Abstract
The microtubule associated protein tau (MAPT) H1 haplotype shows a strong association to the sporadic neurodegenerative diseases progressive supranuclear palsy and corticobasal degeneration. The functional biological mechanisms behind the genetic association have started to emerge with differences recently shown in haplotype splicing of the neuropathologically relevant exon 10. Here we investigate the hypothesis that expression of the alternatively spliced N-terminal exons also differs between the two MAPT haplotypes. We performed allele-specific gene expression analysis on a H1/H2 heterozygous human neuronal cell line model and 14 H1/H2 heterozygous human post-mortem brain tissues from two brain regions. In both cell culture and post-mortem brain tissue, we show that the protective MAPT H2 haplotype significantly expresses two-fold more 2N (exons 2+ 3+) MAPT transcripts than the disease-associated H1 haplotype. We suggest that inclusion of exon 3 in MAPT transcripts may contribute to protecting H2 carries from neurodegeneration.
1. Introduction
The microtubule associated protein tau (MAPT) gene locus is located on chromosome 17q21 and consists of 16 exons. In the adult human central nervous system, six protein isoforms are generated by alternative splicing of exons 2, 3 and 10. Exons 2 and 3 exhibit alternative splicing resulting in transcripts that contain 0, 1 or 2 N-terminal inserts (0N, exons 2−3−; 1N, exons 2+3−; 2N, exons 2+3+) [12, 20]. In the adult brain, the relative abundance of the N-terminal isoforms is 1N > 0N > 2N [3, 15].
The interaction of tau with microtubules is mediated by the microtubule binding domains encoded by exons 9 – 12 [13, 14]. Exons 2 and 3 code for short amino terminal inserts that form part of the acidic projection domain that may interact with the plasma membrane [4] and regulate spacing between microtubules [8].
Two major haplotypes have been defined spanning the MAPT genomic locus, H1 and H2, of which the H1 haplotype is associated with progressive supranuclear palsy (PSP) and corticobasal degeneration (CBD) [2, 17]. In addition, H1 variants have been found associated with Alzheimer's disease (AD) [27, 28].
Consistent with strong association of the H1 haplotype with tauopathies several H1 polymorphisms which may confer susceptibility to neurodegenerative disease have been proposed [22, 27, 28, 30-32]. Recent studies have started to look at the functional effects of the H1 haplotype, focusing particularly on overall tau expression and the splicing of the neuropathologically relevant exon 10 [7, 22, 28, 32]. These have yet to consider haplotype effect on splicing of the MAPT N-terminal exons. In this study we analyze MAPT haplotype-specific N-terminal exon expression in a heterozygous H1/H2 human neuronal cell culture model and in post-mortem human brain tissue, where we can directly compare the level of transcript expression from the H1 and H2 chromosomes.
2. Methods
2.1 Cell culture
SK-N-F1 cells were differentiated into neuronal cells following a two-stage protocol developed for SH-SY5Y cells [9] and previously used to differentiate SK-N-F1 cells [7]. Total RNA was extracted from differentiated cells using Trizol reagent (Invitrogen) followed by an additional purification step with the RNeasy Mini Kit (Qiagen).
2.2 Post-mortem brain material
Frontal cortex (BA 46) and globus pallidus brain tissue was obtained from 14 previously-identified heterozygous H1/H2 individuals [7]. Genotyping of rs1467967, rs242557/htSNP167, rs3785883, rs2471738 and rs7521 was done as previously described [30]. The fresh-frozen brain tissues were obtained from control subjects with no evident pathological signs of neurodegenerative disease from the brain banks of the Oxford Project to Investigate Memory and Ageing (OPTIMA) and the Thomas Willis Oxford Brain Collection. Brain tissue samples are collected with full consent of the patient and with the approval of the local Ethics Committee (COREC approval number 1656). Expression analysis has been approved by local Ethics Committee review (ref 06/Q1605/8). Total RNA was extracted with an initial homogenization in Trizol Reagent (Invitrogen) followed by a column purification using the RNeasy Mini Kit (Qiagen).
