Abstract
Thirteen field isolates of infectious bronchitis virus (IBV) were isolated from broiler flocks in Thailand between January and June 2008. The 878-bp of the S1 gene covering a hypervariable region was amplified and sequenced. Phylogenetic analysis based on that region revealed that these viruses were separated into two groups (I and II). IBV isolates in group I were not related to other IBV strains published in the GenBank database. Group 1 nucleotide sequence identities were less than 85% and amino acid sequence identities less than 84% in common with IBVs published in the GenBank database. This group likely represents the strains indigenous to Thailand. The isolates in group II showed a close relationship with Chinese IBVs. They had nucleotide sequence identities of 97-98% and amino acid sequence identities 96-98% in common with Chinese IBVs (strain A2, SH and QXIBV). This finding indicated that the recent Thai IBVs evolved separately and at least two groups of viruses are circulating in Thailand.
Keywords: infectious bronchitis virus, phylogenetic analysis, S1 gene, sequencing, Thailand
Introduction
Infectious bronchitis (IB), caused by the IB virus (IBV), is a highly contagious disease and results in a significant economic loss to the commercial chicken industry. The disease frequently causes respiratory signs including gasping, coughing, sneezing, tracheal rales, and nasal discharge. In layer fowls, respiratory distress and a decrease in egg production have been reported [10]. In addition, some strains have been associated with kidney lesions [18,19,22].
IBV, the causative agent of IB, belongs to the genus Coronavirus in the family Coronaviridae [6]. It is an enveloped virus and has a positive-sense, single-stranded, RNA genome, approximately 27 kb in length. The virion has three major structural proteins namely the nucleocapsid (N) protein, the membrane (M) protein, and the spike (S) glycoprotein. The S glycoprotein is post-translationally cleaved into the S1 and S2 subunits [6]. The S1 subunit, located on the outside of the virion, is responsible for the fusion between the virus envelope and the host cell membrane. Moreover, it is responsible for neutralizing serotype-specific antibodies in chickens [4]. The S1 subunit demonstrates more sequence variability than S2 [14]. Neutralizing and serotype-specific epitopes are associated with the defined hypervariable region (HVR) in the S1 subunit; therefore, the molecular characterization of IBV is based on analysis of the S1 gene [12].
During 1953-1954, the first IBV outbreak was reported in Thailand [7]. Although many IB vaccine strains such as Connecticut, H120, Ma5, M41 and Armidale A3, have been used in Thailand for many years, IBV outbreaks have been ongoing [1]. Moreover, the relationships between recent Thai IBV isolates and foreign IBV isolates are not known.
The objective of this study was to characterize IBV field isolates in the recent outbreak of the disease in Thailand by analyzing the S1 gene and compared them with those that have been published previously.
Materials and Methods
Viruses
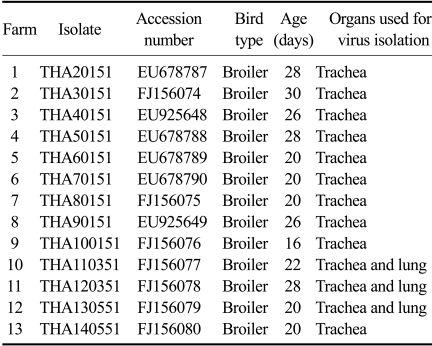
Between January and June, 2008, thirteen poultry farms in the eastern part of Thailand had an outbreak of a mild-to-moderate respiratory disease (Table 1). All flocks had been vaccinated against IB with commercial live attenuated H120. Chickens showed respiratory symptoms including gasping, coughing, sneezing, and tracheal rales. Sick chickens were selected and sent to the Department of Veterinary Medicine, Faculty of Veterinary Science, Chulalongkorn University. Necropsy was performed and gross lesions were evaluated. Gross lesions showed mild- to moderated tracheitis and non-purulent airsacculitis. No gross lesion were found in the kidneys. The trachea and lung samples were taken as pools of chickens from the same farm. The samples were prepared as 10% w/v suspensions in phosphate-buffered saline (pH 7.4) and clarified at 1,800 × g for 10 min; the supernatants were then collected for analysis.
Table 1.
