Abstract
Methionyl-tRNA synthetase (MetRS) specifically binds its methionine substrate in an induced fit mechanism, with methionine binding causing large rearrangements. Mutated MetRS able to efficiently aminoacylate the methionine (Met) analog azidonorleucine (Anl) have been identified by saturation mutagenesis combined with in vivo screening procedures. Here, to reveal the structural basis for the altered specificity, the crystal structure of such a mutated MetRS was determined in the apo form as well as complexed with Met or Anl (1.4 to 1.7 Å resolution). The mutations result in both the loss of important contacts with Met, and in the creation of new contacts with Anl, thereby explaining the specificity shift. Surprisingly, the conformation induced by Met binding in wild-type MetRS already occurs in the apo form of the mutant enzyme. Therefore, the mutations cause the enzyme to switch from an induced fit mechanism to a lock and key one, thereby enhancing its catalytic efficiency.
Keywords: aminoacyl-tRNA synthetase, methionine, azidonorleucine, induced fit, X-ray crystallography
INTRODUCTION
Enzymes are highly efficient and specific catalysts that use the substrate binding energy to stabilize the transition state of the reaction 1; 2; 3; 4. There are two main molecular mechanisms that underlie the utilization of substrate binding energy. In the lock and key mechanism, the binding pockets for the substrates exist in full form in the free enzyme. The substrates readily bind and the reaction is driven because the active site has better complementarity to the transition state of the reaction than to the substrates at the ground state. In the induced fit mechanism, complementarity to the transition state is not achieved before substrate binding. Instead, a part of the binding energy is used to distort the enzyme in such a way that complementarity is obtained only after binding. Therefore, although induced fit can provide some advantages in the functioning of an enzyme, this mechanism is intrinsically less efficient since a smaller amount of the substrate binding energy remains available for reaching the transition state 3.
Each aminoacyl-tRNA synthetase (aaRS) catalyzes the attachment of an amino acid to its cognate tRNAs. These enzymes have been thoroughly studied for many years because of their pivotal role in faithful decoding of the genetic message (reviewed in 5; 6; 7; 8). Catalysis by aaRS is a two-step process. The amino acid is first activated into aminoacyl-adenylate in an ATP-dependent manner, and then transferred onto the 3′ end of tRNA. The problem of the discrimination by aaRS of amino acids depends on the considered system. In some cases, the correct substrate is sufficiently different from incorrect ones to be specifically recognized at the level of enzyme binding 9. However, some amino acids such as valine and isoleucine differ only by a methylene group. In these cases, the corresponding aaRS have evolved proofreading mechanisms in order to achieve a sufficient level of discrimination in vivo. Although methionyl-tRNA synthetase (MetRS) displays a high level of specificity for the binding and activation of methionine, it also can activate homocysteine at a significant rate. To avoid erroneous homocysteylation of tRNAMet, MetRS-bound homocysteinyl adenylate is rapidly converted into homocysteine thiolactone by the enzyme 10; 11. E. coli MetRS is a homodimer from which a fully active monomer can be generated upon truncation of nearly 120 C-terminal residues 12; 13; 14. Several X-ray structures of this active monomer, corresponding to the free enzyme as well as to complexes with methionine or methionyl-adenylate analogs, are known 15; 16; 17. These studies have concluded that methionine binds the enzyme in an induced fit mechanism, with a large rearrangement of aromatic residues enabling the protein to form a hydrophobic pocket around the ligand side-chain. Among the key residues involved in Met binding, the main chain NH of L13 as well as with the side chains of Y260 and H301 facilitate the recognition of the sulphur atom of Met 16; 17; 18.
Saturation mutagenesis targeting such important residues coupled with high-throughput screening procedures have led to the isolation of several MetRS mutants able to efficiently charge methionine analogs onto tRNAsMetin vivo 19; 20; 21; 22. We recently reported the optimization of these screening procedures, and their use to isolate MetRS variants with both improved activity towards the Met-analog azidonorleucine (Anl) and higher selectivity against Met 22. The aim of the present work is to decipher the structural basis for remodelling the amino acid specificity of such variants. The high-resolution X-ray structures of a representative variant, MetRS-SLL, carrying Ser at position 13 and Leu at positions 260 and 301, as well as of its complexes with Met or Anl are described. Modification of the amino acid cavity results in both the loss of important contacts with Met, and in the creation of new contacts with Anl, thereby shifting the specificity of the enzyme. Interestingly, we observe that the conformation induced by Met binding in wild-type MetRS occurs in the free form of the mutant enzyme. Therefore, the mutations cause the enzyme to switch from an induced fit to a lock and key mechanism, increasing its catalytic efficiency. This effect is important to render the mutant enzyme usable for in vivo incorporation of non-canonical amino acids into proteins.