2.3 Haplotype-specific expression analysis
RNA was reverse transcribed into cDNA as previously described [7]. PCR primers to amplify the genomic DNA were: forward 5′-CTCTCTTCACCCCCACTCTG-3′ and reverse 5′-CGTGATCTTCCATCACTTCG-3′. Primers to amplify the expressed N-terminal isoforms use the forward primer 5′-CTTCTCCTCCTCCGCTGTC-3′ and the following reverse primers: 0N MAPT transcripts (exons 2− 3−) 5′-CTGCTTCTTCAGCTTTCAGG-3′; 1N MAPT transcripts (exons 2+ 3−) 5′- ATGCCTGCTTCTTCAGCTTC-3′; 2N MAPT transcripts (exons 2+ 3+) 5′- GAGCTCCCTCATCCACTAAGG-3′.
2.4 Quantitation of haplotype-specific gene expression
PCR products were analyzed by MALDI-ToF MS using the MassArray system on the Sequenom platform as previously described [7]. Each extension primer assay was repeated eight times, comprising four independent RT-PCR reactions analyzed in duplicate for each RNA sample. For the SK-N-F1 cells, expression data was obtained from the reverse assay with 8 MALDI-ToF outputs. For each brain sample, SNP 1 was assayed in both the forward and reverse direction and expression data was obtained from 12 - 16 successful MALDI-ToF outputs.
All results were normalized to genomic DNA which has a one to one ratio of SNP 1 alleles in heterozygotes (mean genomic H2:H1 = 1.34, SD = 0.05, p = 0.0971 by one-way ANOVA). SNP 1 is located at exon 1 +5 therefore the forward extension primer spans the exon boundary in cDNA, a sequence that does not exist in genomic DNA. To normalize the forward primer extension assay, 80-mer oligos containing both SNP 1 alleles in a one to one ratio were cloned into pBluscript (Promega) and used as a PCR template.
Statistical analysis for normally distributed data was performed as previously described [7, 29] using a one sample t-test that calculates a P-value indicating the significance of the observed allelic expression ratios from the expected ratio of one. The GP 1N data does not fulfil the formal requirements for normality and significance has been tested using the Wilcoxon signed rank test.
3. Results
3.1 Haplotype-specific expression assay design
Polymorphisms present in coding sequences can be used to tag the MAPT haplotypes and distinguish between the H1 and H2 allelic transcripts. These tagging SNPs are required for assaying MAPT allele-specific expression in heterozygous samples by measuring the ratio of MAPT haplotype transcripts within any PCR product pool defined by the initial PCR primers used. This methodology has advantages over quantitative PCR as it does not require normalization to a housekeeping gene. In addition, assaying both haplotypes within one sample controls for underlying genetic or environmental factors that vary between individuals, as well as controlling for sample quality including RNA quality and post-mortem interval.
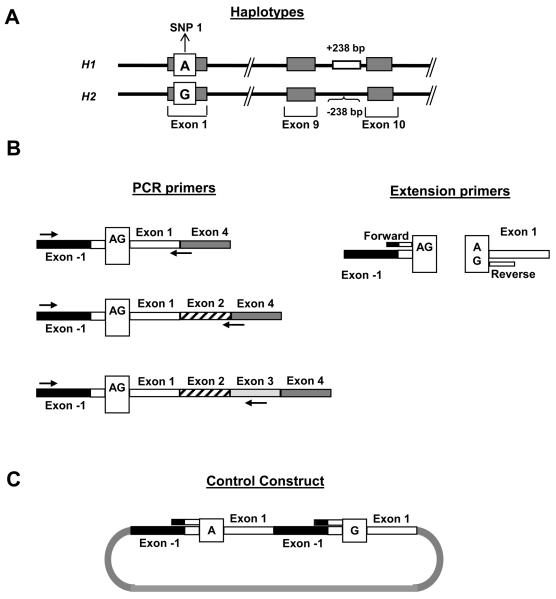
We used a previously defined coding SNP in MAPT exon 1 (SNP 1; rs17650901) [2] as the basis for the haplotype-specific, N-terminal exon expression assay (Fig. 1A). We designed PCR primers to specifically amplify either the 0N, 1N or 2N MAPT transcripts by placing the reverse primers spanning the exon 1 – 4 boundary, 2 – 4 boundary or within exon 3, respectively (Fig. 1B).
