Abstract
Microorganisms must sense their environment and rapidly tune their metabolism to ambient conditions to efficiently use available resources. We have identified a gene encoding a response regulator, NblR, that complements a cyanobacterial mutant unable to degrade its light-harvesting complex (phycobilisome), in response to nutrient deprivation. Cells of the nblR mutant (i) have more phycobilisomes than wild-type cells during nutrient-replete growth, (ii) do not degrade phycobilisomes during sulfur, nitrogen, or phosphorus limitation, (iii) cannot properly modulate the phycobilisome level during exposure to high light, and (iv) die rapidly when starved for either sulfur or nitrogen, or when exposed to high light. Apart from regulation of phycobilisome degradation, NblR modulates additional functions critical for cell survival during nutrient-limited and high-light conditions. NblR does not appear to be involved in acclimation responses that occur only during a specific nutrient limitation. In contrast, it controls at least some of the general acclimation responses; those that occur during any of a number of different stress conditions. NblR plays a pivotal role in integrating different environmental signals that link the metabolism of the cell to light harvesting capabilities and the activities of the photosynthetic apparatus; this modulation is critical for cell survival.
Microorganisms have developed diverse mechanisms for sensing and acclimating to changes in their environment (for reviews see refs. 1–4). Acclimation responses observed in photosynthetic organisms include the alteration of light harvesting complex synthesis and degradation in response to changes in light quality (5–7), light intensity (8), and nutrient availability (9–11). Such alterations help these organisms efficiently balance the absorption of excitation energy and the production of NADPH and ATP with their utilization for cell maintenance and growth. It is critical to maintain this balance because excess excitation of the photosynthetic reaction centers could result in the production of toxic oxygen species that would be damaging to many cellular processes (12).
Acclimation responses to nutrient availability can be characterized as those that are specific to the nutrient that is limiting, and those that are general and occur during any of a number of different nutrient limitation conditions. The specific acclimation responses encompass a number of metabolic changes that help the organism better scavenge the limiting nutrient from the environment. These responses include increased synthesis of high affinity transport systems (13–16) and the production of hydrolytic enzymes that allow utilization of alternate forms of the nutrient (e.g., synthesis of extracellular phosphatases and sulfatases in response to phosphorus and sulfur limitations, respectively) (17–19). The general acclimation responses that occur during nutrient starvation include the cessation of cell division and dramatic alterations in cellular structure and metabolism (10, 11, 20, 21). One of the general responses that has been studied in the cyanobacterium Synechococcus sp. strain PCC 7942 involves the degradation of the phycobilisomes (PBS), pigment-protein complexes that harvest most of the light energy for cyanobacteria (9–11, 22, 23).
The PBS give cyanobacterial cells their typical blue-green color. Imposing nitrogen or sulfur deprivation on cultures of Synechococcus sp. strain PCC 7942 triggers the rapid and complete degradation of the PBS, which causes the cells to look yellow or bleached. Therefore, mutants that fail to degrade the PBS can be identified visually. Analysis of such mutants should define the mechanism of PBS degradation and identify regulatory elements that control proteolysis or other modifications that may occur during nutrient-limited growth.
Mutants of Synechococcus sp. strain PCC 7942 that were unable to degrade their PBS during sulfur- or nitrogen-limited growth previously have been isolated (22). The mutant phenotype was complemented with the nblA (nonbleaching) gene, which encodes a polypeptide of 59 amino acids. The transcription of nblA was dramatically induced by sulfur and nitrogen starvation and, to a lesser extent, by phosphorus starvation. NblA does not show homology to a polypeptide of known function and its role in the mechanism of degradation has not been clarified.
To further study the mechanism of PBS degradation and its regulation by environmental conditions we isolated additional nbl mutants of Synechococcus sp. strain PCC 7942. Because bleaching during nutrient stress is a component of the general responses (see above), characterization of such mutants provides insights into regulatory pathways that are governed by diverse environmental factors. In this study we characterized an nbl mutant that has a lesion in a gene encoding a response regulator. This response regulator is involved in the integration of the cells responses to both nutrient deprivation and light intensity and is essential for survival during stress conditions.
MATERIALS AND METHODS
Strain, Culture Conditions, and Quantitation of Pigments.
Synechococcus sp. strain PCC 7942 was cultured and starved for sulfur or nitrogen essentially as previously described (10). Briefly, cells were harvested by centrifugation (5,000 × g, 10 min) during exponential growth, resuspended in an equal volume of wash medium (BG-11 medium lacking nitrogen and sulfur), repelleted by centrifugation, and resuspended in wash medium to 1/10 the original culture volume. This concentrated cell culture was used to inoculate medium lacking either sulfur or nitrogen to a cell density of approximately 5 × 107 cells/ml. Except for high light conditions (500 μmol photons m−2⋅s−1), the cells were illuminated at an intensity of 70 μmol photons m−2⋅s−1 from incandescent bulbs. Low-sulfur agar plates were prepared as described previously (22). When appropriate, antibiotics were added to final concentrations of 2 μg/ml of ampicillin, 25 μg/ml of spectinomycin, and 25 μg/ml of kanamycin. Chlorophyll (chl) and phycocyanin (PC) were quantified as described previously (10).
Mutagenesis.
