Abstract
Serine palmitoyltransferase (SPT) has been localized to the endoplasmic reticulum (ER) by subcellular fractionation and enzymatic assays, and fluorescence microscopy of epitope-tagged SPT; however, our studies have suggested that SPT subunit 1 might be present also in focal adhesions and the nucleus. These additional locations have been confirmed by confocal microscopy using HEK293 and HeLa cells, and for focal adhesions by the demonstration that SPT1 co-immunoprecipitates with vinculin, a focal adhesion marker protein. The focal adhesion localization of SPT1 is associated with cell morphology, and possibly cell migration, because it is seen in most cells before they reach confluence but disappears when then become confluent, and is restored by a standard scratch-wound healing assay. Conversely, elimination of SPT1 using SPTLC1 siRNA causes cell rounding. Thus, in addition to its “traditional” localization in the ER for de novo sphingolipid biosynthesis, SPT1 is present in other cellular compartments, including focal adhesions where it is associated with cell morphology.
Keywords: Sphingolipid metabolizing enzymes, Subcellular localization, Focal adhesions
1. Introduction
Sphingolipids are a complex family of compounds that play important roles in membrane structure, biological recognition, cell adhesion, and cell regulation as participants in signaling, membrane trafficking, autophagy and other processes [1–5]. In some cases, complex sphingolipids (glycosphingolipids and sphingomyelins) affect cell behavior by associating with membrane microdomains that contain receptors, transporters, and other signal transducers such as Src family kinases, small G-proteins (e.g., RhoA, Ras), and focal adhesion kinase [5]. In other cases, the lipid backbones and related metabolites (e.g., ceramide, ceramide 1-phosphate, sphingosine 1-phosphate, and sphingosine) serve as first and second messengers [1, 2, 6] for behaviors such as cell migration and the associated actin stress fiber formation and focal adhesion formation [7–9]. These are often orchestrated by changes in the membrane dynamics of the pertinent sphingolipids as well as their trafficking, metabolic remodeling and de novo sphingolipid biosynthesis [2].
Sphingolipid biosynthesis is initiated by condensation of L-serine with palmitoyl-CoA by serine palmitoyltransferase (SPT), therefore, this is a critical enzyme for this pathway. In mammalian cells, SPT is thought to be a membrane bound heterodimer comprised of two subunits, SPT1 and SPT2, with molecular weights of 53 kDa and 63 kDa, respectively [10, 11], but in some cells, an additional subunit, SPT3, has also been noted [12]. SPT is often described as an enzyme of the endoplasmic reticulum (ER) because enzymatic activity has been found primarily in microsomes [13, 14], and hemaglutinin (HA)-tagged SPT1 has been localized to the ER when expressed in Chinese hamster ovary cells [11]. Nonetheless, immunohistochemical analyses of different tissues and cells using the antibodies for SPT1 have found cross-reacting species in not only the cytoplasm (i.e., ER at this resolution) but also the nucleus [15, 16] and focal adhesions [17]. Furthermore, a recent study [18] has detected an interaction of SPT1 with ABCA1.
This report establishes that SPT1 is, indeed, found in multiple subcellular locations using confocal microscopy and siRNA suppression of SPT to confirm the specificity of the antibody staining. The localization of SPT1 with focal adhesions was further confirmed by its co-immunoprecipitation with vinculin in HEK293 cells. The localization of SPT1 with focal adhesions is likely to play a role in cell morphology because it is seen in growing but not confluent cells in culture, and reappears when cell migration is induced by a scratch-wound healing assay, and suppression of SPT1 with siRNA results in cell rounding and detachment.
2. Materials and methods
2.1 Materials
The affinity purified polyclonal rabbit anti-human primary antibodies raised against SPT1 peptides have been previously described [15]. Anti-BiP antibody was from StressGen, Inc. (Victoria, BC, Canada). Anti-vinculin antibody was from Sigma Aldrich (Saint Louis, MO) and anti-ABCA1 antibody was from Novus Biologicals. The secondary antibodies were Alexa Fluor-conjugated goat anti-rabbit and anti-mouse obtained from Molecular Probes, Inc. (Eugene, OR). The nucleic acid dye Hoechst 33342 (Invitrogen, Carlsbad, CA). SPT1 epitope peptides and another rabbit anti-SPT1 antibody (catalog # AP2534b) were from Abgent (San Diego, CA).
2.2 Cells and cell culture
The HEK293, HEK293T and HeLa cells were obtained from the American Type Culture Collection (Manassas, VA). HEK293 cells were grown in DMEM/F12 medium (1:1) (Gibco BRL, MD) supplemented with 10% fetal bovine serum (FBS), penicillin (100 U / ml) and streptomycin (100 µg /ml) at 37 °C in a humidified 5 % CO2 atmosphere. HeLa cells and HEK293T cells were grown in Dulbecco's minimum essential medium (DMEM; Gibco BRL, MD) with the same supplements.
2.3 DNA constructs
SPTLC1 cDNA was obtained from Origene. To generate N-terminal GFP-tagged SPT1 the SPTLC1 gene was amplified by using the following primers: 5’-ATGGCGACCGCCACGGAGCAGTGG-3’ and 5’-GCCTAGAGCAGGACGGCCTGGGCT-3’. The PCR fragment corresponding to the coding sequence (CDS) of SPTLC1 was cloned into pEGFP-C2/SmaI (Clontech, Mountain View, CA) resulting in pEGFP-C2-SPT1. For C-terminal GFP-tagged SPT1, SPTLC1 gene was first amplified using the primers: 5’-ATGGCGACCGCCACGGAGCAGTGG-3’ and 5’-GAGCAGGACGGCCTGGGCTACCTC-3’. The PCR fragment of SPTLC1 was then subcloned into the pUC18 cloning vector. It was next digested from the pUC18 vector by using BamHI and EcoRI restriction enzymes and subcloned into the same sites in pEGFP-N1 to generate pEGFPN1-SPTLC1.
To create small peptide-tagged SPT1, pCMV-MAT-FLAG vector was used (Sigma Aldrich, Saint Louis, MO). SPTLC1 was first amplified by the following primers: 5’-ATGGCGACCGCCACGGAGCAGTGG-3’ and 5’-TAAGCGTAATCTGGAACATCGTATGGGTAGAGCAGGACGGCCTGGGCTACCTC-3’, the latter contains a HA sequence. The PCR fragment corresponding to the CDS of SPTLC1 was then cloned into pCMV-MAT-FLAG/EcoRV resulting in pCMVMAT-FLAG-SPTLC1-HA.
To construct internal tagged SPT1, the 5’-end of SPTLC1 gene was first amplified by 5’-GGATCCGCCACCATGGCGACCGCCACGGAGCAGTG-3’ and 5’-GATATCAGCGTAATCTGGAACATCGTATGGGTAAGCGTAATCTGGAACATCGTATGGGTAAGCGTAATCTGGAACATCGTATGGGTATCCATTCACCACAGTTTTGTGGCTTG-3’. The forward primer contains BamHI site and the reverse primer has three HA sequences. The 3’-end of SPTLC1 was amplified by the following primers: 5’-CATACGATGTTCCAGATTACGCTGATATCAAAGAATGTATAAACTTCGCCTCATTTAATTTTC-3’ and 5’-CTAGAGCAGGACGGCCTGGGC-3’. The forward primer has an overlap sequence with the previous reverse primer. The overlap extension was completed by using the same forward primer from the first pair and the same reverse primer from the second pair to amplify SPTLC1 gene from the beginning to the end. Then the amplified CDS was cleaved by BamHI and inserted into pcDNA 3.1 vector (Invitrogen, Carlsbad, CA).
All of the above have been sequenced to confirm the fidelity of the constructs.
2.4 Generation of SPT1 and SPT2 over-expressing cell lines
SPTLC1 and SPTLC2 were cloned from human monocytes. Briefly, total RNA from cells treated for 4 h with 1 µM dexamethasone was isolated using the RNeasy RNA isolation kit (Qiagen, Valencia, CA). The following oligos were used as amplification primers: SPTLC1 5’-CCGGAATTCATGGCGACCGCCACGGAGCAG, SPTLC1 3’-CCGGAATTCGACTCTGCCTAGAGCAGGAC, SPTLC2 5’-CCGCTCGAGATGCGGCCGGAGCCCGGAGGCTG, SPTLC2 3’-CTAGTCTAGAGGCTCAGTCTTCTGTTTGTTC. The SPTLC1 gene was cloned into pcDNA3.1NEO and SPTLC2 was cloned into pcDNA3.1ZEO. The expression plasmids were co-transfected into HEK293 cells using Superfect (Qiagen, Valencia, CA). 400 µg/ml geneticin or 200 µg/ml zeocin were added to the culture media 48 h after transfection to select for cells stably expressing SPT1 and SPT2. The co-transfection was selected in media containing both geneticin and zeocin. The media was changed every 4 days. After 2 weeks, surviving colonies were selected and grown in individual wells of a 6-well plate. 3 colonies were selected and checked by RT-PCR for the transfected gene transcript and by western blot for recombinant protein expression. The highest expressing cell line was selected for further study.