RESULTS AND DISCUSSION
Crystallization of MetRS-SLL
Crystallization of the active truncated monomeric forms of E. coli MetRS (M547 or M551; 13) using ammonium citrate as precipitant yields two closely related types of crystals. Most of the crystals form thin plates, belonging to the P21 space group with cell parameters a=78.2 Å, b=46.3 Å, c=87.7 Å, β=109.1°. Beside this major species called type 1, a few percent of rod-shaped crystals can be observed. These type 2 crystals belong to the same P21 space group but are characterized by a slightly smaller c parameter (c=86.3 Å) and a slightly smaller β angle (β=107.9°). Crystallization of M547 or M551 in the presence of methionine yields type 2 crystals, even when crystallization is triggered by microseeding with type 1 crystals (16 and our unpublished observations). MetRS-SLL was crystallized at 6°C in the same conditions as the wild-type M551 enzyme (3 mg/mL protein in 1.08 M ammonium citrate, 30 mM potassium phosphate, pH 7.0) by microseeding with type 1 crystals of the M551 enzyme. Notably, MetRS-SLL crystals, visible after a few days, were all rod-shaped. Their cell parameters (a=78.2 Å, b=45.3 Å, c=86.0 Å, β=107.3°; Table 1) corresponded to those of type 2 crystals.
Table 1.
Crystallographic and refinement data.
| Crystal | MetRS-SLL | MetRS-SLL:Met | MetRS-SLL:Anl | M547 |
|---|---|---|---|---|
| X-ray source | ESRF-id14eh2 | ESRF-id14eh2 | ESRF-id14eh2 | SOLEIL-Proxima1 |
| Data collection wavelength (Å) | 0.933 | 0.933 | 0.933 | 0.980 |
| Space Group | P21 | P21 | P21 | P21 |
| Cell Parameters: a,b,c,(Å); β (°) | 78.20, 45.26, 85.99; 107.28 | 78.17, 45.29, 85.87; 107.25 | 78.26, 45.38, 85.89; 107.24 | 78.89, 45.40, 86.21; 107.30 |
| Unique reflections | 59 541 | 111 291 | 86282 | 109028 |
| Resolution (Å) | 1.7 | 1.4 | 1.5 | 1.4 |
| Completeness (%)/Redundancy | 93.4/2.5 | 98.0/2.8 | 93.1/3.1 | 94.7/3.1 |
| Rsym (I) (%)1 | 2.3 (4.2) | 2.5 (7.5) | 2.6 (9.4) | 5.7 (30.0) |
| R/Free-R2 (%) | 15.9/18.2 | 16.8/18.0 | 16.5/18.8 | 16.5/19.1 |
| rmsd bonds (Å)/angles (°) | 0.0046/1.2 | 0.0043/1.2 | 0.0044/1.2 | 0.006/1.1 |
| Residues in model | 4–547, 983 waters, 2 citrates, 1 Zn | 4–547, 1047 waters, 2 citrates, 1 Zn, 1 Met | 4–547, 1161 waters, 2 citrates, 1 Zn, 1 Anl | 4–547, 1095 waters, 1 citrate, 1 Zn |
| Mean Bvalues enzyme residues/Zn/solvent/ligand | 8.0/6.0/23.9/- | 8.0/5.8/20.7/9.7 | 8.6/6.0/24.8/11.2 | 12.5/26.5/9.8/- |
Values in parentheses are Rsym (I) in the highest shell of resolution.
Rfree was calculated with 6% of the reflections.
Overall structure of the MetRS-SLL enzyme
The structure was solved by rigid body refinement of the M551 enzyme model (PDB 1QQT; 15. After several round of energy minimization and manual rebuilding, the model was refined to 1.7 Å resolution. The structure is similar to that of the wild-type truncated monomer and is composed of four parts. (i) The Rossmann fold domain contains the active site. (ii) The connective polypeptide 1 (CP1) contains a zinc-binding knuckle and is inserted between the two halves of the catalytic domain 23; 24. (iii) The KMSKS domain borders the catalytic crevice, and contains residues critical for catalysis 25; 26. (iv) The C-terminal helix bundle domain is responsible for recognition of the CAU anticodon of tRNAsMet 27; 28; 29; 30.
As compared to the wild-type M551 enzyme, both the zinc-binding module and the anticodon-binding region display a slight rotation toward the active site crevice (Fig. 1). Such changes were already observed in the structure of the M551:Met complex. Accordingly, the MetRS-SLL structure can be superimposed to that of the M551:Met complex (PDB id 1F4L; 16 with an rmsd of 0.22 Å for all Cα atoms, while the rmsd is 0.76 Å upon superimposition of MetRS-SLL to M551.
Figure 1.
