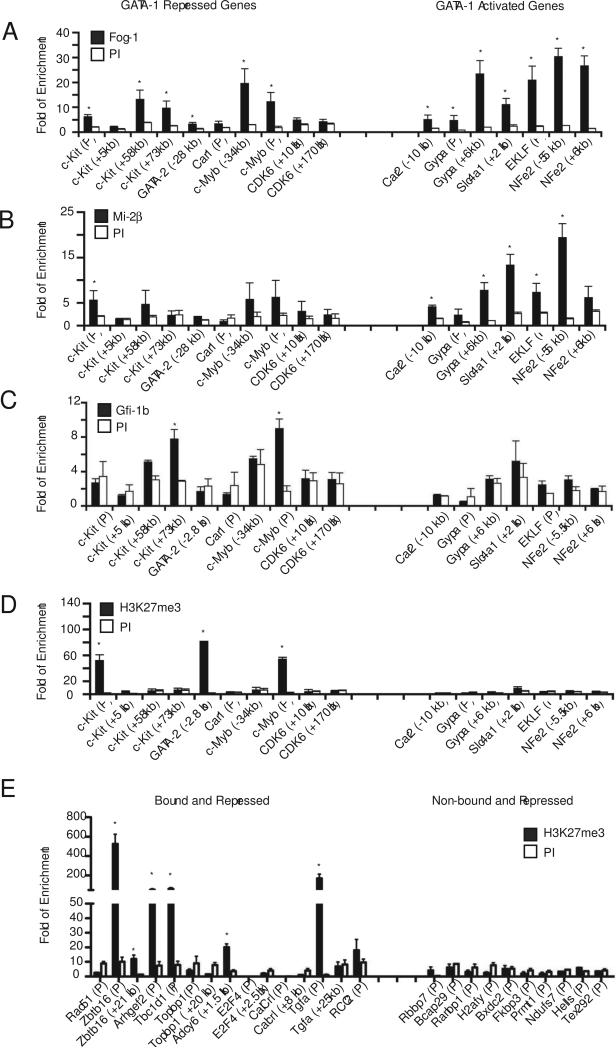
ChIP analysis for occupancy of candidate repressive factors and histone modifications at GATA-1 bound regions of activated and repressed genes in sorted CD71+/lowTer119+ primary e13.5-14.5 fetal liver cells. (A) FOG-1, (B) Mi-2 , (C) Gfi-1b, and (D,E) H3K27me3. Details of the ChIP assays are as described in Figure 3.
We also examined occupancy of EKLF and ZBP-89, two other erythroid Krüppel-type transcription factors that bind similar sequences to Zbtb7a and interact with GATA-1 (Perkins et al., 1995; Woo et al., 2008). These Krüppel factors were moderately enriched at the Gypa and/or NF-E-p45 genes, but not at any of the repressed gene examined (Fig. S7).
GATA-1 Mediated Gene Repression
A large number of motifs were statistically enriched at GATA-1 occupancy sites of repressed genes compared to activated genes (Table 1). However, none of these factors has previously been reported to have functional roles in erythroid development.
In vivo occupancy of several candidate factors was then investigated. The GATA cofactor Friend of GAT-1 (FOG-1) interacts with the co-repressor complexes NuRD and CtBP. Interestingly, we found both FOG-1 and the key NuRD component Mi-2β at GATA-1 occupancy sites of both repressed and activated genes (Fig. 4A,B). In fact, enrichment for Mi-2β was generally stronger at activated compared to repressed genes, suggesting that it may have more complex functions than simple transcriptional repression.
Gfi-1b is a repressor that associates with GATA-1 and directly interacts with CoREST and the histone demethylase LSD1 (Saleque et al., 2007). Although we did not find statistically significant enrichment for Gfi-1b binding motifs in our ChIP-seq dataset, occupancy by Gfi-1b was found at GATA-1 sites of several of the repressed genes tested, including a distal regulatory element of the c-kit gene and the c-myb promoter (Fig. 4C).
Role of Polycomb Repressive Complex 2 (PRC2) in GATA-1 Mediated Gene Silencing
Polycomb proteins play critical roles in epigenetic gene silencing. Most studies in mammalian systems have focused on their involvement in embryonic stem cell pluripotency. However, recent work shows that they also play roles during maturation of lineage-restricted tissues, such as skin, in adult animals (Ezhkova et al., 2009). CpG rich sequences are associated with occupancy by PRC2, one of the two Polycomb Repressive Complexes (Ku et al., 2008). Analysis of GATA-1 occupancy peaks in our dataset revealed significant enrichment of CpG-rich sequences at repressed versus activated genes (Fisher's exact test p=1.6E-005), raising the possibility that PRC2 may be involved in epigenetic silencing at some GATA-1 repressed loci.