2.5 Immunofluorescence Confocal Microscopy
Cells were cultured on collagen (BD, San Jose, CA) coated glass coverslips (VWR, Inc., West Chester, PA) in a 24-well plate and fixed with 4 % formaldehyde in PBS at room temperature for 15 min. Fixed cells were permeabilized with 0.1 % Triton X-100 for 5 min, blocked in 10 % fetal calf serum in PBS (serum-PBS) for 30 min and then subjected to indirect immunofluorescence staining. Cells were incubated for 1 h at room temperature, with primary antibody diluted in PBS-serum, then the cells were washed 3 times with PBS-serum for 5 min each, and incubated for 1 h at room temperature with Alexa Fluor-conjugated secondary antibody. Nucleic acids and actin were stained by incubating fixed cells with PBS containing 1 µg/ml Hoechst 33342 dye and rhodamine phalloidin (Invitrogen, Carlsbad, CA). Stained cells were rinsed in PBS and mounted in Fluoromount G (Southern Biotechnology Associates, Inc., Bermingham, AL) before observing under a Zeiss LSM 510 inverted laser scanning confocal microscope (Heidelberg, Germany) equipped with a Zeiss Plan-Apochromat 43 × oil immersion objective lens, and controllers for setting the band excitation and emission of wavelengths in the green, red and blue regions. Slides were scanned in “line mode” and then singled at an average of 16 scans to eliminate background noise. Images were collected with the resident Zeiss confocal microscope software.
2.6 Western blotting
Equal amounts of protein (30 µg) from each fraction and 50 µg of total cell lysate were loaded on a 12 % SDS-PAGE gel (Pierce, Rockford, IL). For Western blotting, the gel was transferred to the nitrocellulose membrane using a Tris-glycine buffer with 20 % methanol as the transfer medium, for 1 h at 100 volts (constant), in a transfer unit (BIO RAD, Hercules, CA). The blot was blocked overnight at 4 °C with SuperBlock blocking buffer (Pierce, Rockford, IL). The membrane was then incubated with rabbit anti-SPT1 antibody (or other antibodies, where noted) diluted in SuperBlock and incubated for 2 h at room temperature. The membrane was rinsed three times in Tris buffered saline with 0.05 % Tween 20 (TBST) and incubated for 1 h at room temperature, with a peroxidase-conjugated secondary antibody (Pierce, Rockford, IL). Finally, the membrane was rinsed in TBST and incubated in Supersignal West Pico chemiluminiscent reagent (Pierce, Rockford, IL).
2.7 Peptide competition assay
The SPT1 epitope peptides KLQERSDLTVKEKEEC (45~59 aa) and KEQEIEDQKNPRKARC (222~236 aa) were synthesized by Abgent (San Diego, CA). Each peptide was dissolved in PBS/10 % serum to a final concentration of 20 µg/ml for the competition experiments. The antibody-peptide mixture was incubated overnight at 4°C and then applied for immunostaining following the standard protocol.
2.8 Co-immunoprecipitation
Immunoprecipitation procedure was modified from the standard protocols [19, 20]. Cells that were 60% confluent in 100-mm dishes were rinsed with ice-cold PBS and then scraped in 0.6 ml of ice- cold RIPA buffer (50 mM Tris-HCl, pH 7.4; 1% NP-40; 0.25% Na-deoxycholate; 150 mM NaCl; and 1 mM sodium orthovanadate) with protease inhibitor cocktail (Roche, Indianapolis, IN), and the cell lysate was centrifuge at 16,000 × g at 4°C for 15 min to prepare the supernatant for the co-immunoprecipitation studies. The cell lysate was cleared of proteins that bind non-specifically by mixing 0.5 ml of the supernatant with 50 µl of a 50% protein G-sepharose bead slurry (Sigma Aldrich, Saint Louis, MO) for 1 h, then centrifuged at 16,000 × g for 5 min. In another microcentrifuge tube, 50 µl of the 50% protein G-sepharose bead slurry was incubated with the antibody of interest (4 µg of mouse anti-vinculin antibody, Sigma Aldrich, Saint Louis, MO, or mouse anti-V5 antibody, Invitrogen, Carlsbad, CA, as a control) for 2 h at 4°C and centrifuged at 16,000 × g for 5 sec (the following centrifugations were at the same condition). The antibody-conjugated beads were washed 3 times with 1 ml of ice-cold RIPA buffer and the supernatant was removed. Then the antibody-conjugated beads were mixed with 0.5 ml of PBS and 0.5 ml of the pre-cleared cell lysate, and incubated with gentle mixing overnight at 4°C. After washing with the beads three times with ice-cold RIPA buffer and once with ice-cold PBS, they were recovered by centrifugation and the bound proteins were released by adding 40 µl of Laemmli sample buffer (Biorad, Hercules, CA), immersion of the tubes in boiling water for 5 min, cooling for the tube and centrifugation. Approximately 25% of the released protein was used for SDS-PAGE and Western blotting.
2.9 siRNA transfection
The day before transfection, HEK293 cells were seeded on collagen-coated coverslips in a 24-well plate at 4 × 104 cells per well. The three different SPTLC1 siRNA probes from Ambion (Austin, TX) targeting to exon 4 or 5 were pooled. The cells were transfected with the SPTLC1 siRNA pool or non-specific control siRNA (Ambion, Austin, TX) at 100 nM using DharmaFECT 1 transfection reagent (Dharmacon, Lafayette, CO). Mock transfection was performed with transfection reagent alone. Cell culture medium was replaced after 48 h of transfection. Cells were fixed for immunofluorescence staining after 72 h of transfection.
2.10 Subcellular fractionation
For a crude subcellular fractionation of the siRNA transfected cells, three 100-mm dishes were transfected with each kind of siRNA as above and after 72 h the cells were washed in PBS then scraped from the dishes in hypotonic buffer (20 mM Hepes, pH 7.9, at 4 °C, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT) plus protease inhibitor cocktail, Complete Mini (Roche, Indianapolis, IN) and homogenized by 30 strokes of a Dounce homogenizer. Nuclei were recovered by centrifugation at 1000 × g for 15 min and washed once using Nuclei Wash Buffer (10 mM Hepes, pH 7.9 at 4 °C, 0.2 mM MgCl2, 10 mM KCl, 0.5 mM DTT, protease inhibitor cocktail) and centrifuged as the curde nuclear fraction. The post-nuclear fraction was then centrifuged for 1 h at 100,000 × g and the supernatant was defined as the cytosoluble fraction and the pellets resuspended in the same buffer as the membrane fraction. All the fractions were stored as aliquots at −80°C [21, 22].
2.11 Cell viability assay
The floating cells were collected from three 100-mm dishes 48 h after siRNA transfection, centrifuged, suspended in 0.1 % trypan blue. Both the viable cells and the unviable cells were counted using a hemocytometer.
2.12 Lipid extraction and analysis by LC-ESI MS/MS
The method has been thoroughly described previously [23]. Briefly, the cells were collected in PBS and lipids were extracted in methanol/chloroform. The mass spectrometry data were collected using a PE Sciex API 3000 triple quadrupole mass spectrometer equipped with a turbo ion-spray source. As the first step of the analyses, the cells were examined by precursor ion scans for m/z 264.4 and 266.4 through the ceramide and monohexosylceramide range, and by precursor ion scans of m/z 184.4 for sphingomyelins to identify the major subspecies. Then for quantitative analysis, samples that had been spiked with an internal standard cocktail (from Avanti Polar Lipids, Alabaster, AL) were analyzed by LC ESI-MS/MS using a multiple reaction monitoring program built to encompass the subspecies of these sphingolipids found in the extracts [23].
2.13 Wound healing assay
HeLa cells were grown on collagen coated glass coverslips until confluent and a pipet tip was used to scratch the plate and the medium was replaced to remove the floating cells [24]. Three h after scratching the cells were fixed and processed for immunofluorescence staining and confocal microscopy.