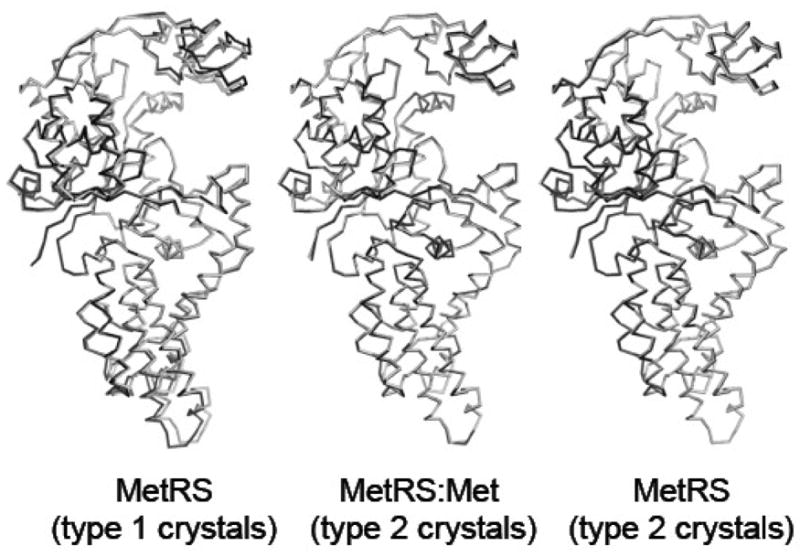
Overall shape of the mutant enzyme. The three views compare the C alpha backbone of MetRS-SLL (grey) to those of unliganded MetRS from type 1 crystals (PDB id 1QQT; black, left view), of the MetRS:met complex (PDB id 1F4L; black, central view) and of unliganded MetRS from type 2 crystals (black, right view). Figures 1, 2 and 3 were drawn using PyMOL (www.pymol.org). In the left view, only residues 4–121 and 189–345 were used for the superimposition of the two structures. In the center and right views, all C alpha atoms were superimposed.
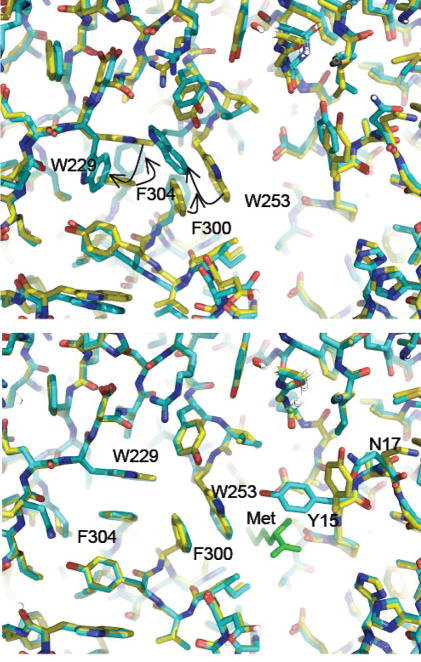
Closer examination of the active site of MetRS-SLL shows important rearrangements of aromatic residues. More precisely, the side chains of W229, W253, F300 and F304 largely rotate, in such a way that W229 stacks on F304 and W253 stacks on F300. Such interactions do not occur in the free wild-type enzyme, where stacking contacts imply Y251 with W253 on the one hand, and F300 with F304 on the other hand (Fig. 2). A notable consequence of the motions of W253 and F300 is to dramatically reduce the size of the active site cavity. Remarkably, these rearrangements exactly match those occurring in the wild-type enzyme upon methionine binding, where the enzyme switches from a “met-off” to a “met-on” conformation 16; Fig. 2). The only exception is the side chain of Y15, which is in a conformation leaving the methionine binding cavity open in the MetRS-SLL and M551 structures, while Y15 is in a conformation that locks methionine in the cavity in the M551:Met structure 17; Fig. 2). As noted above, both MetRS-SLL and M551:Met crystallize to form type 2 crystals. This raises the question of the relationships between the type of the crystals and the crystalline conformation of MetRS. In order to analyze this issue, the structure of the wild-type M547 enzyme within type 2 crystals was determined. On the whole, the same overall shape as that obtained in other type 2 crystals is observed, with the zinc-binding module and the anticodon-binding region slightly rotated toward the active site. However, the conformations of side chains around the methionine-binding crevice clearly match those in the unmutated enzyme in the type 1 crystals, but not those in MetRS-SLL. Therefore, it can be concluded that the observed reorganization of aromatic side chains in MetRS-SLL is a consequence of one or several of the L13S, Y260L and H301L mutations.
Figure 2.
Unliganded MetRS-SLL is in the “met-on” conformation. Top panel: the active site region of unliganded MetRS-SLL (yellow) was superimposed to that of unliganded MetRS (PDB 1QQT; blue). Motions of aromatic residues are indicated by arrows. Bottom panel: same as top panel but unliganded MetRS was replaced by MetRS:Met (PDB 1F4L; blue). The bound methionine is shown in green.
Binding of methionine and of azidonorleucine to MetRS-SLL
MetRS-SLL is known to activate both Anl and 6,6,6-trifluoronorleucine (Tfn) 21; 22. In order to study the binding of these methionine analogues to the MetRS-SLL enzyme, crystallization experiments of the mutant enzyme were carried out in the presence of either 35 mM Anl, 10 mM Tfn or 35 mM methionine. The corresponding structures clearly showed the presence of Anl or of Met in the expected cavity. No density corresponding to Tfn was however visible. Higher concentrations of the unnatural amino acid could not be used because of its limited solubility in the crystallization medium. Binding of Tfn was therefore not further studied.