Figure 1.
Primer design for the MALDI-ToF MS assays for expression of MAPT N-terminal exons. The methodology requires a reverse transcription PCR (RT-PCR) reaction, followed by a primer extension reaction and analysis by MALDI-ToF mass spectrometry. (A) The haplotype tagging variant SNP 1, used to assay expression of the alternatively splice N-terminal exons, is in complete linkage disequilibrium with the haplotype defined 238 bp deletion located in intron 9. The SNP 1 alleles are indicated in the white box, with the H1 shown above the H2 allele. (B). The RT-PCR primers used to amplify fragments containing SNP 1 are shown on the left, and the locations of the extension primers used for the single base extension reaction extending to the SNP are shown on the right. Reverse RT-PCR primers were strategically placed to amplify either the 0N, 1N or 2N MAPT transcripts. On the right, the forward and reverse base-extension primers are indicated by short bars. (C) Structure of the artificial construct built for the forward base-extension assay at SNP 1 to normalize the MALDI-ToF data to the expected 1:1 ratio. The SNP 1 forward assay uses an extension primer that spans the exon −1/exon 1 boundary, a sequence that does not exist in genomic DNA.
3.2 Haplotype-specific expression in cell culture models
Previously, we identified two H1/H2 heterozygous human neuronal cell lines, SK-N-F1 and SK-N-MC, from a panel of 14 genetically independent neuronal cell lines [7]. Upon differentiation, SK-N-F1 cells develop the better neuronal morphology and phenotypic expression of neuronal markers [7] and was thus chosen for further expression analysis. cDNA prepared from the differentiated SK-N-F1 RNA was used as a template for PCR amplification of the three N-terminal MAPT transcripts (0N, 1N, 2N) and analyzed using the haplotype-specific coding SNP in exon 1.
The greatest difference in expression from the two chromosomes is seen in the 2N MAPT transcript expression (exons 2+ 3+) where there is more than a two-fold greater expression of transcripts containing exon 3 from the H2 chromosome (Fig. 2A). The 1N transcript isoform (exons 2+ 3−) also has a significantly greater expression from the H2 chromosome, although to a much lesser extent than the 2N transcripts. The 0N MAPT transcript isoform shows an expression ratio much closer to one with only a 12% greater expression from the H1 chromosome. This deviation is less than the 1.2-fold level of change generally regarded as biologically significant [5, 36].
Figure 2.
Allele-specific MAPT expression of N-terminal exons in a neuronal cell culture model and human post-mortem brain tissue. The expression data is normalised to the equivalent of each SNP 1 allele being present in one copy on genomic DNA to produce a standardized transcript ratio. A ratio of > 1 at SNP 1 indicates more MAPT transcript isoform from H2 than H1. (A) Allele-specific expression of N-terminal MAPT exons in differentiated SK-N-F1 cells. Mean values and standard deviations from eight measurements using the reverse primer are shown. 0N H2:H1 MAPT transcript ratio = 0.88, S.D. = 0.02, p = 0.0002. 1N H2:H1 MAPT transcript ratio = 1.33, S.D. = 0.04, p < 0.0001. 2N H2:H1 MAPT transcript ratio = 2.30, S.D. = 0.28, p = 0.0007. (B) Allele-specific expression of N-terminal MAPT exons in each of the fourteen H1/H2 heterozygotes measured using SNP 1. Mean values and standard deviations from up to sixteen measurements (eight measurements using the forward primer and eight measurements using the reverse primer) are shown. (C) Mean values and standard deviations of relative H2:H1 expression averaged from all fourteen H1/H2 heterozygotes.
3.3 Haplotype-specific expression in post-mortem brain tissue
RNA was extracted from the frontal cortex (FC) and globus pallidus (GP) of 14 H1/H2 heterozygous post-mortem brains samples. Haplotype-specific isoform expression analysis was performed on these two regions, selected as regions severely affected by tau pathology in the sporadic tauopathies CBD and PSP, respectively.