3. Results
3.1 Native cell lines display novel subcellular localizations for endogenous SPT1
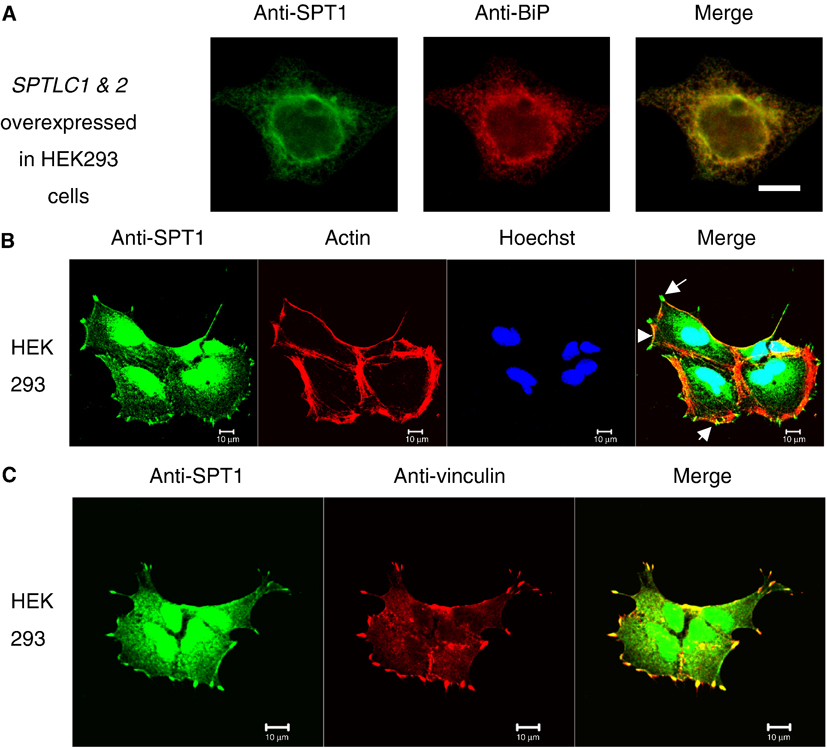
As has been done before [11], we initially examined SPT localization using a cell line that overexpresses this enzyme (in our case, a SPT1/2 cells, a HEK293 cell stably overexpresses SPTLC1 and SPTLC2 as described under “materials and methods”. The localization of SPT1 is shown in Figure 1A, revealing that most of the immunofluorescence produced by the anti-SPT1 antibodies [15, 16] overlapped with BiP, a marker for the ER [25], which agreed with the previous localization of a stably overexpressed SPT1 with epitope tags at either the N or C terminus [11]. However, when untransfected HEK293 cells were examined, the results (shown in Figure 1B) revealed that some of the immunostaining is in the cytoplasm (consistent with SPT1 in the ER), but a substantial portion is in the nucleus (based on co-localization with the DNA intercalating dye Hoechst 33342) and at the periphery of the cell. The SPT1 immunofluorescence at the periphery appears as punctuate dots at the convergence points for actin stress fibers (stained red with phalloidin), which are usually identified as focal adhesions [26]. This was confirmed by comparing the localization of SPT1 with the focal adhesion marker vinculin (Fig. 1C).
Fig. 1.
Intracellular localization of SPT1 in SPTLC1&SPTLC2 overexpressing HEK293 cells and native HEK293 cells by immunofluorescence staining and confocal microscopy. Cells were grown on glass coverslips for 24 h before collecting for immunostaining. Rabbit anti-SPT1 antibody was used as primary labeling and Alexa Fluor 488 anti-rabbit antibody (green) as secondary labeling. (A) Localization of SPT1 in HEK293/SPTLC1/2 stable cells. SPT1 protein was stained in green and the ER marker protein, Bip, was labeled with Alexa Flour 568 (red). (B) Localization of endogenous SPT1 proteins in HEK293 cells. SPT1 was labeled in green. Actin was labeled by rhodamin phalloidin (red) and the nucleus was by Hoescht 33342 (blue). The arrows point to the focal adhesion localization of SPT1 at the tips of actin stress fibers. (C) Co-localization of SPT1 and vinculin in HEK293 cells. Mouse anti-vinculin antibody was used as a primary labeling followed by Alexa Flour 568 anti-mouse antibody (red). The images were taken by Zeiss confocal microscope. Bar, 10 µm.
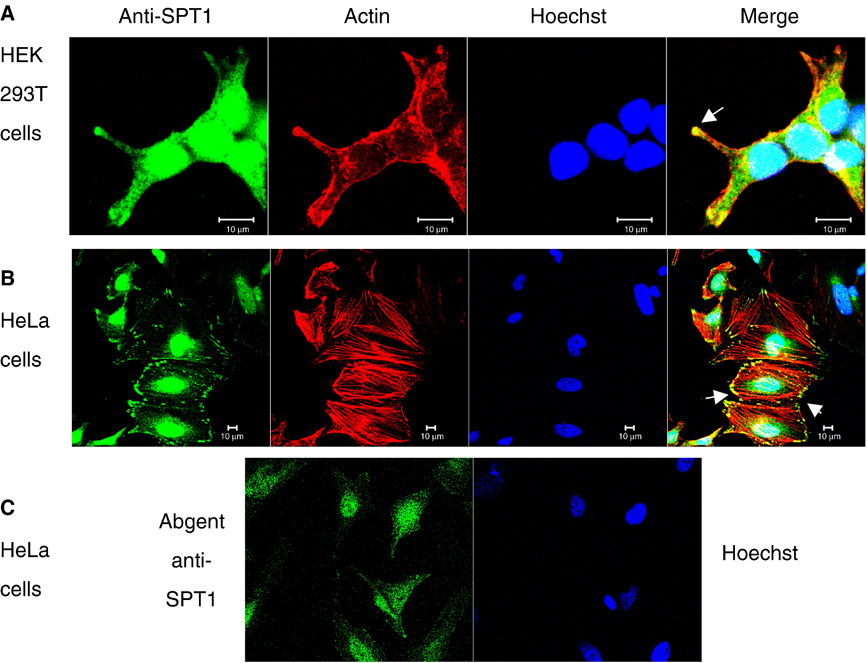
Nuclear and focal adhesion staining were also seen using this anti-SPT1 antibody to examine HEK293T cells (Figure 2A) and HeLa cells (Figure 2B). We have also seen nuclear staining with numerous other anti-SPT1 antibodies (as shown for one example in Figure 2C); however, none of the other antibodies have visualized the focal adhesion staining. This might indicate that the focal adhesion staining is an artifact, however, neither it nor the nuclear staining were seen with pre-immune serum (as shown in supplemental Fig. 1A) in agreement with the original characterization of these antibodies [15, 16]. Furthermore, the majority of the immunostaining was eliminated by pre-incubating the antibody with the peptides that were used for the immunization (Supplemental Fig. 1B).
Fig. 2.
SPT1 staining in HEK293T cells and HeLa cells. Confocal image of SPT1 in HEK293T cells (A) and HeLa cells (B) by rabbit anti-SPT1 antibody labeled by Alexa Fluor 488 anti-rabbit antibody (green), and (C) for HeLa cells using another rabbit anti-SPT1 antibody (catalog # AP2534b from Abgent, San Diego, CA). Actin was labeled by rhodamine phalloidin (red) and the nucleus was by Hoescht 33342 (blue).
These observations were consistent with the absence of these polypeptides in any other proteins (including SPT2 and SPT3, which have substantial homology with SPT1), as determined by protein blast analysis. Nonetheless, while the localization of SPT1 in the nucleus appears conclusive since it is seen with antibodies to several SPT1 epitopes, it is of concern that the only evidence for the presence of SPT1 in focal adhesions has been obtained using a single antibody.
3.2 Co-immunoprecipitation of SPT1 and vinculin in HEK293 cells
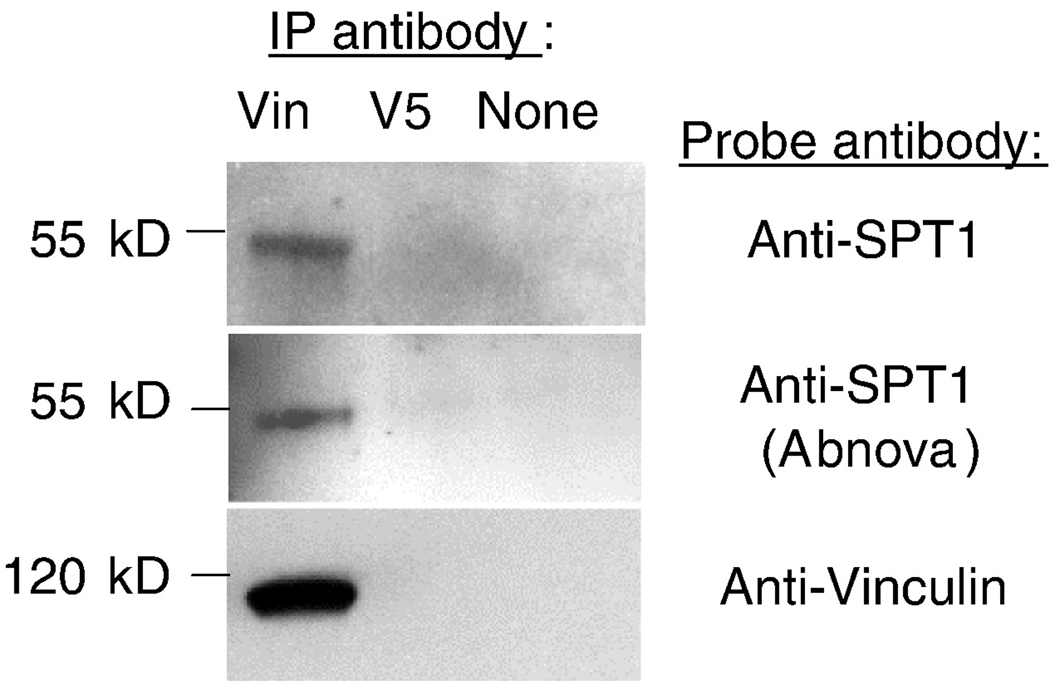
A method that is often used to identify proteins that are associated with focal adhesions is to determine whether they are co-immunoprecipitated by antibodies to the focal adhesion protein vinculin [27, 28]. Shown in Figure 3 are the results of treatment of HEK293 cell lysates with an antibodies to vinculin (Vin) or an unrelated epitope (V5) or no antibody (the latter two as negative controls), followed by protein G sepharose beads and Western blotting with the anti-SPT1 antibody (upper image), an anti-SPT1 antibody from a commercial source (Abnova) (middle), or an anti-vinculin antibody (lower). It is evident from this analysis that SPT1 was only precipitated from the lysates by the anti-vinculin antibody, and as importantly, that the SPT1 in the immunoprecipitate was detectable on the Western blot by both the anti-SPT1 antibody that visualizes SPT1 in focal adhesions (upper image) and the unrelated anti-SPT1 antibody from a commercial source (Abnova) (middle). These results confirm the association of SPT1 with focal adhesions and suggest that the inability of some antibodies to detect this association in fluorescence microscopy may be due to masking of the epitope in the complex.