The nonbleaching mutant nblR (for nonbleaching regulatory) was fortuitously isolated from a screen for genes involved in Ca+2 homeostasis. Synechococcus sp. strain PCC 7942 in which pacL (a gene encoding Ca+2 ATPase) had been inactivated by insertion of a spectinomycin cassette (24) was mutagenized with N-methyl-N′-nitro-N-nitrosoguanidine (22). The mutagenized cells were screened for a growth minus phenotype in the presence of an elevated concentration of Ca+2 (20 mM). However, it was noted that the pacL mutant, as well as wild-type cells, appeared significantly bleached when grown on solid medium (but not liquid) containing 20 mM Ca+2. This bleaching was shown to be a consequence of elevated expression of nblA, as determined by measuring the levels of β-glucuronidase (GUS) activity expressed from an nblA-promoter-GUS fusion in wild-type cells exposed to high levels of Ca+2 (unpublished data). Elevated Ca+2 levels in solid growth medium may decrease the availability of sulfate to the cells, which in turn would activate nblA and result in partial bleaching. While screening for mutants of the pacL strain that were unable to grow on 20 mM Ca+2, we obtained the nblR mutant, which exhibited an nbl phenotype during sulfur- or nitrogen-limited growth.
Chimeric GUS Reporter Gene and Quantitation of GUS Activity.
Wild-type and mutant strains were transformed with an autonomously replicating vector, pCB4′ (22), containing a chimeric gene in which the nblA promoter was fused to a promoterless GUS gene (uidA, but designated GUS throughout). This chimeric gene contained 420 bp upstream of the ATG of nblA, the first three codons of nblA (for Met, Leu, and Pro) and a nine-codon linker region (encoding for Arg, Ile, Pro, Gly, Tyr, Gly, Gln, Ser, and Leu) derived from the polylinker of pCB4′ that precedes the coding region of GUS. Expression from this chimeric gene was demonstrated to strongly depend on the nitrogen and sulfur levels of the medium. Cultures grown in nutrient-repleted medium were washed as described above and inoculated into replete, sulfur-depleted, or nitrogen-depleted medium. GUS activity was measured after 24 h of starvation, essentially as described elsewhere (25).
RNA Isolation and Northern Hybridization Analysis.
RNA was isolated as previously described (22) except that the cell suspension was vortexed for 1 min in the presence of glass beads (about 100 mg, 100–300 microns, Sigma) before adding the hot phenol. This significantly improved the yield of RNA. Northern blot hybridizations and the preparation of radiolabeled RNA and DNA probes were performed as previously described (10, 22, 26).
DNA Manipulation, Complementation of Cyanobacterial Mutants, and Sequence Analysis.
Molecular techniques were performed according to standard procedures (27). Genomic DNA isolated from Synechococcus sp. strain PCC 7942 in which a spectinomycin cassette was inserted at the BglII site of nblA (22) was used to construct a genomic library. A Sau3AI partial digest of this genomic DNA was fractionated on a 0.7% agarose gel and 7- to 10-kbp fragments were isolated and ligated into the BamHI of pUC118. This plasmid cannot replicate autonomously in Synechococcus sp. strain PCC 7942, but can confer ampicillin resistance to the cyanobacterium upon homologous integration into the genome. After integration, a complemented strain contains both a wild-type and a mutated copy of the complementing sequence. Complementation of the nblR mutant was achieved by transformation with the plasmid library and screening for ampicillin-resistant colonies that bleached normally when grown on low-sulfur plates (yellow colonies in a background of blue-green colonies). After several rounds of rescreening to ensure genetic homogeneity, genomic DNA was isolated from the complemented strain, digested with restriction enzymes for which there are sites on the polylinker of pUC118, and analyzed by Southern blot hybridizations using pUC118 as a probe. The EcoRI and PstI digests yielded 8.0- and 5.5-kbp fragments, respectively, each containing pUC118 and flanking genomic DNA. To clone the DNA flanking the vector sequence, total genomic DNA isolated from complemented nblR was digested with EcoRI or PstI, the restriction fragments were diluted and incubated with T4 DNA ligase, and the ligation products were transformed into Escherichia coli cells that were selected on solid medium containing 100 μg/ml of ampicillin. Each of the rescued plasmids was tested for its ability to complement nblR. The nblR strain transformed with the plasmid rescued from PstI digested genomic DNA bleached on solid medium containing low levels of sulfur. The insert in this plasmid (2.5 kbp) was sequenced by using the Applied Biosystems PRISM system (Perkin–Elmer) and was used to generate subclones tested in the complementation assay. The sequence data have been submitted to the GenBank database under accession number AF049128.
Interposon Inactivation.
nblR was interrupted by insertion of a 1.3-kbp kanamycin resistance gene at the XcmI site located 87 nucleotides from the beginning of the gene. The plasmids with the interrupted gene were introduced into wild-type Synechococcus sp. strain PCC 7942, and in vivo gene disruptions were verified by PCR analysis of genomic DNA isolated from the transformant.
RESULTS
Characterization of the nbl Mutant.
