Abstract
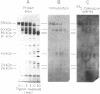
A cDNA that encodes what appears to be the inhibitory domain of the plasma membrane calcium-pumping ATPase (Ca2+-ATPase) has been isolated by screening a lambda gt11 bovine brain cDNA library with antibodies prepared against the human erythrocyte membrane Ca2+-ATPase. This screening resulted in isolation of a bacteriophage containing a 1.5-kilobase cDNA insert encoding a 71-residue polypeptide, the remainder being a large 3' terminal noncoding region. A portion of this deduced peptide sequence was identical to that of a peptide isolated from a V8 protease digest of the human erythrocyte Ca2+-ATPase except for 1 residue. Antibodies purified by immunoabsorption to the fusion protein containing this cDNA-encoded polypeptide reacted only with those fragments of a limited trypsin digest of the human erythrocyte Ca2+-ATPase that contain the inhibitory domain. Moreover, these antibodies were able to partially stimulate basal enzyme activity and block further activation by calmodulin. The encoded polypeptide bears homology to the glutamic acid-rich regions N-terminal to the Ca2+-binding loops of calmodulin and to a lesser extent with the loops themselves. This encoded polypeptide also represents the C terminus of the Ca2+-ATPase. Portions of the isolated cDNA were homologous to the 3' noncoding region of the sarcoplasmic reticulum Ca2+-ATPase cDNA, indicating a possible mechanism for the evolution of these distinct membrane Ca2+ pumps.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Babu Y. S., Sack J. S., Greenhough T. J., Bugg C. E., Means A. R., Cook W. J. Three-dimensional structure of calmodulin. Nature. 1985 May 2;315(6014):37–40. doi: 10.1038/315037a0. [DOI] [PubMed] [Google Scholar]
- Benaim G., Zurini M., Carafoli E. Different conformational states of the purified Ca2+-ATPase of the erythrocyte plasma membrane revealed by controlled trypsin proteolysis. J Biol Chem. 1984 Jul 10;259(13):8471–8477. [PubMed] [Google Scholar]
- Biggin M. D., Gibson T. J., Hong G. F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenthal D. K., Takio K., Edelman A. M., Charbonneau H., Titani K., Walsh K. A., Krebs E. G. Identification of the calmodulin-binding domain of skeletal muscle myosin light chain kinase. Proc Natl Acad Sci U S A. 1985 May;82(10):3187–3191. doi: 10.1073/pnas.82.10.3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandl C. J., Green N. M., Korczak B., MacLennan D. H. Two Ca2+ ATPase genes: homologies and mechanistic implications of deduced amino acid sequences. Cell. 1986 Feb 28;44(4):597–607. doi: 10.1016/0092-8674(86)90269-2. [DOI] [PubMed] [Google Scholar]
- Craig T. A., Watterson D. M., Prendergast F. G., Haiech J., Roberts D. M. Site-specific mutagenesis of the alpha-helices of calmodulin. Effects of altering a charge cluster in the helix that links the two halves of calmodulin. J Biol Chem. 1987 Mar 5;262(7):3278–3284. [PubMed] [Google Scholar]
- Edelman A. M., Takio K., Blumenthal D. K., Hansen R. S., Walsh K. A., Titani K., Krebs E. G. Characterization of the calmodulin-binding and catalytic domains in skeletal muscle myosin light chain kinase. J Biol Chem. 1985 Sep 15;260(20):11275–11285. [PubMed] [Google Scholar]
- Garnier J., Osguthorpe D. J., Robson B. Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. J Mol Biol. 1978 Mar 25;120(1):97–120. doi: 10.1016/0022-2836(78)90297-8. [DOI] [PubMed] [Google Scholar]
- Hanley R. M., Means A. R., Ono T., Kemp B. E., Burgin K. E., Waxham N., Kelly P. T. Functional analysis of a complementary DNA for the 50-kilodalton subunit of calmodulin kinase II. Science. 1987 Jul 17;237(4812):293–297. doi: 10.1126/science.3037704. [DOI] [PubMed] [Google Scholar]
- Hiremath L. S., Webb N. R., Rhoads R. E. Immunological detection of the messenger RNA cap-binding protein. J Biol Chem. 1985 Jul 5;260(13):7843–7849. [PubMed] [Google Scholar]