We measured levels of H3K27me3, a chromatin mark associated with PRC2-mediated epigenetic silencing, at GATA-1 repressed versus activated genes in R3-4 fetal liver cells. We found significant enrichment at three of the repressed genes tested, including the c-kit promoter, GATA-2 −2.8 kb, and c-myb promoter, but at none of the activated genes (Fig. 4D). We then extended our analysis to 10 more GATA-1 bound and repressed genes (Fig. 4E). Of these, 5 also contain significant enrichment for H3K27me3. Surprisingly, examination of 10 repressed genes that were not bound by GATA-1 failed to show enrichment for H3K27me3 (Fig. 4E).
In order to further investigate a role of PRC2 in epigenetic gene silencing of GATA-1 repressed genes, chromatin occupancy by Suz12, a core subunit of PRC2, was examined in the R3-4 fetal liver cells. Seven of the 8 genes containing H3K27me3 also contained Suz12, whereas none of the genes without H3K27me3 were enriched for Suz12 (Fig. 5A).
Figure 5.
Role of PRC2 at direct GATA-1 repressed genes. (A) ChIP assays for occupancy of Suz12 at the indicated GATA-1 repressed genes. Results are expressed as fold change compared to negative control region located 2 kb 5’ to GATA-1 HS1, and represent the mean of 3 independent experiments +/− SEM. Enrichment differences between specific and control antibodies > 2-fold and with p-values < 0.05 (Student's t-test) are indicated with an asterisk. (B) Association of Suz12 with FLAG-BioGATA-1 in induced MEL cells. Western blot analysis using anti-Suz12 or anti-FLAG antibodies of eluted material after SA affinity pull-down from MEL cells expressing birA alone or birA and FLAG-BioGATA-1. 10% input is shown. (C) Co-IP of endogenous Suz12 and EZH2 with GATA-1 from induced MEL cells. 20% input is shown. (D) Co-IP of endogenous GATA-1, Gfi-1b and FOG-1, with Suz12 from induced MEL cells. 20% input is shown. (E) ChIP analysis for Suz12 occupancy and H3K27me3 at the indicated sites in G1ER-4 cells before and 48 hrs after addition of β-estradiol. (F) Left panel, genomic PCR analysis from FACS-sorted “R2” (lin− CD71+Ter119−) and “R3” (lin− (except Ter119) CD71+Ter119+) cell populations for EED allele deletion of EEDfl/fl, EpoR-Cre− and EEDfl/fl, EpoR-Cre+ mice. The expected PCR product positions for the deleted and floxed alleles are indicated. A non-specific band is present in all lanes. Right panel, ChIP assay for H3K27me3 at the indicated sites in FACS-sorted “R3” cells from e13.5 fetal livers of EEDfl/fl, EpoR-Cre− or EEDfl/fl, EpoR-Cre+ embryos. Details of the ChIP assay are as in “A”. The mean value is indicated −/+ SEM (n=3). (G) Effect of loss of EED on erythroid maturation in vivo. Left panel, flow cytometric analysis for CD71 and Ter119 expression of EYFP+ cells from representative EEDfl/wt, Rosa26-floxed stopper EYFP+, Epo-R Cre+ (left) versus EEDfl/fl, Rosa26-floxed stopper EYFP+, Epo-R Cre+ (right) embryonic e13.5 fetal livers. Right panel, compilation of data from 4 independent animals each, showing the percentage of cells in the “R2” and “R3” populations −/+ SEM.
Given the association of Suz12 with GATA-1 repressed genes, we next performed co-IP experiments to determine if GATA-1 and Suz12 physically associate. As shown in Fig. 5B, Suz12 was co-purified after SA-affinity purification of FLAG-bioGATA-1 in induced MEL cells, but not in control cells expressing birA alone. Physical association was also demonstrated by co-IP of endogenous proteins in induced MEL cells (Fig. 5C,D). Immunoprecipitation using an anti-GATA-1 antibody, but not control IgG, co-precipitates Suz12, as well as EZH2, another component of the PRC2 complex. Conversely, anti-Suz12 antibody, but not control IgG, co-precipitates GATA-1. Interestingly, the Suz12 IP also pulls down Gfi-1b, but not FOG-1, suggesting that the Suz12/GATA-1 complex is distinct from GATA-1/FOG-1 complexes, and likely includes Gfi-1b.