Fig. 3.
Coimmunoprecipitation of SPT1 by an anti-vinculin antibody. A HEK293 cell lysate was treated with a mouse anti-vinculin antibody (left lane), an antibody against a protein not found in mammalian cells (mouse anti-V5) (middle lane), or no antibody (right lane) followed by recovery of the capture antibodies using protein G sepharose beads, then analysis of the precipitates by Western blotting (right lane). The immunoprecipitates were applied for SDS-PAGE and immunoblotted with the rabbit anti-SPT1 antibody used for the confocal imaging of SPT1 with focal adhesions (top panel), a commercial rabbit SPT1 antibody (Abnova, middle panel) and an anti-vinculin antibody (bottom panel).
3.3 Confirmation of the novel localization of SPT1 by siRNA silencing of the expression of SPT1
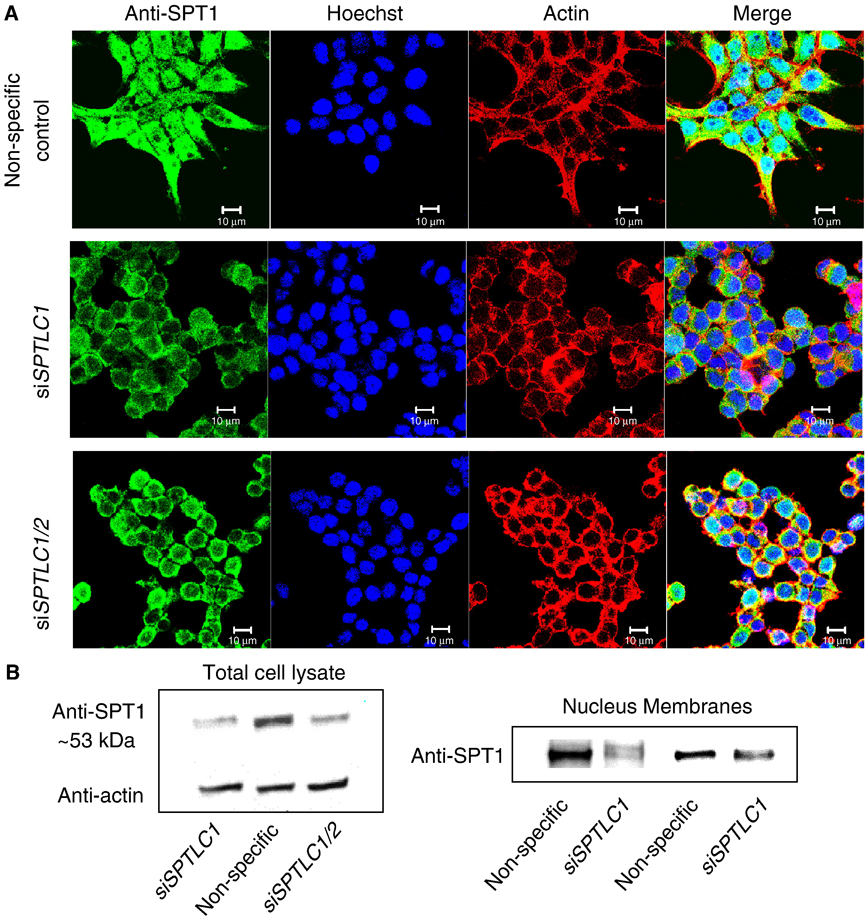
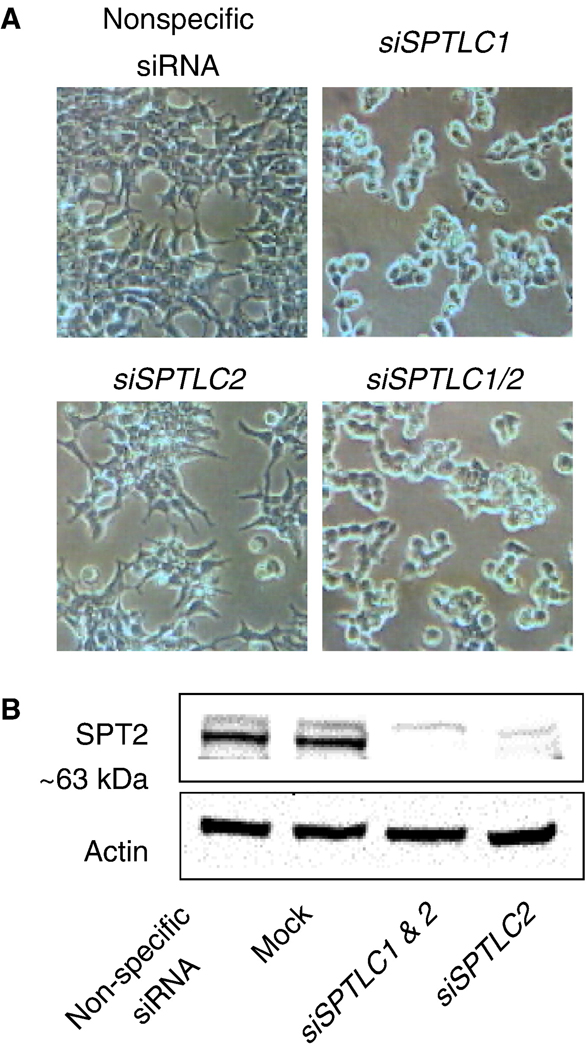
As shown in Figure 4, when HEK293T cells were transfected with siRNA against SPTLC1 or a combination of siSPTLC1 and siSPTLC2 (i.e., against both subunits of SPT), then immunostained with the anti-SPT1 antibody, there was a substantial decrease in the overall fluorescence of the cells (middle and lower panels of Fig. 4A) versus cells treated with a non-specific siRNA control (top panel of Fig. 4A). Indeed, fluorescence in the nucleus and focal adhesions appeared to be suppressed the most by siSPTLC1 and siSPTLC1/siSPTLC2; however, the SPT specific siRNAs caused cell rounding, which obscures some of the decrease in SPT1 in the cytoplasm/ER. SPTLC1 siRNA also caused loss of vinculin-staining in focal adhesions (appearing only as faint, diffuse fluorescence in the cytoplasm) (not shown), which is also suggestive that SPT1 plays a role in the organization or stability of focal adhesions in these cells.
Fig. 4.
Confocal imaging of SPT1 in HEK293T cells transfected with siRNA to silence SPTLC1. (A) HEK293T cells grown on collagen coated coverslips were transfected with non-specific siRNA, siSPTLC1 and siSPTLC1/siSPTLC2 separately for 72 h and then fixed for immunofluorescent staining using anti-SPT1 antibody and Alexa Fluor 488 anti-rabbit antibody (green) as well as for actin fibers (phalloidin) and nuclei (Hoechst 3342) (Bar, 10 µm); or (B) analyzed by western blotting for SPT1 normalized by actin. The right immunoblot is for the nuclear and membrane fractions isolated from HEK293T cells transfected with either non-specific siRNA or siSPTLC1 as described in experimental procedures.
Western blotting of total cell lysates confirmed that the amount of ~53 kDa SPT1 protein decreased significantly in the siSPTLC1 and siSPTLC1/siSPTLC2 transfected cells compared with the non-specific siRNA transfected cells (Fig. 4B left) (this was also confirmed using another SPT1 antibody, as shown in supplemental Figure. 2). Decreases were also seen when the Western blotting was conducted using crude nuclei and membranes (Fig. 4B right), with the nuclear immunostaining appearing to be depleted the most, which is similar to what is seen by confocal microscopy (c.f. middle panels of Fig. 4A). Thus, these observations confirm that the SPT1 immunostaining in focal adhesions and the nucleus is due to SPT1 in those regions of the cell.
3.4 SPT1 suppression using siSPTLC1, but not SPTLC2 silencing, induces cell rounding followed by detachment
Suppression of SPT1 by siRNA for SPTLC1 alone, or the combination of siRNAs for SPTLC1 and SPTLC2, causes cell rounding as observed by confocal (Figure 4) or light microscopy (Figure 5A). Since it is not clear whether the changes in morphology are due to loss of SPT1 per se or to its indirect effect on SPT2, which is known to be bound and stabilized by SPT1 [29], the effect of silencing SPT2 alone was evaluated. When HEK293T cells were transfected with siSPTLC2 alone, there was no noticeable cell rounding (Figure 5A), despite the almost complete disappearance of the ~63 kDa SPT2 polypeptide (Figure 5B). Therefore, suppression of SPT1 but not SPT2 has a much greater impact on cell morphology.