Thai infectious bronchitis virus isolates examined in this study
RNA extraction
Viral RNA was extracted by using Viral Nucleic Acid Extraction Kit (Real Biotech, Taiwan) following the manufacturer's instructions directly from the supernatants of 10% w/v sample suspensions and from the allantoic fluid of embryonated chicken eggs used for virus isolation.
Virus screening with nested RT-PCR
Viral RNA, extracted directly from the supernatants of 10% w/v sample suspensions, was screened for the presence of IBV by using a nested RT-PCR. The reaction was performed with AccessQuick RT-PCR System (Promega, USA). The first amplification reaction was carried out with one-step RT-PCR using the primer sets of FOR1 (5'-CTT TTG TTT GCA CTA TGT AG-3') and RE3 (5'-TAA TAA CCA CTC TGA GCT GT-3'). The second amplification reaction was carried out using the primer sets of FOR2 (5'-CAG TGT TTG TCA CAC ATT GT-3') and RE2 (5'-CCA TCT GAA AAA TTG CCA GT-3'). Amplification products were analyzed in 1.5% agarose gel. The predicted size of nested RT-PCR product was about 400-bp.
Virus isolation and propagation
For virus isolation, the supernatants of IBV-positive samples determined by RT-PCR were inoculated into 10-day-old embryonated chicken eggs. For each sample to be examined, five embryonated chicken eggs were used. The eggs were inoculated with 0.2 mL of the sample into the allantoic cavity. The inoculated eggs were incubated at 37℃ and candled daily. Allantoic fluids were harvested at 96 h postinoculation. A further blind serial passage was performed in a similar way. All of the allantoic fluids were harvested and stored at -70℃.
RT-PCR amplification for sequencing
The allantoic fluids from the second passage of each sample positive for virus screening was submitted to another RT-PCR for amplification of a segment of 878-bp of the S1 gene coding region using a primer combination of FOR1 and RE3. The amplification reaction was carried out with one-step RT-PCR and the reaction conditions were the same as the first amplification of nested RT-PCR described above.
Product purification and sequencing
The RT-PCR products were cut from the gel and purified using the Wizard SV Gel and PCR Clean-Up system (Promega, USA) according to the manufacturer's protocol. Purified RT-PCR products were sequenced in a forward direction using primer FOR1 and in a reverse direction using primer RE3. Sequencing reactions were performed with the ABI Prism BigDye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems, USA) as described by the manufacturer. Sequencing reactions were run on an ABI Prism 310 Genetic Analyzer.
Sequences and phylogenetic analysis
To identify the Thai IBV isolates, sequences of the S1 gene of the Thai IBV isolates were compared with published IBV sequences deposited in the GenBank database using a BLAST search via the National Center of Biotechnology Information (USA). Sequence identities by BLAST analysis were included in alignment and phylogenetic construction. A phylogenetic tree of the nucleotide sequences was constructed using MEGA version 3.1 [13]. The S1 gene sequences of the thirteen IBV isolates were submitted to the GenBank database (Table 1). The other S1 gene sequences from the GenBank database were used for comparison or phylogenetic analysis in this study included M41 (AY561711), Ma5 (AY561713), H120 (M21970), IBN (AAW83034), W93 (AY427818), Connecticut 46 (L18990), Florida 18288 (AF027512), JMK (L14070), Spain/99/319 (DQ064810), Spain/00/337 (DQ064813), J2 (AF286303), BJQ (DQ070839), QXIBV (AF193423), LC2 (DQ480154), A2 (AY043312), SH (DQ480156), K069-01 (AY257061), 4/91 (AF093794), UK2/91 (Z83976), Ark DPI (AF006624), Australian T (AY775779), N1/62 (AIU29522), Armidale (DQ490205), GA/7994/99 (AF338717), GA/8077/99 (AF338718), DLD (EU589323), THA001 (DQ449628).
Results
Virus screening and isolation
For virus screening, pooled trachea and lung samples from each flocks suspected of IBV infection were determined to be positive for IBV by screening with nested RT-PCR. A 400-bp fragment of the S1 gene was amplified in all 13 samples tested (data not shown). The allantoic fluid from the second passage of each sample screened positive for the virus was also determined to be positive with RT-PCR amplification and a segment of 878-bp of the S1 gene was obtained (data not shown).