Suz12 recruitment and H3K27 trimethylation levels were next examined at several GATA-1 repressed genes before and after 48-hr induction of G1-ER4 cells with β-estradiol. As shown in Fig. 5E, a significant increase in Suz12 occupancy and H3K27me3 was observed at the c-kit promoter (1.9 and 2.7 fold, respectively) and GATA-2 −2.8 kb region (2.4 and 5.9 fold, respectively). A smaller trend was seen at the c-myb promoter, although this was not statistically significant. The Car1 promoter did not change significantly after induction, although it has relatively high levels of H3K27 trimethylation in uninduced G1-ER4 cells.
The role of PRC2 in GATA-1 mediated gene regulation was next examined in vivo. Mice containing targeted deletion of PRC2 core component genes, such as Suz12, EED, and EZH2, are embryonic lethal due to gastrulation defects (Faust et al., 1995; O'Carroll et al., 2001; Pasini et al., 2004). We therefore utilized conditional EED knock out mice (EEDfl/fl) containing EpoR-Cre for erythroid-specific inactivation of EED. These mice also contain the cDNA encoding enhanced yellow fluorescence protein (EYFP), preceded by a stopper cassette flanked by loxP sites, inserted into the ubiquitously expressed Rosa26 locus. This allows tracking of cells that express (or at one time expressed) Cre by gating on YFP+ cells. A full description of these mice will be reported elsewhere, but they are viable and grow to adulthood (H. Xie and S.H. Orkin, manuscript in preparation). We first examined excision of the EED allele in sorted lin− CD71+Ter119− (“R2” population) and lin− (excluding Ter119) CD71+Ter119+ cells (“R3” population), and found that excision of the EED allele is incomplete at the R2 stage, but is nearly complete at the R3 stage (Fig. 5F, left panel). Examination of R3 sorted cells for H3K27me3 levels shows a significant reduction in H3K27 trimethylation at the c-kit promoter, GATA-2 −2.8 kb, and c-myb promoter in sorted CD71+Ter119+ cells compared to control EEDfl/fl, EpoR-Cre− mice (Fig. 5F, right panel), consistent with involvement of PRC2 at these GATA-1 direct target genes.
Lastly, we examined the consequences of EED loss on erythroid maturation. Fetal livers from e13.5 EEDfl/fl, EpoR-Cre+, Rosa26EYFPfl/fl or EEDfl/wt, EpoR-Cre+, Rosa26EYFPfl/fl embryos were harvested and processed into single cell suspensions. Surface expression of CD71 and Ter119 was then measured by flow cytometry after gating for YFP+ cells. This revealed an increase in the percentage of R2 cells and reduction in the percentage of R3 cells in conditional knock out compared to control animals (1.4 ± 0.30 % vs. 0.43 +/− 0.035%, p=0.017; and 89 ± 2.2% vs. 94 ± 2.3%, p=0.15, respectively; N=4 each), indicating a partial block in erythroid maturation.
Discussion
In this study, we provide genome-wide chromatin occupancy analysis of the erythroid MTF GATA-1. Comparison to a new comprehensive dataset of GATA-1 induced gene expression changes allowed us to characterize features of GATA-1 in vivo occupancy that correlate with its site selectivity and gene context-dependent transcriptional activity.
We combined transcription factor metabolic biotinylation with streptavidin-based ChIP-seq, and were able to confirm 97% and 91% of our peak calls by independent standard ChIP assays in MEL or primary erythroid cells, respectively. Comparison of our dataset with 63 validated enrichment peaks identified in a recent GATA-1 ChIP-chip study of 66 Mb of mouse chromosome 7 in induced G1-ER4 cells (Cheng et al., 2008) shows an overlap of 21 peaks (33%) (Tables S6 and S7). Of 59 sites identified in the ChIP-chip study that did not validate, none were called as peaks in our dataset. If we relax our threshold call from 14 to 8, then the ChIP-seq dataset picks up 30 of the 63 ChIP-chip validated peaks (48%), and only one of the 59 non-validated sites. Thus, the SA-biotin ChIP-seq technique as applied here has relatively high specificity, but perhaps limited sensitivity, at least based on comparison to this one prior study.