Fig. 5.
Morphology of HEK293T cells after transfection with control and SPTLC siRNAs. (A) Phase contrast images of HEK293T cells transfected with non-specific siRNA, siSPTLC1, siSPTLC2 and siSPTLC1/siSPTLC2 separately. (B) The western blots that show that SPT2 protein has been suppressed by siSPTLC2 alone and in combination with siSPTLC1, with normalization using actin.
In addition to causing cell rounding, siSPTLC1 and the combination of siSPTLC1 & 2 caused a noticeable increase in the number of cells that had detached from the dish after ~48 h (data not shown). This did not appear to be due to cell death because the floating cells were ~70% viable based on the Trypan blue exclusion assay; however, when the floating cells were collected and replated in new medium minus the siRNA(s), they did not survive. These observations suggest that the cell rounding upon suppression of SPT1 facilitates cell detachment and death, presumably due to anoikis after loss of cell-matrix interactions [30].
3.5 The change in cell morphology induced by siSPTLC1 is not due to reduction of the sphingolipid amounts of the cells
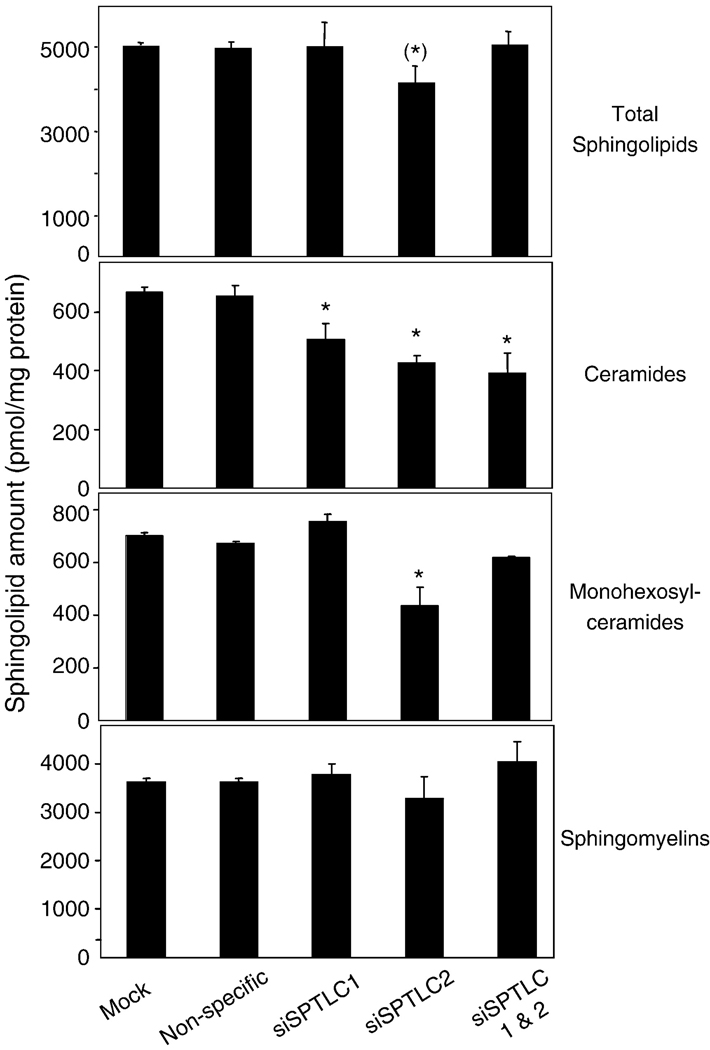
To determine how siRNAs for SPT1 and 2 affect the sphingolipid content of the cells, HEK293T cells were treated with transfection reagent alone, non-specific siRNA or siSPTLC1 and/or 2 then analyzed by liquid chromatography-electrospray tandem mass spectrometry (LCESI MS/MS) (Fig. 6). Compared to the non-specific siRNA, siSPTLC1 did not reduce the amount of total sphingolipids (defined as the major species sphingomyelin + monohexosylceramide + ceramide), but the total sphingolipid content of the siSPTLC2 treated cells was somewhat lower (~17%, P = 0.06). Ceramides were lower for all of the cells treated with for siSPTLC1 (13%), siSPTLC2 (35%) or both (40%), as might be anticipated to occur upon suppression of de novo biosynthesis of ceramide. These findings are consistent with previous findings with LY-B cells that when de novo biosynthesis is eliminated, cells can still survive when supplied medium containing serum sphingolipids [29].1
Fig. 6.
Amounts of ceramide, monohesoxylceramides and sphingomyelins in HEK293T cells after transfection with control vectors or siSPTLC1 and/or siSPTLC2. HEK293T cells were transfected with non-specific siRNA, siSPTLC1, siSPTLC2 and siSPTLC1/SPTLC2 separately for 72 h and then collected for lipid extraction and analysis by LC ESI MS/MS. The amounts are given as the mean ± SD for analysis of 2 dishes.
Thus, since siSPTLC2 caused a greater change in the sphingolipid content of the cells than siSPTLC1 alone, which is the opposite of the effects of these siRNAs on cell morphology, it is unlikely that the cell rounding is induced by changes in sphingolipid metabolism versus a reduction in SPT1 per se. Consistent with this conclusion, treatment of HEK293 cells with myriocin, a potent inhibitor of SPT that essentially eliminated de novo sphingolipid biosynthesis, did not change cell morphology for up to 48 h (data not shown).
3.6 Appearance of SPT1 in focal adhesions decreases as cells in culture reach confluence and increases as cells refill the gap induced by the scratch/wound procedure
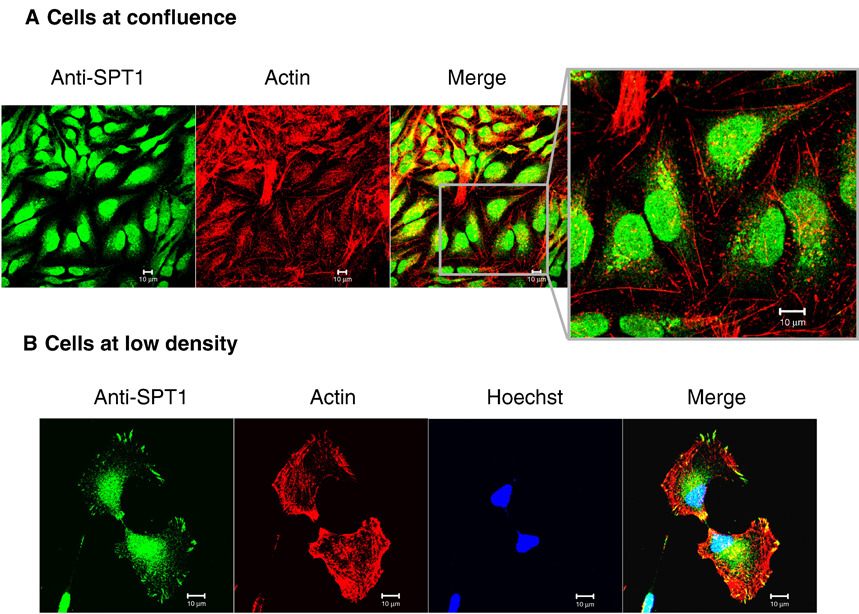
As illustrated in Figure 7A, the focal adhesion staining of SPT1 disappears when the cells grow to confluence (with little difference in the staining of the nucleus and cytoplasm/ER) compared to cells at lower density (Figure 7B). The percentage of cells with focal adhesion staining for SPT1 before confluence (i.e., before the cells are completely surrounded by other cells) was essentially the same for HEK293 cells (95%) and HeLa cells (90%), whereas, neither cell type had any detectable SPT1 with focal adhesions after they were confluent.
Fig. 7.
Focal adhesion staining of SPT1 in low density cells and confluent cells. (A) Confocal fluorescence imaging of SPT1 in confluent HeLa cells. HeLa cells were grown on collagen coated coverslips until confluent and then fixed for immunofluorescent stain with anti-SPT1 antibody followed by Alexa Fluor 488 anti-rabbit antibody (note enlarged image). (B) SPT1 in low density cells. Cells were cultured at low density before applied for immunofluorescence staining. The two cells shown here appear to be at a late stage of mitosis. Actin fibers and nuclei were labeled by rhodamine phalloidin and Hoechst 3342 separately. Bar, 10 µm.