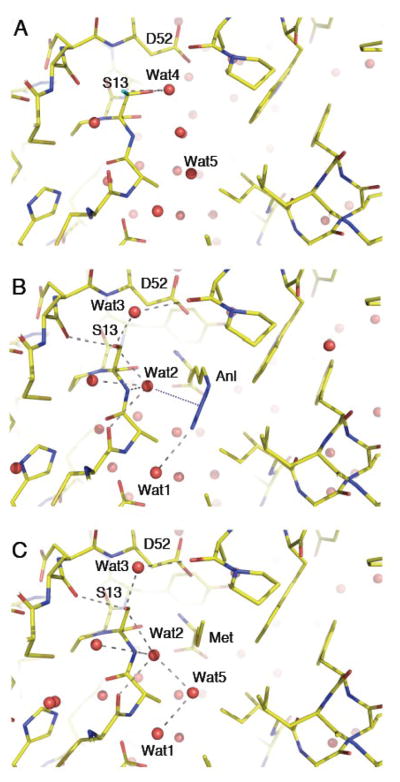
The methionine substrate binds MetRS-SLL exactly at the same site as in unmutated MetRS. However, the three mutations lead to an increase of the size of the cavity on the side of the methyl group, and to the loss of favorable electrostatic contacts of the sulfur atom of bound Met with the OH of Y260 and NεH of H301. Contrasting with the situation in wild-type MetRS, the methionine binding pocket is preformed in MetRS-SLL, in such a way that very few motions occur upon complex formation. The most obvious one concerns Y15, which rotates to lock Met in its cavity like it does in the wild-type MetRS:Met structure. This motion occurs together with those of neighboring residues, in such a way that the C11-N17 peptide is slightly displaced upon methionine binding (Figure 3). Finally, the side chain of Y91 rotates by 30° to make a hydrogen bond with the sulfur atom of C11. Concomitantly, the main chain of I48 is slightly displaced, with its alpha and carbonyl carbons making van der Waals contacts with Y91.
Figure 3.
Binding of azidonorleucine and methionine to MetRS-SLL. The three panels show stick representations of the amino acid binding sites of free or liganded MetRS-SLL, all in the same orientation. Carbon atoms are in yellow, nitrogen atoms in blue and oxygen atoms in red. Water molecules are drawn as red spheres. Relevant hydrogen bonds are indicated by grey dotted lines. Panel A: unliganded MetRS-SLL; the two conformations of the side chain of S13 are drawn, with conformation 1 in standard colours and conformation 2 with the hydroxyl oxygen in green. Panel B: MetRS-SLL:Anl complex. Only the major conformation of S13 is represented. The electrostatic interaction between Wat2 and the N2 atom of Anl is symbolized by a blue dotted line. Panel C: MetRS-SLL:Met complex. Only the major conformation of S13 is represented.
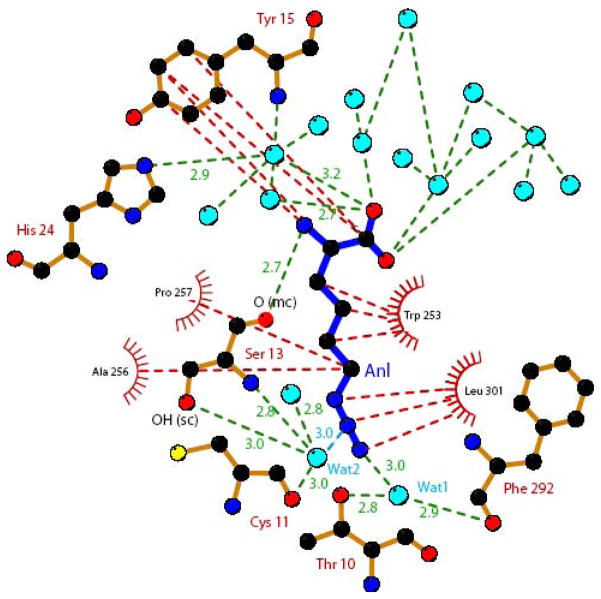
Binding of the non-natural amino acid Anl occurs at the same site as that of Met. For both Met and Anl, the amino group is held by the carboxylate of D52 and by the main chain carbonyl of S13. The carboxylate group of the ligand interacts with 5 water molecules. Enlargement of the cavity caused by the Y260L and H301L mutations allows to accommodate the longer side chain of Anl. Indeed, a Y at position 260 as well as a H at position 301 would cause severe steric clashes with the azido moiety of Anl, thereby precluding its binding. The N3 atom of the azido group of Anl occupies the exact position of a water molecule (Wat5 in Fig. 3C) in the MetRS-SLL unliganded structure. Notably, Wat5 occupies the same position as the OH group of Y260 in the wild-type MetRS. The N3 of Anl interacts with a water molecule (Wat1 in Figs. 3 and 4), also present in the structure of the wild-type enzyme. Wat1 is held by the hydroxyl of T10 and by the carbonyl of F292. Despite the removal of several water molecules from the binding pocket upon Anl binding, Wat2 remains in the vicinity of Anl in the complex (Figs. 3B and 4). Wat2 is tightly held by (i) the hydroxyl and amino groups of S13, (ii) the carbonyl of C11 and (iii) a water molecule. Notably, Wat2 is still present when the active site is occupied by Met (Fig. 3C). This water molecule is likely to enhance the Anl binding affinity by making a favorable electrostatic contact with the N2 nitrogen in the azido group of the ligand (distance 2.95 Å). Furthermore, the presence of Wat2 in the MetRS-SLL:Anl complex may also indirectly contribute to Anl binding by diminishing the number of water molecules to be removed from the active site for the amino acid ligand to bind. In this context, the case of S13 deserves particular attention. In the unliganded structure, the side chain of this residue can adopt two equally populated conformations. In one conformation (conformation 1), the hydroxyl group interacts with a water molecule (Wat4) held by the carbonyl of A256. In the second conformation (conformation 2), the hydroxyl of S13 interacts with the carbonyl of A50 and with Wat2. Upon Anl or Met binding, S13 becomes predominantly restrained to conformation 2, where it tightens the binding of the Wat2 molecule.