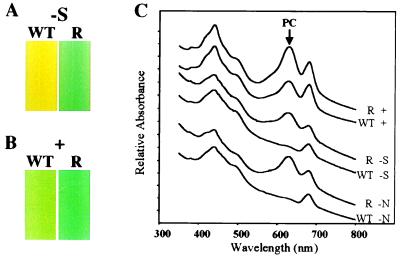
Fig. 1A shows cultures of Synechococcus sp. strain PCC 7942 and the nbl mutant, nblR, after growth for 48 h in medium lacking sulfur. The wild-type culture exhibited the typical yellow color that indicates the degradation of the PBS (Fig. 1A). This observation is reflected in the absorbance spectra (Fig. 1C) as a dramatic decrease in the absorbance at 620 nm, a consequence of the loss of PC, the major pigment-protein of the PBS (6, 9, 10, 28). In contrast, mutant nblR exhibited a blue-green color (Fig. 1A) and retained most of the absorbance peak at 620 nm after starvation for sulfur or nitrogen (Fig. 1C). Furthermore, the nblR mutant did not exhibit (not shown) the partial loss of PBS constituents that is observed in wild-type cells during phosphorus limitation (10).
Figure 1.
Cultures of wild-type cells (WT) and the nblR mutant (R) after (A) 48 h in medium lacking sulfur (−S) or (B) after growth in complete medium (+). (C) Whole-cell absorbance spectra of WT and the nblR mutant after growth in replete medium (+) and after 24 h in medium lacking sulfur (−S) or nitrogen (−N). An arrow marks the 620 nm absorbance peak of PC. Cultures were adjusted to the same OD750 before determining the spectra. Spectra were offset along the y axis, and relative heights of absorbance peaks at 620 nm can be directly compared.
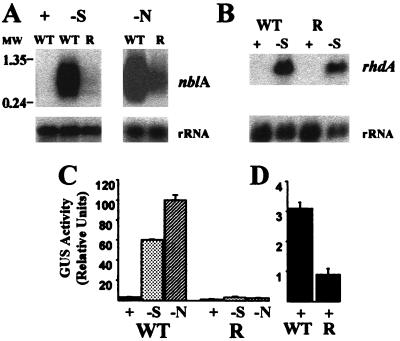
To determine whether the accumulation of the nblA transcript was altered in the mutant, we examined the transcript levels by Northern blot hybridizations. As shown in Fig. 2A, mutant nblR had dramatically reduced levels of the nblA transcripts during sulfur and nitrogen limitation. In contrast, as shown in Fig. 2B, expression of genes such as rhdA that are activated by specific nutrient limitation conditions (26) do not appear to be altered in the nblR mutant.
Figure 2.
(A) Northern blot hybridization of a riboprobe specific for nblA to RNA from wild type (WT) grown in complete medium (+) and WT and the nblR mutant (R) grown in medium devoid of sulfur (−S) or nitrogen (−N) for 24 h. Molecular weight markers (MW) are in kbp. (B) Northern blot hybridization of a probe specific for rhdA to RNA from WT and the nblR mutant grown in complete medium (+) and medium devoid of sulfur (−S) for 4 h. Hybridization to rRNA served as loading controls. (C) GUS activity in WT and the nblR mutant harboring a plasmid with the chimeric nblA (promoter)-GUS reporter gene. GUS activity was measured in cells grown in complete medium (+) or after 24 h of starvation for sulfur (−S) or nitrogen (−N). (D) GUS activity determined for WT and the nblR mutant during growth on complete medium (note the expanded scale on the y axis).
To quantitate expression from the nblA promoter, we transformed both the wild-type and the nblR mutant cells with a plasmid bearing a chimeric gene consisting of the nblA promoter fused to the promoterless uidA gene encoding GUS (see Materials and Methods). GUS activity in the strains harboring the chimeric gene was measured after growth in replete medium or after growth for 24 h in medium devoid of sulfur or nitrogen. For wild-type cells, sulfur and nitrogen starvation resulted in a 20- and 30-fold increase in GUS activity, respectively, whereas mutant nblR showed only a small increase in GUS activity after starvation (Fig. 2C). Reduced expression from the chimeric gene in mutant nblR suggested that the nblA promoter was less active in the mutant than in the wild-type genetic backgrounds. Hence the nblR strain probably harbors a lesion in a gene required for transcriptional control of nblA. Furthermore, GUS activity in the nblR mutant was lower compared with that of wild-type cells even in cultures grown in nutrient-replete medium (Fig. 2D). These findings are consistent with the observation that the nblR mutant grown in complete medium exhibited a more “bluish” pigmentation (Fig. 1B) and contained 30–50% more PC/cell than wild-type cells, as calculated from whole-cell absorption spectra (Fig. 1C).
Complementation of nblR and Analysis of the Mutated Gene.