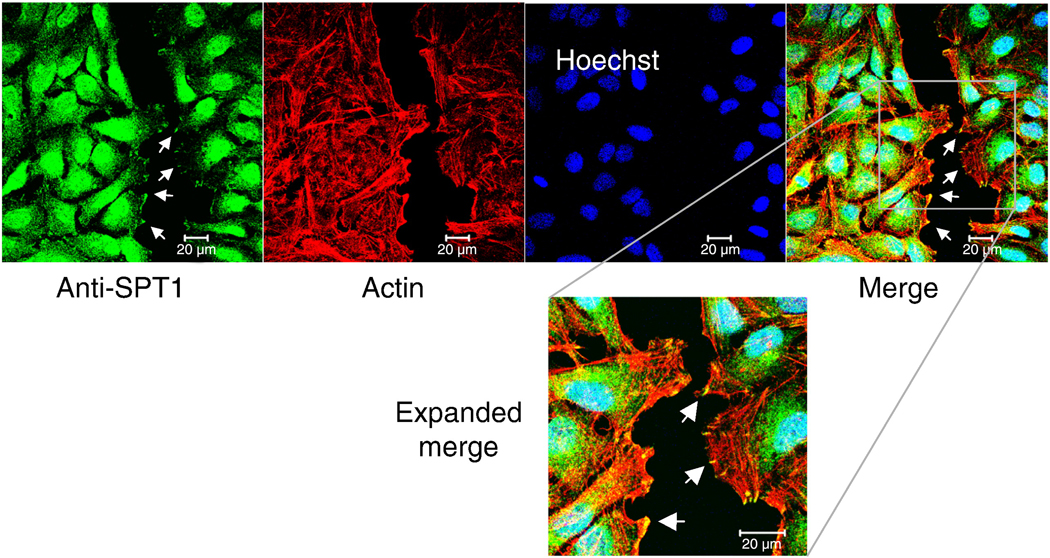
The dynamic of SPT1 localization was also examined using the scratch-wound healing assay to induce cell migration [24]. HeLa cells were grown to confluence on collagen coated glass coverslips, scratched to produce an open furrow using a pipet tip, and then the cells were incubated for 3 h before being fixed and stained. Confocal imaging of the cells (Fig. 8) showed that the focal adhesion staining of SPT1 appears mainly at the tips of actin stress fibers at the edges of the cells that refill the wound gap (Fig. 8), whereas little or none was seen in cell-cell junctions away from the wound edge, nor in any of the confluent cells away from the site of the scratch.
Fig. 8.
Focal adhesion staining of SPT1 during the scratch wound-healing assay. HeLa cells were grown to confluent on collagen coated glass coverslips, scratched by a pipet tip, and processed for immunofluorescence staining using anti-SPT1 antibody after 3 h. The bottom image is an enlargement of the shown part of the merged image. The arrows point to some of the focal adhesions. Bar, 10 µm.
These images provide compelling evidence that the SPT1 in focal adhesions is associated with, and appears to play a role in, these changes in cell morphology. Expression of SPT1 may not be essential for formation of focal adhesions in all cell types, however, because LY-B cells show no SPT1 immunostaining in focal adhesions, but are able to form these structures (Supplemental Fig. 3A).
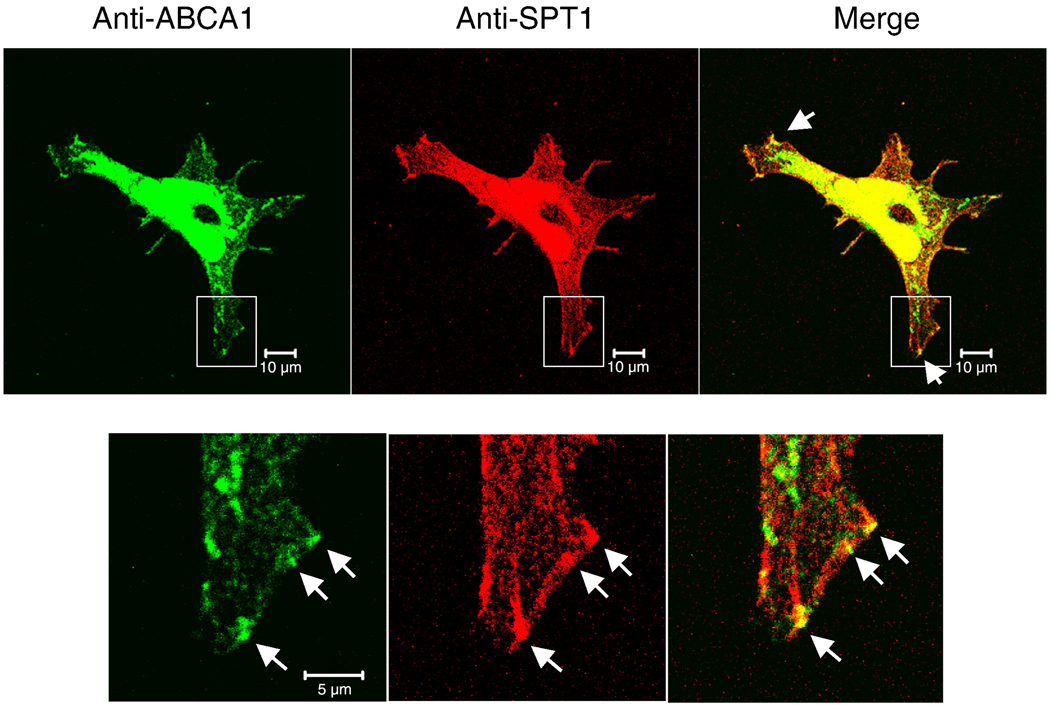
3.7 Partial co-localization of ABCA1 and SPT1 in the cell periphery
It has recently been reported that SPT1 can be found in ABCA1 immunoprecipitates [18]; therefore, the localization of ABCA1 was compared to that of SPT1 in HEK293 cells (Fig. 9). Most of the ABCA1 staining appeared to be associated with intracellular membranes (and in a pattern similar to that for SPT1), and a portion (see expanded panel in the lower half of Fig. 9 and merge) was also detected in focal adhesion-like assemblies in the same vicinity as SPT1. These results are consistent with the proposal that SPT1 and ABCA1 interact [18], although it is not clear if this is direct or perhaps by binding to the same scaffold protein system, as will be discussed later.
Fig. 9.
Localization of SPT1 and ABCA1 in HEK cells. HEK293 cells grown on collagen coated coverslips were stained with rabbit anti-SPT1 antibody and mouse anti-ABCA1 antibody followed by Alexa Fluor 488 anti-rabbit antibody (green) and Alexa Flour 568 anti-mouse antibody (red). The enlarged images show the localization similarity of the two proteins at the leading edge of the cell. Upper panel: bar, 10 µm; lower panel: bar, 5 µm.
3.8 Unlike native SPT1, overexpressed SPT1s do not appear to localize in the nucleus nor focal adhesions
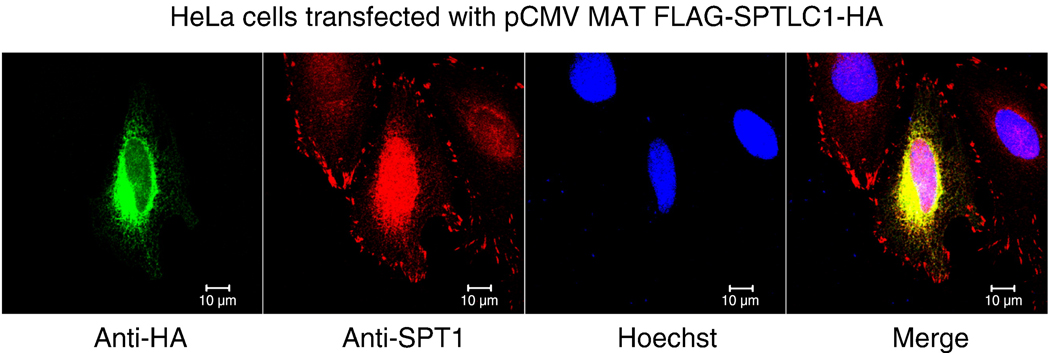
It is puzzling that SPT1 does not appear in focal adhesions and the nucleus in SPT1 overexpressing cells (i.e., only ER staining can be seen clearly in the anti-SPT1 stained cells in Fig. 1A, and a similar observation has been made with other types of SPT1 constructs, see supplemental Fig. 4). This raises the possibility that the ectopic SPT1 polypeptide might inhibit the localization of endogenous SPT1 in focal adhesions and the nucleus. To test this possibility, HeLa cells were transfected with pCMV-MAT-FLAG-SPTLC1-HA and immunostained for the overexpressed SPT1 using a mouse anti-HA antibody and for both the overexpressed and endogenous SPT1 using a rabbit anti-SPT1 antibody (Fig. 10). This analysis showed that the localization of the epitope tagged SPT1 was similar to that seen with the other overexpressed forms (c.f., Fig. 10 with Fig. 1A); however, the anti-SPT1 antibodies in this case detected SPT1 in these locations and in the nucleus and focal adhesions. Therefore, it does not appear that overexpression of a recombinant SPT1 interferes with the nuclear and focal adhesion (and presumably also ER) localization of endogenous SPT1.
Fig. 10.
Localization of SPT1 in SPT1-overexpressing HeLa cells. HeLa cells were transfected with pCMV MAT FLAG-SPTLC1-HA and then stained by mouse anti-HA and rabbit anti-SPT1 antibody followed by anti-mouse Alexa Flour 488 (green) and anti-rabbit Alexa Flour 568 (red). Bar, 10 µm.