Figure 4.
Schematic representation of hydrophobic (red dotted lines) and electrostatic (green dotted lines) interactions made between MetRS-SLL and azidonorleucine. Anl is represented with blue bonds. Carbons are in black, nitrogens in dark blue, oxygens in red and sulfurs in yellow. Water molecules are symbolized as light blue spheres. Relevant hydrogen bonding distances are indicated in Å. The electrostatic interaction between Wat2 and the N2 atom of Anl is indicated by a blue dotted line. The figure was drawn with Ligplot 38.
Role of L13S, Y260L and H301L mutations in modifying the substrate specificity of MetRS
The most remarkable feature of MetRS-SLL is its “ready to bind” conformation. This facilitates the binding of new ligands at the amino acid site, by lowering the part of the binding energy spent for changing the conformation of the enzyme 3. How do the three mutations cause MetRS-SLL to be in the met-on conformation, even in the absence of a bound amino acid? A first reason is that a Leu at position 301 is not compatible with the met-off conformation of F300, which puts the CD2 of F300 at only 2.8 Å away from the CD2 of L301. This might favor its displacement and its subsequent stacking on W253. Second, the H301L and Y260L mutations create a hydrophobic environment favoring the position of W253 corresponding to the met-on conformation. Such stacking of F300 on W253 is likely to trigger the whole met-on conformation. Indeed, previous structural studies of the binding of Met analogs to MetRS led to the idea that motion of W253 is a primary event in the conformational transition 17. Its stacking on F300 would release F304 and open the new position for W229 stacked on F304.
In vivo screening for MetRS variants able to incorporate Anl or Tfn strongly selected Leu at position 301 21; 22. Moreover, the most efficient mutants, both in vivo and in vitro, also carry Leu at position 260. More precisely, in the high stringency screen for mutants charging Anl, more than 80 % of the selected MetRS carry the H301L mutation, the other ones having either H301I or H301V 22. At position 260, all mutants but one harbor a hydrophobic residue, with more than 50% having Leu. According to the above analysis of the structure, it is likely that most, if not all, of the mutants selected for their catalytic efficiency also display the met-on conformation. This observation favors the idea that the met-on conformation, as observed in MetRS-SLL, is important to achieve a high catalytic efficiency with non-methionine substrates.
Notably, L260 and L301 side chains make van der Waals contacts (3.9 to 4 Å) with the three azido nitrogens of Anl. This observation might explain the selection of Y260L and H301L, beyond the importance of the occurrence of hydrophobic residues at this position for the met-on conformation, as discussed above. Concerning mutations at position 13, either small polar residues (Ser, Asn or Cys) or proline lead to the highest efficiencies for Anl activation and to reduced efficiencies for Met activation 22. Actually, the nature of the amino acid at position 13 has a greater effect on the activation of Met than on the activation of Anl 22. The deleterious effect of the proline mutation toward Met binding can easily be explained by the involvement of the main chain NH of L13 in a hydrogen bond with the sulfur of the Met ligand in wild-type MetRS 16; 17. In the structure of the MetRS-SLL mutant, both the side chain and the main chain NH of S13 interact with Wat2. By stabilizing this water molecule, the side chain of S13 is likely to render the NH main chain less available for interacting with the S group of the methionine substrate. This may explain why the L13S mutation disfavors Met binding. Similarly, the different behaviors regarding Met binding of mutants with Asn or Cys at position 13 may also be correlated with the ability of the corresponding side chains to favor masking of the main chain NH of residue 13 21; 22. Finally, the side chain of residue at position 13 also influences the catalytic efficiency of Anl activation 22. A favorable electrostatic contact of the Wat2 molecule with the azido group of Anl (Figs. 3B and 4) may at least partly explain the favorable effect toward Anl binding of small polar side chains at this position. In addition, several water molecules must be displaced to allow Anl binding (Fig. 3). The possibility for the enzyme to tightly bind a water molecule such as Wat2 in the vicinity of the substrate may lower the cost for such desolvation of the amino acid binding cavity. In the case of Pro, favorable van der Waals contacts with the Anl side chain may substitute for the favorable effect of Wat2.