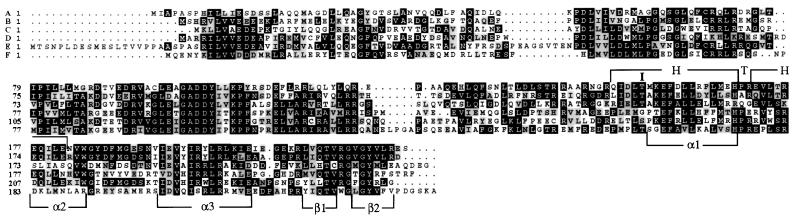
To identify the lesion in nblR, this strain was transformed with a recombinant library of Synechococcus sp. strain PCC 7942 genomic DNA, and transformants were screened for bleaching during sulfur limitation. Transformation of mutant nblR with the nblA gene (integrated into the genome by homologous recombination) resulted in partial suppression of the nbl phenotype, although the nblA gene was not altered in the mutants as determined by sequence analysis. Therefore the recombinant library was constructed with genomic DNA from a strain in which nblA had been inactivated (nblAΩ). A plasmid rescued from a transformant of nblR that was phenotypically complemented was able to recomplement the mutant phenotype; absorbance spectra of the strain transformed with this plasmid indicated a dramatic decrease in the PC level after sulfur and nitrogen starvation (not shown). Complementation with subclones of this plasmid (not shown) defined the nblR gene, which encoded a protein with similarities to response regulators of two component signal transduction pathways. Fig. 3 shows the homology between NblR and several response regulators, including sll0396 of Synechocystis sp. strain PCC 6803 (29), CopR from Pseudomonas syringae (30), PhoB from E. coli (31), SphR, the PhoB homologue in Synechococcus sp. strain PCC 7942 (32, 33) and OmpR from E. coli (34, 35). These response regulators, excluding sll0396 that has not been studied in detail, were shown to activate transcription of genes involved in the acclimation of bacterial cells to various stress conditions. For example, PhoB activates transcription of the pho regulon, which is composed of several operons important for scavenging phosphorus from the environment (4). CopR is involved in acclimation of cells to elevated levels of copper (30) and OmpR regulates transcription of the genes encoding the major outer membrane porin proteins in response to changes in osmolarity (36). The sequence of the nblR gene from the mutant strain revealed a point mutation that resulted in the replacement of threonine at position 156 with an isoleucine (T156I, Fig. 3).
Figure 3.
Alignment of NblR with various response regulators of two component signal transduction systems. (A) NblR. (B) sll0396 (putative response regulator in Synechocystis sp. strain PCC 6803). (C) CopR (copper-related response regulator from Pseudomonas syringae). (D) PhoB (phosphorus-related response regulator from E. coli). (E) SphR (PhoB homolog of Synechococcus sp. strain PCC 7942). (F) OmpR (osmolarity-related response regulator of E. coli). Identical and similar residues in at least two of the sequences are on a black or gray background, respectively. The threonine (T) to isoleucine (I) change at amino acid 156 and α-helical regions (α1, α2, α3) and β-sheets (β1, β2) close to the helix–turn–helix (H, T, H) of the predicted DNA binding site are indicated.
Because nblR was obtained by mutagenesis of a strain in which the pacL gene (encoding Ca+2 ATPase), was inactivated (see Materials and Methods), it was possible that the nbl phenotype resulted from the combination of the two defective genes, pacL and nblR. Therefore, nblR of wild-type cells was disrupted by the insertion of a kanamycin-resistance gene. The resulting mutant, nblRΩ, had a pronounced nbl phenotype upon either sulfur or nitrogen starvation and contained even more PC than the nblR mutant during nutrient-replete growth (not shown). Hence, a lesion in nblR alone is sufficient to cause the nbl phenotype.
Survival of the nblR Mutant During Nutrient Limitation and Exposure to High Light.
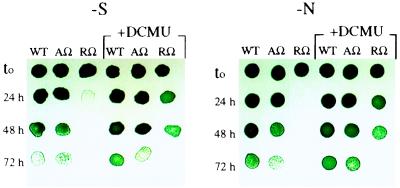
In addition to providing cells with a certain amount of the limiting nutrient, PBS degradation could prevent excess excitation of photosystem II reaction centers, which might be critical for surviving starvation conditions (see Discussion). To explore this question, wild-type cells and the nblRΩ mutant were assayed for their ability to survive nitrogen and sulfur starvation in liquid medium. The nbl nblA-inactivated strain (nblAΩ) also was examined to determine whether retention of the light-harvesting complex during nutrient limitation affects survival in the presence of functional NblR. After various times of starvation, aliquots of cells were removed, spotted onto nutrient-replete solid medium, and allowed to grow. Fig. 4 shows that the nblRΩ mutant dies rapidly during sulfur and nitrogen starvation. In contrast, the nblAΩ mutant survives nutrient deprivation nearly as well as wild-type cells, although there is some decreased viability. Hence, in addition to regulating nblA expression and PBS degradation, NblR functions to regulate other genes during nitrogen and sulfur limitation that prolong cell survival during starvation conditions.
Figure 4.
Wild type (WT), nblRΩ, (RΩ), and nblAΩ (AΩ) were spotted onto nutrient-replete solid medium after various times (24, 48, and 72 h) of starvation for sulfur (−S) or nitrogen (−N) in liquid medium. Each spot consisted of 10 μl from a 2-fold dilution of the original culture. Where indicated, the cultures were made 5 μM of DCMU upon initiation of starvation (t0).
Interestingly, the presence of the photosynthetic electron transport inhibitor 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU) slows the death of all strains examined during both sulfur and nitrogen starvation. The most dramatic affect is on the survival of nblRΩ (Fig. 4).