Again, the most striking allelic difference in expression occurs in the haplotype-expression ratio of the 2N MAPT transcripts consistent with the results from the SK-N-F1 cell culture model. In both the frontal cortex and globus pallidus, there is a two-fold greater expression of 2N MAPT transcripts from the protective H2 haplotype (FC mean H2:H1 2N MAPT transcript ratio = 1.96, SD = 0.22, p < 0.0001; GP mean H2:H1 2N MAPT transcript ratio = 1.99, SD = 0.44, p < 0.0001).
For the 1N isoform, the mean MAPT H2:H1 transcript ratio of the 14 brains showed no allelic difference in the frontal cortex (FC mean H2:H1 1N MAPT transcript ratio = 1.03, SD = 0.10, p = 0.3167). The remaining isoforms show less than the 20% difference in expression considered to be the accepted threshold value for detecting differences in allelic expression [5, 36] (GP mean H2:H1 1N MAPT transcript ratio = 1.17, SD = 0.16, p = 0.0001; FC mean H2:H1 0N MAPT transcript ratio = 1.19, SD = 0.08, p < 0.0001; GP mean 0N MAPT H2:H1 transcript ratio = 1.16, SD = 0.10, p <0.0001). As such, the modest variation in allelic expression of the 0N and 1N isoforms seen in post-mortem brain tissue is unlikely to be of biological significance.
Recent work has uncovered sequence variation within the H1 haplotype and identified H1 sub-haplotypes associated with PSP [30-32], AD [27, 28] and PD [33]. Previously, we genotyped the H1/H2 brain tissue for SNPs rs1467967, rs242557/htSNP167, rs3785883, rs2471738 and rs7521 to determine the H1 sub-haplotype present [7]. We subdivided the brain samples into two H1-haplotype sub-groups based on rs242557, the marker that shows the greatest association with PSP [30, 32]. We found no significant difference in the haplotype-specific expression ratios of alternatively-spliced N-terminal exons between samples with the H1c “A” allele and the “G” allele in the frontal cortex or globus pallidus (all data sets p > 0.05).
4. Discussion
This is the first study to investigate haplotype-specific differences in expression of the N-terminal alternatively spliced exons at the MAPT locus. The data presented show that there are significant haplotype differences in expression of the MAPT N-terminal alternatively spliced exons most notably in the 2N MAPT transcripts, where there is a two-fold greater expression from the H2 chromosome in both a neuronal cell line model and in two regions of post-mortem human brain tissue. A two-fold difference in expression between alleles has been observed at other loci [23] and is likely to have the biological significance expected for a susceptibility loci in a complex disease. The haplotype-specific difference we observe in the low abundance 2N MAPT transcript isoform is greater than the difference observed when total MAPT transcripts or those containing the alternatively spliced exon 10 were considered [7, 28]. Importantly, we show the effect of haplotype variation on N-terminal MAPT exon expression and splicing to be conveyed by the underlying genetic sequence in control brain samples and is therefore independent of disease status.
Recently, it has been shown that there are significant differences in the haplotype expression of MAPT exon 10+ transcripts [7, 28]. Our previous work shows H1 expressing up to 43% more exon 10+ 4R MAPT than H2 [7]. Exon 10 is strongly implicated in the neurodegenerative process, as 4R isoforms are the predominant component of insoluble tau aggregates in PSP, CBD, and FTDP-17 [1, 6]. In addition, the FTDP-17 splice site mutations within MAPT that increase the inclusion of exon 10 in transcripts show that altering the ratio of 3R and 4R tau isoforms is sufficient to cause disease [18, 34]. It has been suggested that the altered tau isoform ratios seen in PSP may also be due in part to a decrease in isoforms lacking both N-terminal inserts [11]. In CBD, some studies have shown the 64 and 69 kDa bands do not contain exon 3 [10, 21, 35].