Concluding remarks
This work describes the origins of amino acid substrate recognition by a methionyl-tRNA synthetase mutant with modified specificity. In MetRS-SLL, modification of the binding cavity results in both the loss of important contacts with Met, and the creation of new contacts with Anl, thereby shifting the specificity of the enzyme. With wild-type MetRS, it has been demonstrated that conformational changes induced by the Met ligand are involved in binding the cognate amino acid 16. Since this “met-on” conformation is maintained in the MetRS:methionyl-adenylate complex 17, this conformation is likely to exist during the transition state of the activation reaction also, and consequently play an important role in driving the enzyme toward catalysis. In this context, a striking consequence of the mutations in MetRS-SLL is that the free enzyme adopts the “met-on” conformation. Consequently, less substrate binding energy is spent to distort the enzyme for reaching the transition state of the reaction. Therefore, MetRS-SLL is likely to owe its enlarged substrate specificity to the possibility to more easily reach the transition state, which increases its catalytic efficiency. This illustrates that changing the mode of substrate binding can be used to engineer an enzyme. Finally, the new amino acid substrate of MetRS-SLL studied here, Anl, contains a polar group at the end of its side chain. It should be pointed out that this polar group is bound by a water molecule. This indicates that solvent molecules must carefully be considered in the course of designing engineered active sites for new ligands.
EXPERIMENTAL PROCEDURES
Expression and purification of the MetRS variants
The unmutated monomeric version of E. coli MetRS truncated at residue 548 (M547) was produced from the pBSM547 plasmid and purified as described 18; 29.
The pMTY21-sll plasmid codes for an N-terminally 6XHis-tagged M547 MetRS regulated by an IPTG inductible T5 promoter 21. This plasmid carries a modified MetRS gene leading to expression of the MetRS-SLL mutant (L13S, Y260L, H301L). E. coli JM101Tr cells 31 transformed with pMTY21-sll were grown at 37°C in 1 L of 2xTY medium containing ampicillin (50 μg/mL) and induced with 1 mM IPTG at an OD650 of 0.6. The culture was then continued for 12 h at 37°C.
Purification of MetRS-SLL
A 1-litre culture of cells overproducing MetRS-SLL were harvested, resuspended in 50 mL of buffer A (50 mM NaCl, 10 mM HEPES pH 7.5, 3 mM 2-mercaptoethanol) and disrupted by sonication. After centrifugation, the supernatant was loaded onto a column (5 mL) containing Talon affinity resin (Clontech). The resin was washed with 50 mL of buffer A and then with 50 mL buffer A supplemented with 10 mM imidazole. The protein was eluted with buffer A containing 125 mM imidazole. The pooled sample was then dialyzed against buffer A. Finally, the protein was loaded onto a column (20 mL) containing Q-Hiload resin (Amersham) equilibrated in buffer A and eluted with a 300 mL gradient from 50 to 450 mM NaCl in 10 mM HEPES pH 7.5, 3 mM 2-mercaptoethanol. Fractions containing the protein were pooled, dialyzed against buffer A and concentrated to 6 mg/mL by using a Centricon-30 concentrator (Amicon). Protein concentration was deduced from A280 measurement using an extinction coefficient of 1.72 cm2/mg 32. Approximately 50 mg of protein per litre of culture was obtained.
Crystallization of MetRS-SLL
Crystals of M547 or of MetRS-SLL were obtained in a solution containing 30 mM KPO4, 4 mM 2-mercaptoethanol,1.08 mM ammonium citrate and 3.6 mg/mL of protein by microseeding with crystals of M551 at pH 7.0 15. For ligand binding studies, the amino acid was added to the crystallization medium prior to microseeding (Table 1). Final concentrations during crystallization were 35 mM for methionine, 35 mM for Anl or 10 mM for Tfn. For data collection, crystals were first stabilized in 1.4 M ammonium citrate plus 30 mM potassium phosphate (pH 7.0), quickly soaked in the same solution plus 25 % v/v of glycerol and then flash-cooled in liquid ethane. Data were collected using synchrotron sources at either ESRF (Grenoble, France) or SOLEIL (Gif sur Yvette, France). Diffraction images were analyzed with the XDS program 33 and the data further processed using programs from the CCP4 package 34. Data statistics are summarized in Table 1.
Structure solution and refinement
Each structure was solved by rigid body refinement of the M551 model (PDB id 1QQT), using the CNS program 35. After calculation of initial density maps, the model was manually adjusted using O 36. Each model was refined by cycles of manual adjustment and addition of water molecules followed by conjugate gradient minimization using either CNS 35 or PHENIX 37. Refinement statistics are summarized in Table 1.
Accession numbers
Coordinates and structure factors have been deposited at the Protein Data Bank with IDs 3H97, 3H99, 3H9B and 3H9C.
Acknowledgments
We acknowledge the European Synchrotron Radiation Facility for providing the X-ray facilities on beamline ID14-2 as well as the synchrotron SOLEIL for providing the X-ray facilities on beamline Proxima I. We thank CNRS and Ecole Polytechnique for financial support to the “Unité Mixte de Recherche 7654”. Work at Caltech was supported by NIH Grant GM62523 and by the Army Research Office through the Institute for Collaborative Biotechnologies. Coordinates have been deposited at the PDB with Ids 3H97, 3H99, 3H9B and 3H9C.