Although our dataset may fail to identify all of the bona fide GATA-1 occupancy sites, ascertainment of a large number of high-confidence sites allowed us to apply statistical methods to further understand GATA-1's transcriptional activity. We found that the binding motif that best predicts global GATA-1 in vivo is slightly more extended and has more sequence preference than the canonical motif defined in previous DNAse I footprinting and in vitro studies, and that palindromic GATA binding motifs are significantly enriched at in vivo occupancy sites. Moreover, a higher overall number of GATA motifs is predictive of in vivo occupancy. While our data may be biased toward high affinity sites, these findings may help explain GATA-1's in vivo site selectivity.
Comparison of our ChIP-seq dataset with GATA-1 induced gene expression changes enabled the identification of global cell processes that are under direct GATA-1 control. As expected, we found marked enrichment for genes involved in hemoglobin synthesis, providing further validation of our dataset. In addition, we found that many GATA-1 direct target genes are involved in cell cycle control and Ras/GTPase signaling.
GATA-1's role in regulating cell proliferation has been previously studied. Rylski et al. showed that GATA-1 represses expression of the cyclin-dependent kinase 6 (Cdk6) and cyclin D2, and activates expression of the Cdk inhibitors p18INK4C and p27Kip1 (Rylski et al., 2003). This occurs, in part, by direct transcriptional repression of the oncogene c-myc. Munogalavadla et al. found that GATA-1 directly regulates c-myb leading to altered cell cycle (Munugalavadla et al., 2005). In addition, GATA-1 deficiency causes marked hyperproliferation of murine megakaryocytes (Vyas et al., 1999a), and exclusive production of a short GATA-1 isoform (GATA-1s) leads to transient myeloproliferative disorder in Down syndrome neonates (Muntean et al., 2006). Our findings add to the list of cell cycle related genes, including E2F4, Cdc6, and Nek6, that are under direct GATA-1 transcriptional control.
Ras signaling has previously been shown to affect erythroid differentiation. K-ras−/− mice die during embryonic development from severe anemia (Johnson et al., 1997), and expression of oncogenic N-ras and H-ras perturbs erythroid differentiation (Darley et al., 1997; Zhang et al., 2003). Since Ras/GTPase signaling is also involved in cell survival, our findings may partly explain GATA-1's anti-apoptotic activity during erythroid development (Weiss and Orkin, 1995).
Motif search analysis of GATA-1 bound regions of direct target genes revealed enrichment for Zbtb7a binding sequences, particularly at activated genes. Zbtb7a occupancy was confirmed at a significant number of GATA-1 enrichment sites in stage-sorted primary fetal liver erythroid cells, although it was found at both activated and repressed genes. Zbtb7a is highly expressed in CD71+ Ter119+ primary erythroid cells and is transcriptionally activated by EKLF (Hodge et al., 2006; Maeda et al., 2007). Zbtb7a−/− mice die at around e16.5 from severe anemia (Maeda et al., 2007). Yet, the mechanisms underlying the anemia have not been reported. Our data suggest that combinatorial transcriptional activity with GATA-1 may be involved in this phenotype. Zbtb7b (Th-Pok), a close family member of Zbtb7a, is a key regulator of CD4/CD8 lineage choice during T-cell development, a process that involves the GATA family member GATA-3 (He et al., 2005; Sun et al., 2005). It is possible that similar combinatorial processes occur between Zbtb7b and GATA-3 during lymphoid development.
Compared to gene activation, less is known about how GATA-1 functions as a transcriptional repressor. Polycomb repressive complexes play critical roles in epigenetic gene silencing during development. Unlike in Drosophila, in which polycomb protein complexes are recruited to Polycomb Response Elements (PREs), the recruitment of polycomb in mammalian cells is poorly understood. In FACS-sorted primary fetal liver erythroblasts, we found significant levels of H3K27me3 at a number of GATA-1 bound and repressed genes, but not activated genes or non-GATA-1 bound repressed genes. Although we cannot conclude that GATA-1 directly recruits PRC2 to these sites, several pieces of data support PRC2 involvement in late stages of GATA-1 mediated gene silencing, at least for a subset of genes. First, GATA-1 physically associates with Suz12 and EZH2 in erythroid cells (Fig. 5B-D). Second, activation of GATA-1 in G1-ER4 cells results in increased Suz12 chromatin occupancy and H3K27me3 levels at some GATA-1 direct target genes within 48 hours. Third, erythroid-specific deletion of the core PRC2 component EED results in reduced H3K27me3 at direct GATA-1 target genes. Fourth, erythroid specific deletion of EED results in impaired erythroid maturation.