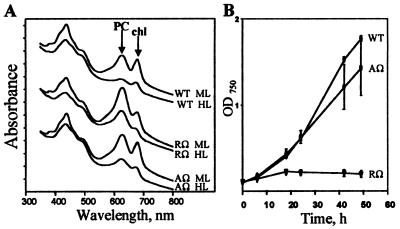
High-intensity irradiation generates a highly reduced photosynthetic electron transport chain and the size of the light-harvesting complex decreases; this helps to prevent overexcitation of the photosynthetic apparatus (8). Therefore, we examined the effect of high light on pigmentation and growth of the nbl strains nblRΩ and nblAΩ. Fig. 5A presents absorbance spectra of cells grown in medium and high light intensities (70 and 500 μmol photons m−2⋅s−1, respectively). Wild-type cells grown in high light contain lower levels of chl and PC than cells grown in medium light (Fig. 5A). The nbl strains nblRΩ and nblAΩ reduce the level of chl in response to high light; however, they maintain more PC relative to wild-type cells. These data suggest that NblA is important for the decrease in PC levels observed in high-light-grown cells and that NblR modulates expression of nblA under these conditions. The nblAΩ mutant has a somewhat lower level of PC relative to nblRΩ (Fig. 5A). The ability of this mutant to grow in high light, in contrast to nblRΩ (see below), might result in dilution of the pigment after cell division.
Figure 5.
(A) Whole-cell absorbance spectra of wild type (WT), nblRΩ (RΩ), and nblAΩ (AΩ) grown at medium light (ML) or high light (HL) (70 and 500 μmol photons m−2⋅s−1, respectively). Cultures were adjusted to the same OD750 before determining the spectra. The spectra were offset along the y axis. Arrows indicate the absorbance peaks of PC and chl. (B) Growth, as measured by OD750, of WT, nblRΩ, and nblAΩ at HL (500 μmol photons m−2⋅s−1).
As shown in Fig. 5B, the growth rates of wild-type cells and the nblAΩ mutant are similar in high light while the nblRΩ strain does not grow. Furthermore, the analysis of nblRΩ viability indicates that this strain dies rapidly upon exposure to high light (not shown).
To examine whether the pleiotrophic nature of the nblR mutation is the result of a polar effect on downstream genes, we inserted nblR into a neutral site in the genome of the nblR mutant. This action resulted in complementation of all aspects of the mutation, including the degradation of the PBS and altered viability during nutrient-limited and high-light growth (not shown). This observation excludes the possibility of polar effects and suggests that NblR modulates acclimation responses in addition to the degradation of the PBS.
DISCUSSION
NblR Integrates Diverse Environmental Signals.
The cyanobacterial mutant, nblR, was isolated based on its inability to degrade PBS during sulfur and nitrogen starvation. The complementing gene encodes a response regulator of two-component signal transduction pathways (Fig. 3), which is essential for the activation of transcription of nblA during sulfur and nitrogen starvation (Fig. 2 A and C). In addition, NblR modulates phycobiliprotein levels and the activity of the nblA gene during nutrient-replete growth (Figs. 1C and 2D). Furthermore, the nblR mutant did not exhibit the partial loss of PC observed when wild-type cells are starved for phosphorus (not shown) or exposed to high light (Fig. 5A). Hence, the nblR signal transduction pathway must integrate a number of different environmental signals.
Specific regulatory proteins have been associated with the acclimation of microbes to nutrient starvation. Genes identified in cyanobacteria that encode such regulators are ntcA and ntcB (37, 38), cysR (39), sphR (32), and phoB (40). NtcA and NtcB are involved in regulation of nitrogen transport and metabolism, CysR regulates genes that respond to sulfur availability, and SphR and PhoB modulate acclimation to phosphorus limitation. In contrast to nutrient-specific responses, there are general responses, such as PBS degradation, that are triggered by changes in any of a number of different environmental parameters.
The sulfur-specific responses, mostly involved in scavenging the limiting nutrient, are not significantly affected by a mutation in nblR. The nblR mutant shows normal induction of transcription of rhdA (Fig. 2B), which encodes a rhodanase-like polypeptide (26), and cysW (data not shown), which encodes one of the pore-forming proteins of the sulfate permease (41), after sulfur starvation. The nitrogen-specific responses are also likely to be normal in the nblR mutant. In Synechococcus sp. strain PCC 7942, most of the genes that respond to environmental levels of nitrogen-containing metabolites become active upon ammonium deprivation in the presence of nitrate (42). The nblR mutant grows normally in medium that contains nitrate as the sole nitrogen source, suggesting that elimination of NblR does not prevent the activation of genes encoding nitrate transport and assimilation functions. In sum, NblR does not appear to affect nutrient-specific responses, but is a pivotal regulatory element in a signal transduction pathway that modulates at least some of the general responses. Aspects of the general pathway controlled by NblR are the complete degradation of the PBS during sulfur and nitrogen starvation (Fig. 1C), the partial loss of pigmentation that accompanies phosphorus limitation (not shown) and exposure to high light (Fig. 5A), and the survival of cells starved for sulfur, nitrogen or exposed to high light (Figs. 4 and 5B, respectively). Because the nutrient-specific acclimation responses appear normal in the nblR mutant, decreased viability during starvation conditions is not a result of the inability of this strain to scavenge the limiting nutrient. Elements that regulate gene expression in response to multiple environmental factor have been found in other organisms; these include SNF1, which controls the responses of yeast to both phosphorus and glucose depletion (43).
NblR Controls Expression of nblA.