Abbreviations
- aaRS
aminoacyl-tRNA synthetase
- MetRS
methionyl-tRNA synthetase
- Anl
Azidonorleucine
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Haldane JBS. Enzymes. Longmans Green; London, UK: 1930. [Google Scholar]
- 2.Pauling L. Molecular architecture and biological reactions. Chem Eng News. 1946;24:1375–1377. [Google Scholar]
- 3.Fersht AR. Structure and mechanism in protein science. W.H. Freeman & Co; San Francisco: 1999. [Google Scholar]
- 4.Blow D. So do we understand how enzymes work? Structure. 2000;8:77–81. doi: 10.1016/s0969-2126(00)00125-8. [DOI] [PubMed] [Google Scholar]
- 5.Mechulam Y, Meinnel T, Blanquet S. A family of RNA-binding enzymes. the aminoacyl-tRNA synthetases. Subcell Biochem. 1995;24:323–76. doi: 10.1007/978-1-4899-1727-0_11. [DOI] [PubMed] [Google Scholar]
- 6.Meinnel T, Mechulam Y, Blanquet S. Aminoacyl-tRNA synthetases: occurrence, structure, and function. In: RajBhandary DSaUL., editor. tRNA: Structure, Biosynthesis, and Function. ASM Press; Washington, DC: 1995. [Google Scholar]
- 7.Ibba M, Soll D. Aminoacyl-tRNA synthesis. Annu Rev Biochem. 2000;69:617–50. doi: 10.1146/annurev.biochem.69.1.617. [DOI] [PubMed] [Google Scholar]
- 8.Schimmel P, Söll D. The world of aminoacyl-tRNA synthetases. In: Ibba M, CFaSC, editors. The aminoacyl-tRNA synthetases. Landes Biosciences; Georgetown, Texas, USA: 2005. [Google Scholar]
- 9.Fersht AR, Dingwall C. Cysteinyl-tRNA synthetase from Escherichia coli does not need an editing mechanism to reject serine and alanine. High binding energy of small groups in specific molecular interactions. Biochemistry. 1979;18:1245–1249. doi: 10.1021/bi00574a020. [DOI] [PubMed] [Google Scholar]
- 10.Fersht AR, Dingwall C. An editing mechanism for the methionyl-tRNA synthetase in the selection of aminoacids in protein synthesis. Biochemistry. 1979;18:1250–1256. doi: 10.1021/bi00574a021. [DOI] [PubMed] [Google Scholar]
- 11.Jakubowski H, Fersht AR. Alternative pathways for editing non-cognate amino acids by aminoacyl-tRNA synthetases. Nucleic Acids Res. 1981;9:3105–3117. doi: 10.1093/nar/9.13.3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cassio D, Waller J. Modification of methionyl-tRNA synthetase by proteolytic cleavage and properties of the trypsin-modified enzyme. Eur J Biochem. 1971;20:283–300. doi: 10.1111/j.1432-1033.1971.tb01393.x. [DOI] [PubMed] [Google Scholar]
- 13.Mellot P, Mechulam Y, Le Corre D, Blanquet S, Fayat G. Identification of an amino acid region supporting specific methionyl- tRNA synthetase: tRNA recognition. J Mol Biol. 1989;208:429–43. doi: 10.1016/0022-2836(89)90507-x. [DOI] [PubMed] [Google Scholar]
- 14.Blanquet S, Crepin T, Mechulam Y, Schmitt E. Class 1 aminoacyl-tRNA synthetases: methionyl-tRNA synthetase. In: Ibba M, Francklin C, Cusack S, editors. Aminoacyl-tRNA synthetase. Landes Bioscience; Georgetown, Texas: 2003. [Google Scholar]
- 15.Mechulam Y, Schmitt E, Maveyraud L, Zelwer C, Nureki O, Yokoyama S, Konno M, Blanquet S. Crystal structure of Escherichia coli methionyl-tRNA synthetase highlights species-specific features. J Mol Biol. 1999;294:1287–97. doi: 10.1006/jmbi.1999.3339. [DOI] [PubMed] [Google Scholar]
- 16.Serre L, Verdon G, Choinowski T, Hervouet N, Risler JL, Zelwer C. How methionyl-tRNA synthetase creates its amino acid recognition pocket upon L-methionine binding. J Mol Biol. 2001;306:863–76. doi: 10.1006/jmbi.2001.4408. [DOI] [PubMed] [Google Scholar]
- 17.Crepin T, Schmitt E, Mechulam Y, Sampson PB, Vaughan MD, Honek JF, Blanquet S. Use of Analogues of Methionine and Methionyl Adenylate to Sample Conformational Changes During Catalysis in Escherichia coli Methionyl-tRNA Synthetase. J Mol Biol. 2003;332:59–72. doi: 10.1016/s0022-2836(03)00917-3. [DOI] [PubMed] [Google Scholar]
- 18.Fourmy D, Mechulam Y, Brunie S, Blanquet S, Fayat G. Identification of residues involved in the binding of methionine by Escherichia coli methionyl-tRNA synthetase. FEBS Lett. 1991;292:259–263. doi: 10.1016/0014-5793(91)80879-8. [DOI] [PubMed] [Google Scholar]
- 19.Kiick KL, Saxon E, Tirrell DA, Bertozzi CR. Incorporation of azides into recombinant proteins for chemoselective modification by the Staudinger ligation. Proc Natl Acad Sci U S A. 2002;99:19–24. doi: 10.1073/pnas.012583299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Link AJ, Vink MK, Agard NJ, Prescher JA, Bertozzi CR, Tirrell DA. Discovery of aminoacyl-tRNA synthetase activity through cell-surface display of noncanonical amino acids. Proc Natl Acad Sci U S A. 2006;103:10180–5. doi: 10.1073/pnas.0601167103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoo TH, Tirrell DA. High-throughput screening for methionyl-tRNA synthetases that enable residue-specific incorporation of noncanonical amino acids into recombinant proteins in bacterial cells. Angew Chem Int Ed Engl. 2007;46:5340–3. doi: 10.1002/anie.200700779. [DOI] [PubMed] [Google Scholar]