The N-terminal exons form part of the acidic projection domain of tau which has been shown to interact with the plasma membrane [4]. In addition, this domain is thought to regulate spacing between microtubules by perhaps acting as a polymer brush or spring between microtubules [8, 24-26]. It has been suggested that tau aggregation might occur if the polymer brush failed [25]. Recent work has focussed on the N-terminal region of the tau protein in studies investigating tau protein folding and aggregation. FRET analysis suggests that tau adopts a paperclip-like conformation in solution, in which the N-terminus comes near the C-terminus as the C-terminus associates with the microtubule-binding repeats [19]. It has been suggested that stabilization of this folded state may have pathological consequences as the proposed confirmation fits well with the epitope reactivities of antibodies that detect abnormal tau in the early stages of AD [19]. The N-terminal has also been suggested to play a role in the regulation of tau solubility, in that N-terminal fragments inhibit tau polymerization [16]. The involvement of the N-terminus in tau aggregation is complex and further experimentation is required to understand what effect differential expression of N-terminal tau isoforms may have on tau conformation and aggregation.
We suggest that it is possible that a two-fold greater expression of the 2N isoform transcripts from H2 chromosomes may contribute to the mechanism of protection against H1-associated tauopathies. Though the haplotype-expression differences are observed in the minor protein isoform (2N MAPT represents 10–15 % of total MAPT expression) we should expect, like in other complex diseases, that the functional influences of common genetic polymorphisms at the MAPT locus to be subtle and cumulative over many years.
Acknowledgements
This work was supported by a Research Career Development Fellowship awarded to RW-M from the Wellcome Trust and by a Clarendon Fund Bursary awarded to TMC. We would like to thank Carolyn Sloane for sample preparation, and Margaret Esiri and David Smith for access to OPTIMA resources and the Thomas Willis Oxford Brain Collection.
Footnotes
Disclosure Statement
The authors declare they have no actual or potential conflicts of interest that could inappropriately influence or bias this manuscript. Brain tissue samples are collected with full consent of the patient and with the approval of the local Ethics Committee (COREC approval number 1656). Expression analysis has been approved by local Ethics Committee review (ref 06/Q1605/8).
References
- 1.Arai T, Ikeda K, Akiyama H, Shikamoto Y, Tsuchiya K, Yagishita S, Beach T, Rogers J, Schwab C, McGeer PL. Distinct isoforms of tau aggregated in neurons and glial cells in brains of patients with Pick's disease, corticobasal degeneration and progressive supranuclear palsy. Acta Neuropathol. (Berl) 2001;101:167–73. doi: 10.1007/s004010000283. [DOI] [PubMed] [Google Scholar]
- 2.Baker M, Litvan I, Houlden H, Adamson J, Dickson D, Perez-Tur J, Hardy J, Lynch T, Bigio E, Hutton M. Association of an extended haplotype in the tau gene with progressive supranuclear palsy. Hum Mol Genet. 1999;8:711–5. doi: 10.1093/hmg/8.4.711. [DOI] [PubMed] [Google Scholar]