Phylogenetic analysis
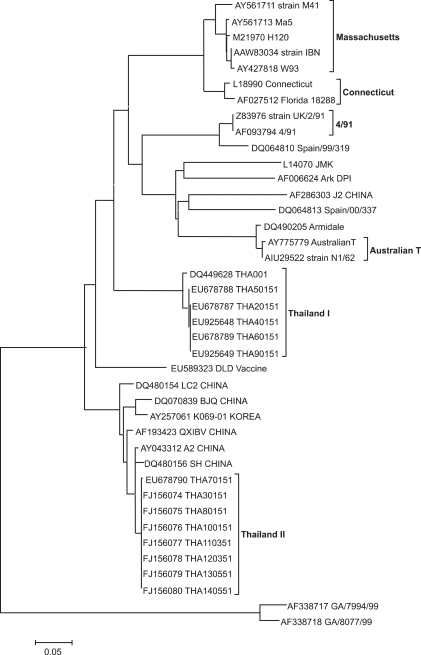
To assess the genetic relationship among the IBV isolates, a phylogenetic tree was constructed from the nucleotide sequences of S1 genes. The results are shown in Fig.1. The thirteen IBV isolates were separated into two distinct groups. Group I consisted of five isolates including THA20151, THA40151, THA50151, THA60151, and THA90151. The isolates in group I showed evolutionary distances from each other. Group II consisted of eight isolates including THA30151, THA 70151, THA 80151, THA100151, THA110351, THA120351, THA130551, and THA140551, which had a close relationship with Chinese IBV isolates (strain A2, SH and QXIBV).
Fig. 1.
Phylogenetic tree based on the nucleotide sequence between aligned S1 sequences from Thai infectious bronchitis virus isolates and published sequences.
Nucleotide and amino acid sequence comparison
The S1 gene of the thirteen IBV isolates was sequenced to characterize the isolates. The nucleotide and deduced amino acid sequences were determined and compared among each other and with other IBV strains published in the GenBank database. Group I Thai IBV isolates had nucleotide and amino acid sequence identities between 99~100% with each other. They had nucleotide sequence identities less than 85% and amino acid sequence identities less than 84% with IBVs published in the GenBank database. Group II Thai IBV isolates had nucleotide and amino acid sequence identities between 99~100% with each other. They had nucleotide sequence identities of 97~98% and amino acid sequence identities of 96~98% with Chinese IBVs (strain A2, SH and QXIBV).
Discussion
One of the major problems of IBV is the frequent emergence of new variants [21]. Different serotypes have been reported world wide and new variant serotypes continue to be recognized [9,18]. Thus, it is necessary and important to be able to diagnose these new serotypes. Furthermore, determining the type as well as field isolates is important to select an appropriate vaccine against IBV infection in the next flock. Therefore, several tests have been employed to identify the isolates into serotypes or genotypes [15]. Typing with RT-PCR and sequencing of the S1 gene is easier and faster than the more traditional virus neutralization methods, but it is difficult to design PCR primers that can be used for detect all of IBV isolates [16]. In this study, IBV isolates in group I could not be detected with the primer set and PCR method described by Gelb et al. [8]. To overcome this problem, the new primer sets were designed and the nested RT-PCR was developed in order to increase the sensitivity and specificity of the test used to detect IBV from the infected samples. Moreover, the region amplified by the outer primer covering the HVR of the S1 gene (nucleotide position 31~908) could be used for typing of IBV.
Genotyping of IBV on the basis of the S1 gene sequence, particularly the HVR region of the S1 gene, is the most common way to classify IBV isolates. It has been shown that the genetic typing based on HVR I (nucleotide position 114~325) of the S1 gene could represent the grouping method based on the whole S1 gene [20]. Furthermore, in a recent study using an approximately 450-bp region covering HVR I and HVR II for IBV typing, it was found that typing by this region correlates with virus neutralization results [17]. In the present study, an approximately 878-bp region of S1 gene covering HVR I and HVR II were amplified and used for typing the field isolates in Thailand.