Response regulators such as NblR often work in conjunction with a sensor protein with histidine kinase activity. Changes in the environment can trigger autophosphorylation of the sensor at a specific histidine residue. The phosphoryl group on the histidine then is transferred to an aspartate residue in the receiver domain of a response regulator. The phosphorylated response regulator then may modify the transcriptional activity of specific groups of genes (for reviews see refs. 1, 3, and 44). The strong similarity between NblR and OmpR provides some insights into the functions of specific domains of NblR. Based on the crystal structure, the C-terminal output domain of OmpR consists of three α-helices packed against two antiparallel β-sheets (indicated in Fig. 3 below OmpR sequence). Helices α2 and α3 of OmpR, plus residues in the loop connecting them, constitute a variation of the helix–turn–helix (HTH) motif, with α3 being the DNA recognition helix (45). A HTH structure is predicted for the analogous region of NblR (Fig. 3, Chou-Fasman analysis). In the nblR mutant, the threonine residue located within the α1 helix, is mutated to an isoleucine (T156I, see Fig. 3). The equivalent mutation in OmpR (T162I) completely eliminates DNA binding (46). Based on the predicted three-dimensional structure of OmpR, T162 is in close proximity to helix α3 and may facilitate interactions of that helix with DNA. A similar role of T156 in NblR would explain why the T156I nblR mutant was unable to fully activate nblA. The isoleucine residue, unlike the threonine residue, does not have a hydroxyl group, which may function in DNA-protein interaction. Furthermore, the isoleucine residue is larger and more hydrophobic, which might result in steric retardation of NblR-DNA association. Additional evidence suggesting the functional importance of amino acid 156 in NblR comes from mutational studies of PhoB from E. coli (47).
NblR Is Critical for Survival During Starvation and Exposure to High Light.
A mutant in nblR dies rapidly during sulfur and nitrogen starvation or upon exposure to high light. There are several reasons that could explain why the nblRΩ strain is more sensitive to nutrient limitation than wild-type cells. Starvation of wild-type cells for sulfur and nitrogen triggers the disconnection of the PBS from photosystem II reaction centers, the degradation of the light-harvesting complex, and the down-regulation of photosystem II activity (22, 48). The finding that the nbl nblAΩ strain, specifically impaired in PBS degradation, dies slightly more rapidly than wild-type cells suggests that PBS degradation has an effect on viability during acclimation to nutrient starvation. However, nblRΩ dies much more rapidly than nblAΩ, indicating that additional processes controlled by NblR are critical for viability during starvation. The rapid death of nblRΩ during starvation makes it difficult to determine which of the general acclimation responses is impaired. However, because the photosynthetic electron transport inhibitor DCMU markedly improved survival of nblRΩ during starvation (Fig. 4), it is likely that the mutant strain cannot down-regulate photosynthetic electron transport during nutrient starvation. Additional evidence for the importance of the down-regulation of photosystem II during nutrient starvation comes from studies of a mutant of Chlamydomonas reinhardtii, sac1, that is unable to acclimate to sulfur limitation (49). This strain cannot modify a number of aspects of photosystem II activity (21) and dies more rapidly in the light during sulfur deprivation than wild-type cells (49). As with nblRΩ, the death of the sac1 strain can be retarded by the addition of DCMU (49).
NblR is critical for the acclimation of Synechococcus sp. strain PCC 7942 to high light. The mutants nblAΩ and nblRΩ show much less of a decline in the levels of light-harvesting pigment during illumination with high light than wild-type cells (Fig. 5A). However, while nblAΩ grows in high light, the nblRΩ mutant does not grow (Fig. 5B) and rapidly dies (not shown). Hence, NblR modulates acclimation responses, in addition to the degradation of the PBS, that are essential for survival of cyanobacterial cells in high light. Although it is unclear what processes are governed by NblR that allow survival in high light, they may include (i) modulation of energy transfer from the light-harvesting complex to the photosystem II reaction center, (ii) dissipation of excess absorbed light energy, (iii) repair of photosystem II reaction centers that suffer photoinhibitory damage, and (iv) detoxification of active oxygen species formed in high light. The inability of the nblRΩ mutant to modulate one or more of these processes during exposure to high light could result in elevated photodamage and death.
The mechanism by which high light and nutrient deprivation control NblR activity is uncertain; however, both would create a highly reduced intracellular environment. During nutrient limitation, when the anabolism of the cell is slowed down or completely arrested, NADP+, the final electron acceptor of the photosynthetic electron transport chain, is not recycled as fast as under nutrient-replete conditions and the electron carriers are maintained in a relatively reduced state. Retention of the PBS and light absorption during nutrient-limited and high-light conditions could further reduce the electron carriers and result in photodamage. Therefore, aside from providing the cell with some of the limiting nutrient, the degradation of the PBS might help minimize the absorption of excess excitation energy.
The redox state of the photosynthetic electron carriers is known to modulate a large number of acclimation responses including induction of transcription (50), regulation of translation (51), state transitions, and changes in the stoichiometry of the photosystems (52). The possibility that NblR activity is linked to the redox state of the photosynthetic electron carriers is supported by the finding that DCMU, which prevents photosynthetic electron flow beyond QA, inhibits the accumulation of nblA mRNA in sulfur-starved cells (not shown).