- 3.Boutajangout A, Boom A, Leroy K, Brion JP. Expression of tau mRNA and soluble tau isoforms in affected and non-affected brain areas in Alzheimer's disease. FEBS Lett. 2004;576:183–9. doi: 10.1016/j.febslet.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 4.Brandt R, Leger J, Lee G. Interaction of tau with the neural plasma membrane mediated by tau's amino-terminal projection domain. J Cell Biol. 1995;131:1327–40. doi: 10.1083/jcb.131.5.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bray NJ, Buckland PR, Owen MJ, O'Donovan MC. Cis-acting variation in the expression of a high proportion of genes in human brain. Hum Genet. 2003;113:149–53. doi: 10.1007/s00439-003-0956-y. [DOI] [PubMed] [Google Scholar]
- 6.Buee Scherrer V, Hof PR, Buee L, Leveugle B, Vermersch P, Perl DP, Olanow CW, Delacourte A. Hyperphosphorylated tau proteins differentiate corticobasal degeneration and Pick's disease. Acta Neuropathol. (Berl) 1996;91:351–9. doi: 10.1007/s004010050436. [DOI] [PubMed] [Google Scholar]
- 7.Caffrey TM, Joachim C, Paracchini S, Esiri MM, Wade-Martins R. Haplotype-specific expression of exon 10 at the human MAPT locus. Hum Mol Genet. 2006;15:3529–37. doi: 10.1093/hmg/ddl429. [DOI] [PubMed] [Google Scholar]
- 8.Chen J, Kanai Y, Cowan NJ, Hirokawa N. Projection domains of MAP2 and tau determine spacings between microtubules in dendrites and axons. Nature. 1992;360:674–7. doi: 10.1038/360674a0. [DOI] [PubMed] [Google Scholar]
- 9.Encinas M, Iglesias M, Liu Y, Wang H, Muhaisen A, Cena V, Gallego C, Comella JX. Sequential treatment of SH-SY5Y cells with retinoic acid and brain-derived neurotrophic factor gives rise to fully differentiated, neurotrophic factor-dependent, human neuron-like cells. J Neurochem. 2000;75:991–1003. doi: 10.1046/j.1471-4159.2000.0750991.x. [DOI] [PubMed] [Google Scholar]
- 10.Feany MB, Ksiezak-Reding H, Liu WK, Vincent I, Yen SH, Dickson DW. Epitope expression and hyperphosphorylation of tau protein in corticobasal degeneration: differentiation from progressive supranuclear palsy. Acta Neuropathol (Berl) 1995;90:37–43. doi: 10.1007/BF00294457. [DOI] [PubMed] [Google Scholar]
- 11.Gibb GM, de Silva R, Revesz T, Lees AJ, Anderton BH, Hanger DP. Differential involvement and heterogeneous phosphorylation of tau isoforms in progressive supranuclear palsy. Brain Res Mol Brain Res. 2004;121:95–101. doi: 10.1016/j.molbrainres.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 12.Goedert M, Spillantini MG, Jakes R, Rutherford D, Crowther RA. Multiple isoforms of human microtubule-associated protein tau: sequences and localization in neurofibrillary tangles of Alzheimer's disease. Neuron. 1989;3:519–26. doi: 10.1016/0896-6273(89)90210-9. [DOI] [PubMed] [Google Scholar]
- 13.Goedert M, Wischik CM, Crowther RA, Walker JE, Klug A. Cloning and sequencing of the cDNA encoding a core protein of the paired helical filament of Alzheimer disease: identification as the microtubule-associated protein tau. Proc Natl Acad Sci U S A. 1988;85:4051–5. doi: 10.1073/pnas.85.11.4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gustke N, Trinczek B, Biernat J, Mandelkow EM, Mandelkow E. Domains of tau protein and interactions with microtubules. Biochemistry. 1994;33:9511–22. doi: 10.1021/bi00198a017. [DOI] [PubMed] [Google Scholar]
- 15.Hong M, Zhukareva V, Vogelsberg-Ragaglia V, Wszolek Z, Reed L, Miller BI, Geschwind DH, Bird TD, McKeel D, Goate A, Morris JC, Wilhelmsen KC, Schellenberg GD, Trojanowski JQ, Lee VM. Mutation-specific functional impairments in distinct tau isoforms of hereditary FTDP-17. Science. 1998;282:1914–7. doi: 10.1126/science.282.5395.1914. [DOI] [PubMed] [Google Scholar]