Although many different strains of live attenuated and inactivated vaccines have been widely used to control IB, the outbreaks of the disease have continued to be a problem in Thailand [1]. In this study, thirteen IBV isolates from different commercial poultry farms in Thailand were analyzed by sequencing of the HVR in S1 gene. The molecular data indicated that the recent IBV isolated in Thailand evolved separately into two groups. The IBV isolates in group I were not related to other IBV strains published in the GenBank database. This genotype likely represents strains indigenous to Thailand. IBV isolates in group II were genetically related to Chinese strains.
The results from both S1 gene comparison and phylogenetic analysis showed that IBV isolates in group I had a distant relation to vaccine strains used in Thailand including Ma5, H120, M41, and Connecticut. New serotypes or variant strains can emerge as a result of only a few changes in the amino acid composition of S1 gene [5]. The rationale for these changes could be due to immunological pressure caused by the wide spread use of vaccines, recombination as a consequence of mixed infections, or the decrease of dominant serotypes as a result of vaccination, allowing other field strains to emerge [19].
Group II Thai isolates and Chinese strains shared more than 97% nucleotide identity and 96% amino acid identity; therefore, they were grouped into the same genotype. Interestingly, group II Thai isolates were closely related to Chinese QXIBV which appeared to become widespread in several countries in the world including the UK [11], Russia [3], and Italy [2]. Although no information is available currently for the introduction of QXIBV from China to other countries, it had been hypothesized that wild birds were the source of introduction based on the evidence that IBV may replicate in Anseriformes [3].
In summary, this finding indicated that the recent Thai IBVs evolved separately and at least two groups of viruses are cocirculating in Thailand. The distinctive dissimilarity between group I Thai isolates and the widely used IBV vaccine indicated that the antigenic drift is likely to occur among some Thai IBVs under the long-term immune pressure. Group II Thai isolates were closely related to Chinese strains, suggesting that the transfers of IBV infection occur among countries.
Acknowledgments
This work was financially supported by The Graduate School and the Faculty of Veterinary Science of Chulalongkorn University and Intervet Schering-Plough Animal Health Ltd., Bangkok, Thailand. We would like to thank all the staff of the Virology Unit, Department of Pathology, for their help throughout this work.
References
- 1.Antarasena C, Sahapong S, Aowcharoen B, Choe-ngern N, Kongkanunt R. Avian infectious bronchitis in the Southern part of Thailand. Songklanakarin J Sci Technol. 1990;12:273–279. [Google Scholar]
- 2.Beato MS, De Battisti C, Terregino C, Drago A, Capua I, Ortali G. Evidence of circulation of a Chinese strain of infectious bronchitis virus (QXIBV) in Italy. Vet Rec. 2005;156:720. doi: 10.1136/vr.156.22.720. [DOI] [PubMed] [Google Scholar]
- 3.Bochkov YA, Batchenko GV, Shcherbakova LO, Borisov AV, Drygin VV. Molecular epizootiology of avian infectious bronchitis in Russia. Avian Pathol. 2006;35:379–393. doi: 10.1080/03079450600921008. [DOI] [PubMed] [Google Scholar]
- 4.Boursnell MEG, Brown TDK, Foulds IJ, Green PF, Tomley FM, Binns MM. Completion of the sequence of the genome of the coronavirus avian infectious bronchitis virus. J Gen Virol. 1987;68:57–77. doi: 10.1099/0022-1317-68-1-57. [DOI] [PubMed] [Google Scholar]
- 5.Cavanagh D, Davis PJ, Cook JKA. Infectious bronchitis virus: evidence for recombination within the Massachusetts serotype. Avian Pathol. 1992;21:401–408. doi: 10.1080/03079459208418858. [DOI] [PubMed] [Google Scholar]