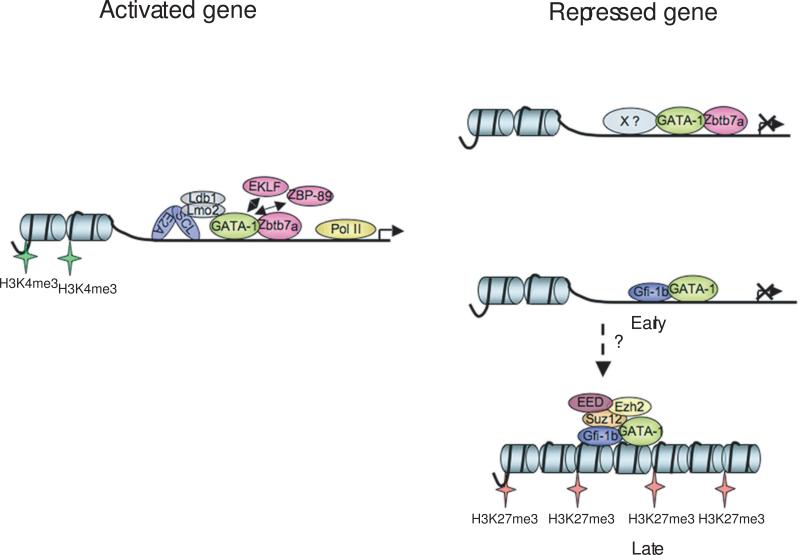
We feel that it is unlikely that PRC2 is involved in the initial steps of GATA-1 mediated gene repression. Rather, we favor the view that it participates in stabilizing epigenetic silencing once the initial decision to turn off the gene is made. Gfi1b is a transcriptional repressor that is required for normal erythroid development (Saleque et al., 2007). It interacts with LSD1, which has specific H3K4 demethylase activity. Interestingly, Gfi-1b co-purifies with Suz12 and GATA-1 in MEL cells (Fig. 5D), and Gfi-1b occupies GATA-1 bound regions of several repressed genes (Fig. 4C). The actions of Gfi-1b, via LSD1, may be the initial step in reversing gene activation by removing H3K4 methylation at genes that are initially on during early erythroid development, such as GATA-2 and c-kit. The H3K27 methyltransferase activity of PRC2 may then act to stabilize the silencing at a subset of genes (Fig. 6). GATA-1 may coordinate these activities, acting as a platform for both Gfi-1b and PRC2. It is also possible that the absence of SCL/TAL1 complexes might enable recruitment of PRC2. Future studies will be needed to address these possibilities.
Figure 6.
Model of transcription factor and cofactor occupancy at in vivo GATA-1 binding sites correlating with gene activation versus repression.
In summary, our data provide a genome-wide analysis of GATA-1 chromatin occupancy, facilitating examination of its transcriptional mechanisms. The findings implicate Zbtb7a as a factor involved in GATA-1 mediated gene regulation, and the PRC2 complex as being involved in late stages of silencing of some GATA-1 repressed genes. This dataset should provide a valuable resource to other investigators studying the transcriptional regulation of terminal cell maturation in mammalian systems.
Supplementary Material
Acknowledgements
A.B.C. is supported by a grant from the NIH (P01 HL32262-25). L.R. is supported by the CSBi Merck-MIT postdoctoral fellowship. E.F. is the recipient of the Eugene Bell Career Development Chair. The authors would like to thank Gerd Blobel, Xiaohua Shen, Jian Xu, and Jonathan Snow for critical review of the manuscript, and Pier Paolo Pandolfi for helpful discussions and for the Zbtb7a antibody. The ChIP-seq (submission GSE16594) and G1-ER4 cDNA microarray data (submission GSE18042) have been deposited in the Gene Expression Omnibus (GEO) database.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Material and Methods
See Supplemental Information online for all Materials and Methods.