Different environmental stimuli govern NblR activity, which in turn, activates nblA transcription and PBS degradation. In addition to the environmental signals discussed above, low ambient CO2 is postulated to affect NblR, based on the partial bleaching observed under this condition (R.S. and A. Kaplan, unpublished results). PBS degradation contributed to a small extent to cell survival during nutrient stress but did not appear to be critical for survival under the high light conditions (500 μmol photons m−2⋅s−1). Reduced PBS levels might become more critical when the cells experience higher light intensities or when exposed to a combination of stress conditions. Independent of its affect on nblA and PBS degradation, NblR must alter the expression of other genes critical for survival in high light or during extended periods of nutrient deprivation. Although it is unknown what specific features of high light and nutrient limitation control NblR activity, both conditions would lead to an increase in the intracellular redox state. Future work will help elucidate how NblR integrates multiple environmental cues and triggers a set of responses that allow the organism to better compete under suboptimal growth conditions.
Acknowledgments
We thank David Kehoe, Nadia Dolganov, Devaki Bhaya, Jean-David Rochaix, Dennis Wykoff, John Davis, and Mikael Schwarz for reading the manuscript and for helpful discussions. We also thank Tom Berkelman for providing us with the nblR mutant. R.S. was supported by postdoctoral scholarships from the Hebrew University of Jerusalem, the Lamas Foundation, and the Human Frontier Science Program Organization. The work also was supported by U.S. Department of Agriculture Grant 94-37306-0344 and National Science Foundation Grant MCB 9727836 awarded to A.R.G. This is Carnegie Institution of Washington Publication No. 1373.
ABBREVIATIONS
- PBS
phycobilisomes
- nbl
nonbleaching
- chl
chlorophyll
- PC
phycocyanin
- GUS
β-glucuronidase
- DCMU
3-(3,4-dichlorophenyl)-1,1-dimethylurea
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF049128).
References
- 1.Parkinson J S. Cell. 1993;73:857–871. doi: 10.1016/0092-8674(93)90267-t. [DOI] [PubMed] [Google Scholar]
- 2.Stock A M, Mowbray S L. Curr Opin Struct Biol. 1995;5:744–751. doi: 10.1016/0959-440x(95)80006-9. [DOI] [PubMed] [Google Scholar]
- 3.Stock J B, Stock A M, Mottonen J M. Nature (London) 1990;344:395–400. doi: 10.1038/344395a0. [DOI] [PubMed] [Google Scholar]
- 4.Wanner B L. J Cell Biochem. 1993;51:47–54. doi: 10.1002/jcb.240510110. [DOI] [PubMed] [Google Scholar]
- 5.Bryant D A. Eur J Biochem. 1981;119:425–429. doi: 10.1111/j.1432-1033.1981.tb05625.x. [DOI] [PubMed] [Google Scholar]
- 6.Grossman A R, Kehoe D A. Photosynth Res. 1997;53:95–108. [Google Scholar]
- 7.Tandeau de Marsac N. J Bacteriol. 1977;130:82–91. doi: 10.1128/jb.130.1.82-91.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Horton P, Ruban A B, Walters R G. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:655–684. doi: 10.1146/annurev.arplant.47.1.655. [DOI] [PubMed] [Google Scholar]
- 9.Allen M M, Smith A J. Arch Microbiol. 1969;69:114–120. doi: 10.1007/BF00409755. [DOI] [PubMed] [Google Scholar]
- 10.Collier J L, Grossman A R. J Bacteriol. 1992;174:4718–4726. doi: 10.1128/jb.174.14.4718-4726.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamanaka G, Glazer A N. Arch Microbiol. 1980;124:39–47. [Google Scholar]
- 12.Asada K. In: Causes of Photooxidative Stress and Amelioration of Defense Systems in Plants. Foyer C H, Mullineaux P M, editors. Boca Raton, FL: CRC; 1994. pp. 77–104. [Google Scholar]
- 13.Green L S, Grossman A R. J Bacteriol. 1988;170:583–587. doi: 10.1128/jb.170.2.583-587.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grillo J F, Gibson J. J Bacteriol. 1979;140:508–517. doi: 10.1128/jb.140.2.508-517.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Madueño F, Vega-Palas M A, Flores E, Herrero A. FEBS Lett. 1988;239:289–291. [Google Scholar]
- 16.Omata T, Andriesse X, Hirano A. Mol Gen Genet. 1993;236:193–202. doi: 10.1007/BF00277112. [DOI] [PubMed] [Google Scholar]