- 22.Tanrikulu IC, Schmitt E, Mechulam Y, Goddard W, III, Tirrell DA. Discovery of E. coli methionyl-tRNA synthetase mutants for efficient labeling of proteins with azidonorleucine in vivo. Proc Natl Acad Sci U S A. 2009;106:15285–15290. doi: 10.1073/pnas.0905735106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fourmy D, Dardel F, Blanquet S. Methionyl-tRNA Synthetase Zinc Binding Domain. Three-dimensional Structure and Homology with Rubredoxine and gag Retroviral Proteins. J Mol Biol. 1993;231:1078–1089. doi: 10.1006/jmbi.1993.1353. [DOI] [PubMed] [Google Scholar]
- 24.Fourmy D, Mechulam Y, Blanquet S. Crucial role of an idiosyncratic insertion in the Rossmann fold of class1 aminoacyl-tRNA synthetases: The case of methionyl-tRNA synthetase. Biochemistry. 1995;34:15681–15688. doi: 10.1021/bi00048a012. [DOI] [PubMed] [Google Scholar]
- 25.Mechulam Y, Dardel F, Le Corre D, Blanquet S, Fayat G. Lysine 335, part of the KMSKS signature sequence, plays a crucial role in the amino acid activation catalysed by the methionyl-tRNA synthetase from Escherichia coli. J Mol Biol. 1991;217:465–75. doi: 10.1016/0022-2836(91)90750-z. [DOI] [PubMed] [Google Scholar]
- 26.Schmitt E, Meinnel T, Blanquet S, Mechulam Y. Methionyl-tRNA synthetase needs an intact and mobile 332KMSKS336 motif in catalysis of methionyl adenylate formation. J Mol Biol. 1994;242:566–577. doi: 10.1006/jmbi.1994.1601. [DOI] [PubMed] [Google Scholar]
- 27.Ghosh G, Pelka H, Schulman LH. Identification of the tRNA anticodon recognition site of Escherichia coli methionyl-tRNA synthetase. Biochemistry. 1990;29:2220–2225. doi: 10.1021/bi00461a003. [DOI] [PubMed] [Google Scholar]
- 28.Meinnel T, Mechulam Y, Le Corre D, Panvert M, Blanquet S, Fayat G. Selection of suppressor methionyl-tRNA synthetases: mapping the tRNA anticodon binding site. Proc Natl Acad Sci U S A. 1991;88:291–5. doi: 10.1073/pnas.88.1.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmitt E, Meinnel T, Panvert M, Mechulam Y, Blanquet S. Two acidic residues of Escherichia coli methionyl-tRNA synthetase are negative discriminants towards the binding of non-cognate tRNA anticodons. J Mol Biol. 1993;233:615–628. doi: 10.1006/jmbi.1993.1540. [DOI] [PubMed] [Google Scholar]
- 30.Nakanishi K, Ogiso Y, Nakama T, Fukai S, Nureki O. Structural basis for anticodon recognition by methionyl-tRNA synthetase. Nat Struct Mol Biol. 2005;12:931–2. doi: 10.1038/nsmb988. [DOI] [PubMed] [Google Scholar]
- 31.Hirel PH, Lévêque F, Mellot P, Dardel F, Panvert M, Mechulam Y, Fayat G. Genetic engineering of methionyl-tRNA synthetase: in vitro regeneration of an active synthetase by proteolytic cleavage of a methionyl-tRNA synthetase-β-galactosidase chimeric protein. Biochimie. 1988;70:773–782. doi: 10.1016/0300-9084(88)90107-1. [DOI] [PubMed] [Google Scholar]
- 32.Blanquet S, Fayat G, Waller JP, Iwatsubo M. The mechanism of action of methionyl-tRNA synthetase from Escherichia coli- Interaction with ligands of the amino-acid-activation reaction. Eur J Biochem. 1972;24:461–469. doi: 10.1111/j.1432-1033.1972.tb19707.x. [DOI] [PubMed] [Google Scholar]
- 33.Kabsch WJ. Evaluation of single crystal X-ray diffraction data from a position sensitive detector. J Appl Crystallogr. 1988;21:916–924. [Google Scholar]
- 34.Collaborative computational project No4. The CCP4 suite: programs from protein crystallography. Acta Cristallogr. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 35.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. 1998;D54:905–21. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 36.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for the building of proteins model in electron density maps and the location of errors in these models. Acta Crystallogr. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 37.Afonine PV, Grosse-Kunstleve RW, Adams PD. A robust bulk-solvent correction and anisotropic scaling procedure. Acta Crystallogr D Biol Crystallogr. 2005;61:850–5. doi: 10.1107/S0907444905007894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wallace AC, Laskowski RA, Thornton JM. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995;8:127–34. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]