4. Discussion
Serine palmitoyltransferase has been long thought to reside in the ER where de novo sphingolipid biosynthesis is initiated [31], and these studies have confirmed that at least a portion of the endogenous SPT1, and essentially all of the ectopically expressed SPT1s, have this localization. The more novel contributions of these studies are: i) the establishment that a portion of the endogenous SPT1 is also located in the nucleus and focal adhesions, as had been suggested but not proven earlier [15–17]; and ii) the demonstration that the localization in focal adhesions is related to, and apparently necessary for, maintenance of a normal cell morphology.
The structural basis for the surprising localization of SPT1 with the nucleus and focal adhesions is not known, however, SPT1 is predicted to have multiple PDZ binding motifs (e.g., a Class II motif at AVLL470–473, and four Class III domains binding motifs at VEMV11–14, KEEL57–60, EELI58–61 and PEPL66–69) using the ELM (Eukaryotic Linear Motif) functional site prediction tool (http://elm.eu.org/) [32] and the UniProtKB/Swiss-Prot identifier for human SPTLC1: O15269. These motifs might interact with PDZ-containing scaffold proteins in the nucleus and plasma membrane because PDZ-containing scaffold proteins are known to assemble multiprotein complexes in such locations to perform specialized local functions such as organization of subcellular structures and signal transduction [33, 34].
Additional evidence for the possibility of such interactions has been provided by the recent report [18] that SPT1 is associated with ABCA1, and our confirmation that at least a portion of both are in the vicinity of focal adhesions (Fig. 9). ABCA1 is thought to interact with β-syntrophin/utrophin complexes through syntrophin PDZ domains [35, 36], which could also account for the SPT1 (and ABCA1) in focal adhesions via the binding of utrophin to the actin cytoskeleton and the dystroglycan complex in the plasma membrane [37, 38]. The reported interaction of ABCA1 and SPT1 [18] might, therefore, be indirect and mediated by a mutual interaction with β-syntrophin or another protein with multiple PDZ domains.
The physiologic significance of this interaction is not known, but might provide a mechanism to link sphingolipid metabolism and S1P delivery and/or removal from the cell because ABCA1 is the major receptor for HDL [39], the major plasma carriers of S1P [40]. S1P can inhibit de novo sphingolipid biosynthesis [41], therefore, perhaps the SPT1 in these atypical localizations is serving as a “sphingoid base status sensor” rather than as a subunit of the catalytically active SPT1-SPT2 dimer [10] or multimer [12].2 At the very least, the focal adhesion association of SPT1 appears to be involved in cell morphology based in the finding that knockdown of SPT1 mRNA caused cell rounding (Figure 4 and 5), and that the association of SPT1 with focal adhesions disappears when cells grow to confluence and reappears when cells are induced to proliferate and spread to refill gaps made by a scratch-wound healing assay (Figure 8).
SPT1 does not have a noticeable nuclear localization signal, however, its appearance in the nucleus might also be a consequence of interactions with other proteins. Many focal adhesion proteins have been found there, and play a role in signaling from focal adhesion to nucleus [42]. In addition, SPT1 has a transcription cofactor motif (LXXLL) that is found in coregulators that shuttle between nucleus and cytoplasm. Considering that new transcriptional coregulators are continually being discovered and these include factors that were not expected to serve such functions [43] and even nuclear receptors that are down-regulated by binding sphingosine [44], these possibilities should be borne in mind as a potential explanation for the nuclear localization of SPT1.
The finding that all of the SPT1 constructs prepared by us and others [11] produce a recombinant protein that appears only in the ER and not in the nucleus and focal adhesions complicates follow-up investigations into these hypotheses (such as by deletion of the PDZ-binding motifs to determine how this affects localization). There is currently no explanation for this distinction, but it is not unprecedented because neutral sphingomyelinase (nSMase1), for example, has also been found in the nucleus of cells, but the over-expressed enzyme does not appear there [45]. Such behavior might be caused by the existence of additional, endogenous isoform(s), such as splice variants, or perhaps co- or post-translational modification(s) that is (are) important for protein folding and/or interaction with binding partners. These are important questions for future study.
Supplementary Material
Acknowledgement
This work is supported by National Institutes of Health grants GM076217 and ES09204.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
To confirm that cells cultured in serum-containing medium can have substantial amounts of sphingolipids despite suppression of SPT1, we have obtained LY-B and LY-B/LCB1 (i.e., LY-B cells stably transfected with SPT1) from Dr. Hanada and measured the SM content by LC ESI-MS/MS. We found that LY-B and LYB-LCB1 cells grown in serum-containing medium have ~3000 and 4000 pmol SM/mg protein, respectively. Therefore, since LY-B cells—which have absolutely no de novo sphingolipid biosynthesis, have 75% of the SM level of the LY-B-LCB1 cells, it is not surprising that our siRNA knock-down in Hek cells did not decrease SM noticably.
Thus far, we have not been able to detect SPT2 in the focal adhesion structures (data not shown), however, that might be due to limitation of the antibodies that are currently available, and should be examined further.
References
- 1.Alvarez SE, Milstien S, Spiegel S. Autocrine and paracrine roles of sphingosine-1-phosphate. Trends Endocrinol. Metab. 2007;18:300–307. doi: 10.1016/j.tem.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 2.Hannun YA, Obeid LM. Principles of bioactive lipid signalling: lessons from sphingolipids. Nat. Rev. 2008;9:139–150. doi: 10.1038/nrm2329. [DOI] [PubMed] [Google Scholar]
- 3.Merrill AH, Jr, Wang MD, Park M, Sullards MC. (Glyco)sphingolipidology: an amazing challenge and opportunity for systems biology. Trends Biochem. Sci. 2007;32:457–468. doi: 10.1016/j.tibs.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 4.Spiegel S, Milstien S. Sphingosine-1-phosphate: an enigmatic signalling lipid. Nat. Rev. 2003;4:397–407. doi: 10.1038/nrm1103. [DOI] [PubMed] [Google Scholar]
- 5.Todeschini AR, Dos Santos JN, Handa K, Hakomori SI. Ganglioside GM2/GM3 complex affixed on silica nanospheres strongly inhibits cell motility through CD82/cMet-mediated pathway. Proc. Natl. Acad. Sci. U S A. 2008;105:1925–1930. doi: 10.1073/pnas.0709619104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chalfant CE, Spiegel S. Sphingosine 1-phosphate and ceramide 1-phosphate: expanding roles in cell signaling. J. Cell Sci. 2005;118:4605–4612. doi: 10.1242/jcs.02637. [DOI] [PubMed] [Google Scholar]
- 7.Seufferlein T, Rozengurt E. Sphingosine induces p125FAK and paxillin tyrosine phosphorylation, actin stress fiber formation, and focal contact assembly in Swiss 3T3 cells. J. Bio. Chem. 1994;269:27610–27617. [PubMed] [Google Scholar]
- 8.Wang F, Nobes CD, Hall A, Spiegel S. Sphingosine 1-phosphate stimulates rho-mediated tyrosine phosphorylation of focal adhesion kinase and paxillin in Swiss 3T3 fibroblasts. J. Bio. Chem. 1997;324(Pt 2):481–488. doi: 10.1042/bj3240481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Formigli L, Meacci E, Sassoli C, Chellini F, Giannini R, Quercioli F, Tiribilli B, Squecco R, Bruni P, Francini F, Zecchi-Orlandini S. Sphingosine 1-phosphate induces cytoskeletal reorganization in C2C12 myoblasts: physiological relevance for stress fibres in the modulation of ion current through stretch-activated channels. J. Cell Sci. 2005;118:1161–1171. doi: 10.1242/jcs.01695. [DOI] [PubMed] [Google Scholar]
- 10.Hanada K, Hara T, Nishijima M. Purification of the serine palmitoyltransferase complex responsible for sphingoid base synthesis by using affinity peptide chromatography techniques. J. Bio. Chem. 2000;275:8409–8415. doi: 10.1074/jbc.275.12.8409. [DOI] [PubMed] [Google Scholar]
- 11.Yasuda S, Nishijima M, Hanada K. Localization, topology, and function of the LCB1 subunit of serine palmitoyltransferase in mammalian cells. J. Bio. Chem. 2003;278:4176–4183. doi: 10.1074/jbc.M209602200. [DOI] [PubMed] [Google Scholar]