References
- Andrews NC, Erdjument-Bromage H, Davidson MB, Tempst P, Orkin SH. Erythroid transcription factor NF-E2 is a haematopoietic-specific basic-leucine zipper protein. Nature. 1993;362:722–728. doi: 10.1038/362722a0. [DOI] [PubMed] [Google Scholar]
- Bresnick EH, Martowicz ML, Pal S, Johnson KD. Developmental control via GATA factor interplay at chromatin domains. J Cell Physiol. 2005;205:1–9. doi: 10.1002/jcp.20393. [DOI] [PubMed] [Google Scholar]
- Cantor AB, Orkin SH. Hematopoietic development: a balancing act. Curr Opin Genet Dev. 2001;11:513–519. doi: 10.1016/s0959-437x(00)00226-4. [DOI] [PubMed] [Google Scholar]
- Cheng Y, King DC, Dore LC, Zhang X, Zhou Y, Zhang Y, Dorman C, Abebe D, Kumar SA, Chiaromonte F, et al. Transcriptional enhancement by GATA1-occupied DNA segments is strongly associated with evolutionary constraint on the binding site motif. Genome Res. 2008;18:1896–1905. doi: 10.1101/gr.083089.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darley RL, Hoy TG, Baines P, Padua RA, Burnett AK. Mutant N-RAS induces erythroid lineage dysplasia in human CD34+ cells. J Exp Med. 1997;185:1337–1347. doi: 10.1084/jem.185.7.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer E, Rodriguez P, Bonte E, Krijgsveld J, Katsantoni E, Heck A, Grosveld F, Strouboulis J. Efficient biotinylation and single-step purification of tagged transcription factors in mammalian cells and transgenic mice. Proc Natl Acad Sci U S A. 2003;100:7480–7485. doi: 10.1073/pnas.1332608100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans T, Felsenfeld G. The erythroid-specific transcription factor Eryf1: a new finger protein. Cell. 1989;58:877–885. doi: 10.1016/0092-8674(89)90940-9. [DOI] [PubMed] [Google Scholar]
- Evans T, Reitman M, Felsenfeld G. An erythrocyte-specific DNA-binding factor recognizes a regulatory sequence common to all chicken globin genes. Proc Natl Acad Sci U S A. 1988;85:5976–5980. doi: 10.1073/pnas.85.16.5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezhkova E, Pasolli HA, Parker JS, Stokes N, Su IH, Hannon G, Tarakhovsky A, Fuchs E. Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell. 2009;136:1122–1135. doi: 10.1016/j.cell.2008.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faust C, Schumacher A, Holdener B, Magnuson T. The eed mutation disrupts anterior mesoderm production in mice. Development. 1995;121:273–285. doi: 10.1242/dev.121.2.273. [DOI] [PubMed] [Google Scholar]
- Fujiwara Y, Browne CP, Cunniff K, Goff SC, Orkin SH. Arrested development of embryonic red cell precursors in mouse embryos lacking transcription factor GATA-1. Proc Natl Acad Sci U S A. 1996;93:12355–12358. doi: 10.1073/pnas.93.22.12355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grass JA, Boyer ME, Pal S, Wu J, Weiss MJ, Bresnick EH. GATA-1-dependent transcriptional repression of GATA-2 via disruption of positive autoregulation and domain-wide chromatin remodeling. Proc Natl Acad Sci U S A. 2003;100:8811–8816. doi: 10.1073/pnas.1432147100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory T, Yu C, Ma A, Orkin SH, Blobel GA, Weiss MJ. GATA-1 and erythropoietin cooperate to promote erythroid cell survival by regulating bcl-xL expression. Blood. 1999;94:87–96. [PubMed] [Google Scholar]
- He X, He X, Dave VP, Zhang Y, Hua X, Nicolas E, Xu W, Roe BA, Kappes DJ. The zinc finger transcription factor Th-POK regulates CD4 versus CD8 T-cell lineage commitment. Nature. 2005;433:826–833. doi: 10.1038/nature03338. [DOI] [PubMed] [Google Scholar]
- Hodge D, Coghill E, Keys J, Maguire T, Hartmann B, McDowall A, Weiss M, Grimmond S, Perkins A. A global role for EKLF in definitive and primitive erythropoiesis. Blood. 2006;107:3359–3370. doi: 10.1182/blood-2005-07-2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson L, Greenbaum D, Cichowski K, Mercer K, Murphy E, Schmitt E, Bronson RT, Umanoff H, Edelmann W, Kucherlapati R, Jacks T. K-ras is an essential gene in the mouse with partial functional overlap with N-ras. Genes Dev. 1997;11:2468–2481. doi: 10.1101/gad.11.19.