- 16.Horowitz PM, LaPointe N, Guillozet-Bongaarts AL, Berry RW, Binder LI. N-terminal fragments of tau inhibit full-length tau polymerization in vitro. Biochemistry. 2006;45:12859–66. doi: 10.1021/bi061325g. [DOI] [PubMed] [Google Scholar]
- 17.Houlden H, Baker M, Morris HR, MacDonald N, Pickering-Brown S, Adamson J, Lees AJ, Rossor MN, Quinn NP, Kertesz A, Khan MN, Hardy J, Lantos PL, George-Hyslop P, Munoz DG, Mann D, Lang AE, Bergeron C, Bigio EH, Litvan I, Bhatia KP, Dickson D, Wood NW, Hutton M. Corticobasal degeneration and progressive supranuclear palsy share a common tau haplotype. Neurology. 2001;56:1702–6. doi: 10.1212/wnl.56.12.1702. [DOI] [PubMed] [Google Scholar]
- 18.Hutton M, Lendon CL, Rizzu P, Baker M, Froelich S, Houlden H, Pickering-Brown S, Chakraverty S, Isaacs A, Grover A, Hackett J, Adamson J, Lincoln S, Dickson D, Davies P, Petersen RC, Stevens M, de Graaff E, Wauters E, van Baren J, Hillebrand M, Joosse M, Kwon JM, Nowotny P, Che LK, Norton J, Morris JC, Reed LA, Trojanowski J, Basun H, Lannfelt L, Neystat M, Fahn S, Dark F, Tannenberg T, Dodd PR, Hayward N, Kwok JB, Schofield PR, Andreadis A, Snowden J, Craufurd D, Neary D, Owen F, Oostra BA, Hardy J, Goate A, van Swieten J, Mann D, Lynch T, Heutink P. Association of missense and 5′-splice-site mutations in tau with the inherited dementia FTDP-17. Nature. 1998;393:702–5. doi: 10.1038/31508. [DOI] [PubMed] [Google Scholar]
- 19.Jeganathan S, von Bergen M, Brutlach H, Steinhoff HJ, Mandelkow E. Global hairpin folding of tau in solution. Biochemistry. 2006;45:2283–93. doi: 10.1021/bi0521543. [DOI] [PubMed] [Google Scholar]
- 20.Kosik KS, Orecchio LD, Bakalis S, Neve RL. Developmentally regulated expression of specific tau sequences. Neuron. 1989;2:1389–97. doi: 10.1016/0896-6273(89)90077-9. [DOI] [PubMed] [Google Scholar]
- 21.Ksiezak-Reding H, Morgan K, Mattiace LA, Davies P, Liu WK, Yen SH, Weidenheim K, Dickson DW. Ultrastructure and biochemical composition of paired helical filaments in corticobasal degeneration. Am J Pathol. 1994;145:1496–508. [PMC free article] [PubMed] [Google Scholar]
- 22.Kwok JB, Teber ET, Loy C, Hallupp M, Nicholson G, Mellick GD, Buchanan DD, Silburn PA, Schofield PR. Tau haplotypes regulate transcription and are associated with Parkinson's disease. Ann. Neurol. 2004;55:329–34. doi: 10.1002/ana.10826. [DOI] [PubMed] [Google Scholar]
- 23.Lo HS, Wang Z, Hu Y, Yang HH, Gere S, Buetow KH, Lee MP. Allelic variation in gene expression is common in the human genome. Genome Res. 2003;13:1855–62. doi: 10.1101/gr.1006603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marx A, Pless J, Mandelkow EM, Mandelkow E. On the rigidity of the cytoskeleton: are MAPs crosslinkers or spacers of microtubules? Cell Mol Biol (Noisy-le-grand) 2000;46:949–65. [PubMed] [Google Scholar]
- 25.Mukhopadhyay R, Hoh JH. AFM force measurements on microtubule-associated proteins: the projection domain exerts a long-range repulsive force. FEBS Lett. 2001;505:374–8. doi: 10.1016/s0014-5793(01)02844-7. [DOI] [PubMed] [Google Scholar]
- 26.Mukhopadhyay R, Kumar S, Hoh JH. Molecular mechanisms for organizing the neuronal cytoskeleton. Bioessays. 2004;26:1017–25. doi: 10.1002/bies.20088. [DOI] [PubMed] [Google Scholar]