- 17.de Hostos E L, Togasaki R K, Grossman A R. J Cell Biol. 1988;106:29–37. doi: 10.1083/jcb.106.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Quisel J, Wykoff D, Grossman A R. Plant Physiol. 1996;111:839–848. doi: 10.1104/pp.111.3.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ray J M, Bhaya D, Block M A, Grossman A R. J Bacteriol. 1991;173:4297–4309. doi: 10.1128/jb.173.14.4297-4309.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sherman D M, Sherman L A. J Bacteriol. 1983;156:393–401. doi: 10.1128/jb.156.1.393-401.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wykoff D D, Davies J P, Melis A, Grossman A R. Plant Physiol. 1998;117:129–139. doi: 10.1104/pp.117.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Collier J L, Grossman A R. EMBO J. 1994;13:1039–1047. doi: 10.1002/j.1460-2075.1994.tb06352.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grossman A R, Bhaya D, Apt K E, Kehoe D M. Annu Rev Genet. 1995;29:231–287. doi: 10.1146/annurev.ge.29.120195.001311. [DOI] [PubMed] [Google Scholar]
- 24.Berkelman T, Garret-Engele P, Hoffman N E. J Bacteriol. 1994;176:4430–4436. doi: 10.1128/jb.176.14.4430-4436.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dolganov N A M, Bhaya D, Grossman A R. Proc Natl Acad Sci USA. 1995;92:636–640. doi: 10.1073/pnas.92.2.636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laudenbach D E, Ehrhardt D, Green L, Grossman A R. J Bacteriol. 1991;173:2751–2760. doi: 10.1128/jb.173.9.2751-2760.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 28.Yamanaka G, Glazer A N, Williams R C. J Biol Chem. 1980;255:11004–11010. [PubMed] [Google Scholar]
- 29.Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura T, Miyajima N, Hirosawa M, Sugiura M, Tabata S. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 30.Mills S D, Jasalavich C A, Cooksey D A. J Bacteriol. 1993;175:1656–1664. doi: 10.1128/jb.175.6.1656-1664.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Makino K, Shinagawa H, Amemura M, Nakata A. J Mol Biol. 1986;190:37–44. doi: 10.1016/0022-2836(86)90073-2. [DOI] [PubMed] [Google Scholar]
- 32.Aiba H, Nagaya M, Mizuno T. Mol Microbiol. 1993;8:81–91. doi: 10.1111/j.1365-2958.1993.tb01205.x. [DOI] [PubMed] [Google Scholar]
- 33.Nagaya M, Aiba H, Mizuno T. J Bacteriol. 1994;176:2210–2215. doi: 10.1128/jb.176.8.2210-2215.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Forst S A, Delgado J, Inouye M. Proc Natl Acad Sci USA. 1989;86:6052–6056. doi: 10.1073/pnas.86.16.6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hall M N, Silhavy T J. J Mol Biol. 1981;146:23–43. doi: 10.1016/0022-2836(81)90364-8. [DOI] [PubMed] [Google Scholar]
- 36.Csonka L N, Hanson A D. Annu Rev Microbiol. 1991;45:569–606. doi: 10.1146/annurev.mi.45.100191.003033. [DOI] [PubMed] [Google Scholar]
- 37.Vega-Palas M A, Flores E, Herrero A. Mol Microbiol. 1992;6:1853–1859. doi: 10.1111/j.1365-2958.1992.tb01357.x. [DOI] [PubMed] [Google Scholar]
- 38.Aichi M, Omata T. J Bacteriol. 1997;179:4671–4675. doi: 10.1128/jb.179.15.4671-4675.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nicholson M L, Laudenbach D E. J Bacteriol. 1995;177:2143–2150. doi: 10.1128/jb.177.8.2143-2150.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Watson G M F, Scanlan D J, Mann N H. FEMS Microbiol Lett. 1996;142:105–109. doi: 10.1111/j.1574-6968.1996.tb08415.x. [DOI] [PubMed] [Google Scholar]
- 41.Laudenbach D E, Grossman A R. J Bacteriol. 1991;173:2739–2750. doi: 10.1128/jb.173.9.2739-2750.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luque I, Flores E, Herrero A. EMBO J. 1994;13:2862–2869. doi: 10.1002/j.1460-2075.1994.tb06580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thompson-Jaeger S, Francois J, Gaughran J P, Tatchell K. Genetics. 1991;129:697–706. doi: 10.1093/genetics/129.3.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Appleby J L, Parkinson J S, Bourret R B. Cell. 1996;86:845–848. doi: 10.1016/s0092-8674(00)80158-0. [DOI] [PubMed] [Google Scholar]
- 45.Martinez-Hackert E, Stock A M. Structure. 1997;5:109–124. doi: 10.1016/s0969-2126(97)00170-6. [DOI] [PubMed] [Google Scholar]
- 46.Kato N, Tsuzuki M, Aiba H, Mizuno T. Mol Gen Genet. 1995;248:399–406. doi: 10.1007/BF02191639. [DOI] [PubMed] [Google Scholar]
- 47.Makino K, Amemura M, Kawamoto T, Kimura S, Shinagawa H, Nakata A, Suzuki M. J Mol Biol. 1996;259:15–26. doi: 10.1006/jmbi.1996.0298. [DOI] [PubMed] [Google Scholar]
- 48.Collier J L, Herbert S K, Fork D C, Grossman A R. Photosynth Res. 1994;42:173–183. doi: 10.1007/BF00018260. [DOI] [PubMed] [Google Scholar]
- 49.Davies J, Yildiz F, Grossman A R. EMBO J. 1996;15:2150–2159. [PMC free article] [PubMed] [Google Scholar]
- 50.Escoubas J-M, Lomas M, LaRoche J, Falkowski P G. Proc Natl Acad Sci USA. 1995;92:10237–10241. doi: 10.1073/pnas.92.22.10237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim J, Mayfield S P. Science. 1997;278:1954–1957. doi: 10.1126/science.278.5345.1954. [DOI] [PubMed] [Google Scholar]
- 52.Fujita Y, Murakami A, Aizawa K, Ohki K. In: The Molecular Biology of Cyanobacteria. Bryant D A, editor. Dordrecht, The Netherlands: Kluwer; 1994. pp. 677–692. [Google Scholar]