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D, Kuhn RM, Baertsch R, Barber GP, Clawson H, Diekhans M, Giardine B, Harte RA, Hinrichs AS, Hsu F, et al. The UCSC Genome Browser Database: 2008 update. Nucleic Acids Res. 2008;36:D773–779. doi: 10.1093/nar/gkm966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, Presser A, Nusbaum C, Xie X, Chi AS, et al. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 2008;4:e1000242. doi: 10.1371/journal.pgen.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macisaac KD, Gordon DB, Nekludova L, Odom DT, Schreiber J, Gifford DK, Young RA, Fraenkel E. A hypothesis-based approach for identifying the binding specificity of regulatory proteins from chromatin immunoprecipitation data. Bioinformatics. 2006;22:423–429. doi: 10.1093/bioinformatics/bti815. [DOI] [PubMed] [Google Scholar]
- Maeda T, Merghoub T, Hobbs RM, Dong L, Maeda M, Zakrzewski J, van den Brink MR, Zelent A, Shigematsu H, Akashi K, et al. Regulation of B versus T lymphoid lineage fate decision by the proto-oncogene LRF. Science. 2007;316:860–866. doi: 10.1126/science.1140881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DI, Tsai SF, Orkin SH. Increased gamma-globin expression in a nondeletion HPFH mediated by an erythroid-specific DNA-binding factor. Nature. 1989;338:435–438. doi: 10.1038/338435a0. [DOI] [PubMed] [Google Scholar]
- Muntean AG, Ge Y, Taub JW, Crispino JD. Transcription factor GATA-1 and Down syndrome leukemogenesis. Leuk Lymphoma. 2006;47:986–997. doi: 10.1080/10428190500485810. [DOI] [PubMed] [Google Scholar]
- Munugalavadla V, Dore LC, Tan BL, Hong L, Vishnu M, Weiss MJ, Kapur R. Repression of c-kit and its downstream substrates by GATA-1 inhibits cell proliferation during erythroid maturation. Mol Cell Biol. 2005;25:6747–6759. doi: 10.1128/MCB.25.15.6747-6759.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton A, Mackay J, Crossley M. The N-terminal zinc finger of the erythroid transcription factor GATA-1 binds GATC motifs in DNA. J Biol Chem. 2001;276:35794–35801. doi: 10.1074/jbc.M106256200. [DOI] [PubMed] [Google Scholar]
- O'Carroll D, Erhardt S, Pagani M, Barton SC, Surani MA, Jenuwein T. The polycomb-group gene Ezh2 is required for early mouse development. Mol Cell Biol. 2001;21:4330–4336. doi: 10.1128/MCB.21.13.4330-4336.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrott MB, Barry MA. Metabolic biotinylation of recombinant proteins in mammalian cells and in mice. Mol Ther. 2000;1:96–104. doi: 10.1006/mthe.1999.0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Bracken AP, Jensen MR, Lazzerini Denchi E, Helin K. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. Embo J. 2004;23:4061–4071. doi: 10.1038/sj.emboj.7600402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedone PV, Omichinski JG, Nony P, Trainor C, Gronenborn AM, Clore GM, Felsenfeld G. The N-terminal fingers of chicken GATA-2 and GATA-3 are independent sequence-specific DNA binding domains. Embo J. 1997;16:2874–2882. doi: 10.1093/emboj/16.10.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins AC, Sharpe AH, Orkin SH. Lethal beta-thalassaemia in mice lacking the erythroid CACCC-transcription factor EKLF. Nature. 1995;375:318–322. doi: 10.1038/375318a0. [DOI] [PubMed] [Google Scholar]
- Phillips JD, Steensma DP, Pulsipher MA, Spangrude GJ, Kushner JP. Congenital erythropoietic porphyria due to a mutation in GATA-1: the first trans-acting mutation causative for a human porphyria. Blood. 2006 doi: 10.1182/blood-2006-06-022848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porcher C, Swat W, Rockwell K, Fujiwara Y, Alt FW, Orkin SH. The T cell leukemia oncoprotein SCL/tal-1 is essential for development of all hematopoietic lineages. Cell. 1996;86:47–57. doi: 10.1016/s0092-8674(00)80076-8. [DOI] [PubMed] [Google Scholar]
- Rylski M, Welch JJ, Chen YY, Letting DL, Diehl JA, Chodosh LA, Blobel GA, Weiss MJ. GATA-1-mediated proliferation arrest during erythroid maturation. Mol Cell Biol. 2003;23:5031–5042. doi: 10.1128/MCB.23.14.5031-5042.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleque S, Kim J, Rooke HM, Orkin SH. Epigenetic regulation of hematopoietic differentiation by Gfi-1 and Gfi-1b is mediated by the cofactors CoREST and LSD1. Mol Cell. 2007;27:562–572. doi: 10.1016/j.molcel.2007.06.039. [DOI] [PubMed] [Google Scholar]