- 12.Hornemann T, Richard S, Rutti MF, Wei Y, von Eckardstein A. Cloning and initial characterization of a new subunit for mammalian serine-palmitoyltransferase. J. Biol. Chem. 2006;281:37275–37281. doi: 10.1074/jbc.M608066200. [DOI] [PubMed] [Google Scholar]
- 13.Mandon EC, Ehses I, Rother J, van Echten G, Sandhoff K. Subcellular localization and membrane topology of serine palmitoyltransferase, 3-dehydrosphinganine reductase, and sphinganine N-acyltransferase in mouse liver. J. Bio. Chem. 1992;267:11144–11148. [PubMed] [Google Scholar]
- 14.Williams RD, Wang E, Merrill AH., Jr Enzymology of long-chain base synthesis by liver: characterization of serine palmitoyltransferase in rat liver microsomes. Arch. Biochem. Biophys. 1984;228:282–291. doi: 10.1016/0003-9861(84)90069-9. [DOI] [PubMed] [Google Scholar]
- 15.Batheja AD, Uhlinger DJ, Carton JM, Ho G, D'Andrea MR. Characterization of serine palmitoyltransferase in normal human tissues. J. Histochem. Cytochem. 2003;51:687–696. doi: 10.1177/002215540305100514. [DOI] [PubMed] [Google Scholar]
- 16.Carton JM, Uhlinger DJ, Batheja AD, Derian C, Ho G, Argenteri D, D'Andrea MR. Enhanced serine palmitoyltransferase expression in proliferating fibroblasts, transformed cell lines, and human tumors. J. Histochem. Cytochem. 2003;51:715–726. doi: 10.1177/002215540305100603. [DOI] [PubMed] [Google Scholar]
- 17.Yerokun T, Stewart J. Novel functional association of serine palmitoyltransferase subunit 1-A peptide in sphingolipid metabolism with cytochrome P4501A1 transactivation and proliferative capacity of the human Glioma LN18 brain tumor cell line. Int. J. Environ. Res. Public. Health. 2006;3:252–261. doi: 10.3390/ijerph2006030030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tamehiro N, Zhou S, Okuhira K, Benita Y, Brown CE, Zhuang DZ, Latz E, Hornemann T, von Eckardstein A, Xavier RJ, Freeman MW, Fitzgerald ML. SPTLC1 binds ABCA1 to negatively regulate trafficking and cholesterol efflux activity of the transporter. Biochemistry. 2008;47:6138–6147. doi: 10.1021/bi800182t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bonifacino JS, Dell'Angelica EC. Immunoprecipitation, Chapter 7. Curr Protoc Cell Biol. 2001 doi: 10.1002/0471143030.cb0702s00. Unit 7 2. [DOI] [PubMed] [Google Scholar]
- 20.Lacoste J, Ma A, Parsons JT. Assay and purification and focal adhesion kinase. Methods Enzymol. 1998;298:89–102. doi: 10.1016/s0076-6879(98)98011-9. [DOI] [PubMed] [Google Scholar]
- 21.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andrin C, Hendzel MJ. F-actin-dependent insolubility of chromatin-modifying components. J. Bio. Chem. 2004;279:25017–25023. doi: 10.1074/jbc.M401805200. [DOI] [PubMed] [Google Scholar]
- 23.Sullards MC, Allegood JC, Kelly S, Wang E, Haynes CA, Park H, Chen Y, Merrill AH., Jr Structure-specific, quantitative methods for analysis of sphingolipids by liquid chromatography-tandem mass spectrometry: "inside-out" sphingolipidomics. Methods Enzymol. 2007;432:83–115. doi: 10.1016/S0076-6879(07)32004-1. [DOI] [PubMed] [Google Scholar]
- 24.Lampugnani MG. Cell migration into a wounded area in vitro. Methods Mol. Biol. 1999;96:177–182. doi: 10.1385/1-59259-258-9:177. [DOI] [PubMed] [Google Scholar]
- 25.Lee AS. The ER chaperone and signaling regulator GRP78/BiP as a monitor of endoplasmic reticulum stress. Methods. 2005;35:373–381. doi: 10.1016/j.ymeth.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 26.Pellegrin S, Mellor H. Actin stress fibres. J. Cell Sci. 2007;120:3491–3499. doi: 10.1242/jcs.018473. [DOI] [PubMed] [Google Scholar]
- 27.de Hoog CL, Foster LJ, Mann M. RNA and RNA binding proteins participate in early stages of cell spreading through spreading initiation centers. Cell. 2004;117:649–662. doi: 10.1016/s0092-8674(04)00456-8. [DOI] [PubMed] [Google Scholar]
- 28.Ziegler WH, Liddington RC, Critchley DR. The structure and regulation of vinculin. Trends Cell Biol. 2006;16:453–460. doi: 10.1016/j.tcb.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 29.Hanada K, Hara T, Fukasawa M, Yamaji A, Umeda M, Nishijima M. Mammalian cell mutants resistant to a sphingomyelin-directed cytolysin. Genetic and biochemical evidence for complex formation of the LCB1 protein with the LCB2 protein for serine palmitoyltransferase. J. Biol. Chem. 1998;273:33787–33794. doi: 10.1074/jbc.273.50.33787. [DOI] [PubMed] [Google Scholar]
- 30.Frisch SM, Screaton RA. Anoikis mechanisms. Curr. Opin. Cell Biol. 2001;13:555–562. doi: 10.1016/s0955-0674(00)00251-9. [DOI] [PubMed] [Google Scholar]
- 31.Futerman AH, Riezman H. The ins and outs of sphingolipid synthesis. Trends Cell Biol. 2005;15:312–318. doi: 10.1016/j.tcb.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 32.Puntervoll P, Linding R, Gemund C, Chabanis-Davidson S, Mattingsdal M, Cameron S, Martin DM, Ausiello G, Brannetti B, Costantini A, Ferre F, Maselli V, Via A, Cesareni G, Diella F, Superti-Furga G, Wyrwicz L, Ramu C, McGuigan C, Gudavalli R, Letunic I, Bork P, Rychlewski L, Kuster B, Helmer-Citterich M, Hunter WN, Aasland R, Gibson TJ. ELM server: A new resource for investigating short functional sites in modular eukaryotic proteins. Nucleic Acids Res. 2003;31:3625–3630. doi: 10.1093/nar/gkg545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sierralta J, Mendoza C. PDZ-containing proteins: alternative splicing as a source of functional diversity. Brain Res. Brain Res. Rev. 2004;47:105–115. doi: 10.1016/j.brainresrev.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 34.Kim E, Sheng M. PDZ domain proteins of synapses. Nat. Rev. Neurosci. 2004;5:771–781. doi: 10.1038/nrn1517. [DOI] [PubMed] [Google Scholar]
- 35.Okuhira K, Fitzgerald ML, Sarracino DA, Manning JJ, Bell SA, Goss JL, Freeman MW. Purification of ATP-binding cassette transporter A1 and associated binding proteins reveals the importance of beta1-syntrophin in cholesterol efflux. J. Biol. Chem. 2005;280:39653–39664. doi: 10.1074/jbc.M510187200. [DOI] [PubMed] [Google Scholar]
- 36.Buechler C, Boettcher A, Bared SM, Probst MC, Schmitz G. The carboxyterminus of the ATP-binding cassette transporter A1 interacts with a beta2-syntrophin/utrophin complex. Biochem. Biophys. Res. Commun. 2002;293:759–765. doi: 10.1016/S0006-291X(02)00303-0. [DOI] [PubMed] [Google Scholar]
- 37.Keep NH. Structural comparison of actin binding in utrophin and dystrophin. Neurol. Sci. 2000;21:S929–S937. doi: 10.1007/s100720070006. [DOI] [PubMed] [Google Scholar]
- 38.Haenggi T, Fritschy JM. Role of dystrophin and utrophin for assembly and function of the dystrophin glycoprotein complex in non-muscle tissue. Cell Mol. Life Sci. 2006;63:1614–1631. doi: 10.1007/s00018-005-5461-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boadu E, Bilbey NJ, Francis GA. Cellular cholesterol substrate pools for adenosine-triphosphate cassette transporter A1-dependent high-density lipoprotein formation. Curr. Opin. Lipidol. 2008;19:270–276. doi: 10.1097/MOL.0b013e3282feea99. [DOI] [PubMed] [Google Scholar]
- 40.Murata N, Sato K, Kon J, Tomura H, Yanagita M, Kuwabara A, Ui M, Okajima F. Interaction of sphingosine 1-phosphate with plasma components, including lipoproteins, regulates the lipid receptor-mediated actions. Biochem. J. 2000;352(Pt 3):809–815. [PMC free article] [PubMed] [Google Scholar]
- 41.van Echten-Deckert G, Zschoche A, Bar T, Schmidt RR, Raths A, Heinemann T, Sandhoff K. cis-4-Methylsphingosine decreases sphingolipid biosynthesis by specifically interfering with serine palmitoyltransferase activity in primary cultured neurons. J. Biol. Chem. 1997;272:15825–15833. doi: 10.1074/jbc.272.25.15825. [DOI] [PubMed] [Google Scholar]
- 42.Kadrmas JL, Beckerle MC. The LIM domain: from the cytoskeleton to the nucleus. Nat. Rev. 2004;5:920–931. doi: 10.1038/nrm1499. [DOI] [PubMed] [Google Scholar]
- 43.Hermanson O, Glass CK, Rosenfeld MG. Nuclear receptor coregulators: multiple modes of modification. Trends Endocrinol. Metab. 2002;13:55–60. doi: 10.1016/s1043-2760(01)00527-6. [DOI] [PubMed] [Google Scholar]
- 44.Urs AN, Dammer E, Kelly S, Wang E, Merrill AH, Jr, Sewer MB. Steroidogenic factor-1 is a sphingolipid binding protein. Mol. Cell Endocrinol. 2007;265–266:174–178. doi: 10.1016/j.mce.2006.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mizutani Y, Tamiya-Koizumi K, Nakamura N, Kobayashi M, Hirabayashi Y, Yoshida S. Nuclear localization of neutral sphingomyelinase 1: biochemical and immunocytochemical analyses. J. Cell Sci. 2001;114:3727–3736. doi: 10.1242/jcs.114.20.3727. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.