- 6.Cavanagh D, Davis PJ, Pappin DJ, Binns MM, Boursnell ME, Brown TD. Coronavirus IBV: partial amino terminal sequencing of spike polypeptide S2 identifies the sequence Arg-Arg-Phe-Arg-Arg at the cleavage site of the spike precursor propolypeptide of IBV strains Beaudette and M41. Virus Res. 1986;4:133–143. doi: 10.1016/0168-1702(86)90037-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chindavanig P. Studies on the attenuation of infectious bronchitis virus. J Thai Vet Med Assoc. 1962;12:1–7. [Google Scholar]
- 8.Gelb J, Jr, Weisman Y, Ladman BS, Meir R. S1 gene characteristics and efficacy of vaccination against infectious bronchitis virus field isolates from the United States and Israel (1996 to 2000) Avian Pathol. 2005;34:194–203. doi: 10.1080/03079450500096539. [DOI] [PubMed] [Google Scholar]
- 9.Gelb J, Jr, Wolff JB, Moran CA. Variant serotypes of infectious bronchitis virus isolated from commercial layer and broiler chickens. Avian Dis. 1991;35:82–87. [PubMed] [Google Scholar]
- 10.Gough RE, Cox WJ, de B, Welchman D, Worthington KJ, Jones RC. Chinese QX strain of infectious bronchitis virus isolated in the UK. Vet Rec. 2008;162:99–100. doi: 10.1136/vr.162.3.99. [DOI] [PubMed] [Google Scholar]
- 11.Gough RE, Randall CJ, Dagless M, Alexander DJ, Cox WJ, Pearson D. A 'new' strain of infectious bronchitis virus infecting domestic fowl in Great Britain. Vet Rec. 1992;130:493–494. doi: 10.1136/vr.130.22.493. [DOI] [PubMed] [Google Scholar]
- 12.Kingham BF, Keeler CL, Jr, Nix WA, Ladman BS, Gelb J., Jr Identification of avian infectious bronchitis virus by direct automated cycle sequencing of the S-1 gene. Avian Dis. 2000;44:325–335. [PubMed] [Google Scholar]
- 13.Kumar S, Tamura K, Nei M. MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- 14.Kusters JG, Niesters HGM, Lenstra JA, Horzinek MC, van der Zeijst BAM. Phylogeny of antigenic variants of avian coronavirus IBV. Virology. 1989;169:217–221. doi: 10.1016/0042-6822(89)90058-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee CW, Hilt DA, Jackwood MW. Redesign of primer and application of the reverse transcriptase-polymerase chain reaction and restriction fragment length polymorphism test to the DE072 strain of infectious bronchitis virus. Avian Dis. 2000;44:650–654. [PubMed] [Google Scholar]
- 16.Lee CW, Hilt DA, Jackwood MW. Identification and analysis of the Georgia 98 serotype, a new serotype of infectious bronchitis virus. Avian Dis. 2001;45:164–172. [PubMed] [Google Scholar]
- 17.Lee CW, Hilt DA, Jackwood MW. Typing of field isolates of infectious bronchitis virus based on the sequence of the hypervariable region in the S1 gene. J Vet Diagn Invest. 2003;15:344–348. doi: 10.1177/104063870301500407. [DOI] [PubMed] [Google Scholar]
- 18.Liu S, Kong X. A new genotype of nephropathogenic infectious bronchitis virus circulating in vaccinated and non-vaccinated flocks in China. Avian Pathol. 2004;33:321–327. doi: 10.1080/0307945042000220697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu SW, Zhang QX, Chen JD, Han ZX, Liu X, Feng L, Shao YH, Rong JG, Kong XG, Tong GZ. Genetic diversity of avian infectious bronchitis coronavirus strains isolated in China between 1995 and 2004. Arch Virol. 2006;151:1133–1148. doi: 10.1007/s00705-005-0695-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang CH, Huang YC. Relationship between serotypes and genotypes based on the hypervariable region of the S1 gene of infectious bronchitis virus. Arch Virol. 2000;145:291–300. doi: 10.1007/s007050050024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu L, Wang Z, Jiang Y, Low S, Kwang J. Molecular epidemiology of infectious bronchitis virus isolates from China and Southeast Asia. Avian Dis. 2001;45:201–209. [PubMed] [Google Scholar]
- 22.Ziegler AF, Ladman BS, Dunn PA, Schneider A, Davison S, Miller PG, Lu H, Weinstock D, Salem M, Eckroade RJ, Gelb J., Jr Nephropathogenic infectious bronchitis in Pennsylvania chickens 1997-2000. Avian Dis. 2002;46:847–858. doi: 10.1637/0005-2086(2002)046[0847:NIBIPC]2.0.CO;2. [DOI] [PubMed] [Google Scholar]