- Sun G, Liu X, Mercado P, Jenkinson SR, Kypriotou M, Feigenbaum L, Galera P, Bosselut R. The zinc finger protein cKrox directs CD4 lineage differentiation during intrathymic T cell positive selection. Nat Immunol. 2005;6:373–381. doi: 10.1038/ni1183. [DOI] [PubMed] [Google Scholar]
- Trainor CD, Ghirlando R, Simpson MA. GATA zinc finger interactions modulate DNA binding and transactivation. J Biol Chem. 2000;275:28157–28166. doi: 10.1074/jbc.M000020200. [DOI] [PubMed] [Google Scholar]
- Trainor CD, Omichinski JG, Vandergon TL, Gronenborn AM, Clore GM, Felsenfeld G. A palindromic regulatory site within vertebrate GATA-1 promoters requires both zinc fingers of the GATA-1 DNA-binding domain for high-affinity interaction. Mol Cell Biol. 1996;16:2238–2247. doi: 10.1128/mcb.16.5.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripic T, Deng W, Cheng Y, Zhang Y, Vakoc CR, Gregory GD, Hardison RC, Blobel GA. SCL and associated proteins distinguish active from repressive GATA transcription factor complexes. Blood. 2008 doi: 10.1182/blood-2008-07-169417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai SF, Martin DI, Zon LI, D'Andrea AD, Wong GG, Orkin SH. Cloning of cDNA for the major DNA-binding protein of the erythroid lineage through expression in mammalian cells. Nature. 1989;339:446–451. doi: 10.1038/339446a0. [DOI] [PubMed] [Google Scholar]
- Tubman VN, Levine JE, Campagna D, R., Fleming MD, Neufeld EJ. X-linked gray platelet syndrome due to a GATA1 Arg261Gln mutation.. Paper presented at: 47th Annual Conference of The American Society of Hematology; Atlanta, Georgia, Blood. 2005. [DOI] [PubMed] [Google Scholar]
- Viens A, Mechold U, Lehrmann H, Harel-Bellan A, Ogryzko V. Use of protein biotinylation in vivo for chromatin immunoprecipitation. Anal Biochem. 2004;325:68–76. doi: 10.1016/j.ab.2003.10.015. [DOI] [PubMed] [Google Scholar]
- Vyas P, Ault K, Jackson CW, Orkin SH, Shivdasani RA. Consequences of GATA-1 deficiency in megakaryocytes and platelets. Blood. 1999a;93:2867–2875. [PubMed] [Google Scholar]
- Vyas P, McDevitt MA, Cantor AB, Katz SG, Fujiwara Y, Orkin SH. Different sequence requirements for expression in erythroid and megakaryocytic cells within a regulatory element upstream of the GATA-1 gene. Development. 1999b;126:2799–2811. doi: 10.1242/dev.126.12.2799. [DOI] [PubMed] [Google Scholar]
- Wall L, deBoer E, Grosveld F. The human beta-globin gene 3' enhancer contains multiple binding sites for an erythroid-specific protein. Genes Dev. 1988;2:1089–1100. doi: 10.1101/gad.2.9.1089. [DOI] [PubMed] [Google Scholar]
- Weiss MJ, Orkin SH. Transcription factor GATA-1 permits survival and maturation of erythroid precursors by preventing apoptosis. Proc Natl Acad Sci U S A. 1995;92:9623–9627. doi: 10.1073/pnas.92.21.9623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch JJ, Watts JA, Vakoc CR, Yao Y, Wang H, Hardison RC, Blobel GA, Chodosh LA, Weiss MJ. Global regulation of erythroid gene expression by transcription factor GATA-1. Blood. 2004;104:3136–3147. doi: 10.1182/blood-2004-04-1603. [DOI] [PubMed] [Google Scholar]
- Woo AJ, Moran TB, Schindler Y, Choe SK, Langer NB, Sullivan MR, Fujiwara Y, Paw BH, Cantor AB. Identification of ZBP-89 as a Novel GATA-1 Associated Transcription Factor Involved in Megakaryocytic and Erythroid Development. Mol Cell Biol. 2008 doi: 10.1128/MCB.01945-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C, Niakan KK, Matsushita M, Stamatoyannopoulos G, Orkin SH, Raskind WH. X-linked thrombocytopenia with thalassemia from a mutation in the amino finger of GATA-1 affecting DNA binding rather than FOG-1 interaction. Blood. 2002;100:2040–2045. doi: 10.1182/blood-2002-02-0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Socolovsky M, Gross AW, Lodish HF. Role of Ras signaling in erythroid differentiation of mouse fetal liver cells: functional analysis by a flow cytometry-based novel culture system. Blood. 2003;102:3938–3946. doi: 10.1182/blood-2003-05-1479. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.