- 27.Myers AJ, Kaleem M, Marlowe L, Pittman AM, Lees AJ, Fung HC, Duckworth J, Leung D, Gibson A, Morris CM, de Silva R, Hardy J. The H1c haplotype at the MAPT locus is associated with Alzheimer's disease. Hum Mol Genet. 2005;14:2399–404. doi: 10.1093/hmg/ddi241. [DOI] [PubMed] [Google Scholar]
- 28.Myers AJ, Pittman AM, Zhao AS, Rohrer K, Kaleem M, Marlowe L, Lees A, Leung D, McKeith IG, Perry RH, Morris CM, Trojanowski JQ, Clark C, Karlawish J, Arnold S, Forman MS, Van Deerlin V, de Silva R, Hardy J. The MAPT H1c risk haplotype is associated with increased expression of tau and especially of 4 repeat containing transcripts. Neurobiol Dis. 2007;25:561–70. doi: 10.1016/j.nbd.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 29.Paracchini S, Thomas A, Castro S, Lai C, Paramasivam M, Wang Y, Keating BJ, Taylor JM, Hacking DF, Scerri T, Francks C, Richardson AJ, Wade-Martins R, Stein JF, Knight JC, Copp AJ, Loturco J, Monaco AP. The chromosome 6p22 haplotype associated with dyslexia reduces the expression of KIAA0319, a novel gene involved in neuronal migration. Hum Mol Genet. 2006;15:1659–66. doi: 10.1093/hmg/ddl089. [DOI] [PubMed] [Google Scholar]
- 30.Pittman AM, Myers AJ, Abou-Sleiman P, Fung HC, Kaleem M, Marlowe L, Duckworth J, Leung D, Williams D, Kilford L, Thomas N, Morris CM, Dickson D, Wood NW, Hardy J, Lees AJ, de Silva R. Linkage disequilibrium fine mapping and haplotype association analysis of the tau gene in progressive supranuclear palsy and corticobasal degeneration. J. Med. Genet. 2005;42:837–46. doi: 10.1136/jmg.2005.031377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pittman AM, Myers AJ, Duckworth J, Bryden L, Hanson M, Abou-Sleiman P, Wood NW, Hardy J, Lees A, de Silva R. The structure of the tau haplotype in controls and in progressive supranuclear palsy. Hum. Mol. Genet. 2004;13:1267–74. doi: 10.1093/hmg/ddh138. [DOI] [PubMed] [Google Scholar]
- 32.Rademakers R, Melquist S, Cruts M, Theuns J, Del-Favero J, Poorkaj P, Baker M, Sleegers K, Crook R, De Pooter T, Bel Kacem S, Adamson J, Van den Bossche D, Van den Broeck M, Gass J, Corsmit E, De Rijk P, Thomas N, Engelborghs S, Heckman M, Litvan I, Crook J, De Deyn PP, Dickson D, Schellenberg GD, Van Broeckhoven C, Hutton ML. High-density SNP haplotyping suggests altered regulation of tau gene expression in progressive supranuclear palsy. Hum. Mol. Genet. 2005;14:3281–92. doi: 10.1093/hmg/ddi361. [DOI] [PubMed] [Google Scholar]
- 33.Skipper L, Wilkes K, Toft M, Baker M, Lincoln S, Hulihan M, Ross OA, Hutton M, Aasly J, Farrer M. Linkage disequilibrium and association of MAPT H1 in Parkinson disease. Am J Hum Genet. 2004;75:669–77. doi: 10.1086/424492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spillantini MG, Murrell JR, Goedert M, Farlow MR, Klug A, Ghetti B. Mutation in the tau gene in familial multiple system tauopathy with presenile dementia. Proc. Natl. Acad. Sci. U. S. A. 1998;95:7737–41. doi: 10.1073/pnas.95.13.7737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Terada S, Ishizu H, Ishiguro K, Tanabe Y, Itoh N, Yasutake K, Furubayashi A, Kitamura Y, Kuroda S. Exon 3 insert of tau protein in neurodegenerative diseases. Acta Neuropathol (Berl) 2005;110:12–8. doi: 10.1007/s00401-005-1012-x. [DOI] [PubMed] [Google Scholar]
- 36.Yan H, Yuan W, Velculescu VE, Vogelstein B, Kinzler KW. Allelic variation in human gene expression. Science. 2002;297:1143. doi: 10.1126/science.1072545. [DOI] [PubMed] [Google Scholar]