- Jamieson G. A., Jr, Vanaman T. C. Calcium-dependent affinity chromatography of calmodulin on an immobilized phenothiazine. Biochem Biophys Res Commun. 1979 Oct 12;90(3):1048–1056. doi: 10.1016/0006-291x(79)91932-6. [DOI] [PubMed] [Google Scholar]
- Kemp B. E., Pearson R. B., Guerriero V., Jr, Bagchi I. C., Means A. R. The calmodulin binding domain of chicken smooth muscle myosin light chain kinase contains a pseudosubstrate sequence. J Biol Chem. 1987 Feb 25;262(6):2542–2548. [PubMed] [Google Scholar]
- Klee C. B., Vanaman T. C. Calmodulin. Adv Protein Chem. 1982;35:213–321. doi: 10.1016/s0065-3233(08)60470-2. [DOI] [PubMed] [Google Scholar]
- Klevit R. E., Blumenthal D. K., Wemmer D. E., Krebs E. G. Interaction of calmodulin and a calmodulin-binding peptide from myosin light chain kinase: major spectral changes in both occur as the result of complex formation. Biochemistry. 1985 Dec 31;24(27):8152–8157. doi: 10.1021/bi00348a047. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- MacLennan D. H., Brandl C. J., Korczak B., Green N. M. Amino-acid sequence of a Ca2+ + Mg2+-dependent ATPase from rabbit muscle sarcoplasmic reticulum, deduced from its complementary DNA sequence. Nature. 1985 Aug 22;316(6030):696–700. doi: 10.1038/316696a0. [DOI] [PubMed] [Google Scholar]
- Mann D., Vanaman T. C. Specific chemical modification as a probe of calmodulin function. Methods Enzymol. 1987;139:417–433. doi: 10.1016/0076-6879(87)39103-7. [DOI] [PubMed] [Google Scholar]
- Neve R. L., Harris P., Kosik K. S., Kurnit D. M., Donlon T. A. Identification of cDNA clones for the human microtubule-associated protein tau and chromosomal localization of the genes for tau and microtubule-associated protein 2. Brain Res. 1986 Dec;387(3):271–280. doi: 10.1016/0169-328x(86)90033-1. [DOI] [PubMed] [Google Scholar]
- Ni W. C., Klee C. B. Selective affinity chromatography with calmodulin fragments coupled to sepharose. J Biol Chem. 1985 Jun 10;260(11):6974–6981. [PubMed] [Google Scholar]
- Niggli V., Penniston J. T., Carafoli E. Purification of the (Ca2+-Mg2+)-ATPase from human erythrocyte membranes using a calmodulin affinity column. J Biol Chem. 1979 Oct 25;254(20):9955–9958. [PubMed] [Google Scholar]
- Sarkadi B., Enyedi A., Földes-Papp Z., Gárdos G. Molecular characterization of the in situ red cell membrane calcium pump by limited proteolysis. J Biol Chem. 1986 Jul 15;261(20):9552–9557. [PubMed] [Google Scholar]
- Slaughter G. R., Means A. R. Use of the 125I-labeled protein gel overlay technique to study calmodulin-binding proteins. Methods Enzymol. 1987;139:433–444. doi: 10.1016/0076-6879(87)39104-9. [DOI] [PubMed] [Google Scholar]
- Smith D. E., Fisher P. A. Identification, developmental regulation, and response to heat shock of two antigenically related forms of a major nuclear envelope protein in Drosophila embryos: application of an improved method for affinity purification of antibodies using polypeptides immobilized on nitrocellulose blots. J Cell Biol. 1984 Jul;99(1 Pt 1):20–28. doi: 10.1083/jcb.99.1.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szyja W., Wrzosek A., Brzeska H., Sarzała M. G. Interaction of calmodulin and its fragments with Ca2+-ATPase and myosin light chain kinase. Cell Calcium. 1986 Apr;7(2):73–88. doi: 10.1016/0143-4160(86)90010-2. [DOI] [PubMed] [Google Scholar]
- Takio K., Blumenthal D. K., Edelman A. M., Walsh K. A., Krebs E. G., Titani K. Amino acid sequence of an active fragment of rabbit skeletal muscle myosin light chain kinase. Biochemistry. 1985 Oct 22;24(22):6028–6037. doi: 10.1021/bi00343a002. [DOI] [PubMed] [Google Scholar]
- Verma A. K., Gorski J. P., Penniston J. T. Antibodies directed toward human erythrocyte Ca2+-ATPase: effect on enzyme function and immunoreactivity of Ca2+-ATPases from other sources. Arch Biochem Biophys. 1982 May;215(2):345–354. doi: 10.1016/0003-9861(82)90095-9. [DOI] [PubMed] [Google Scholar]
- Watterson D. M., Sharief F., Vanaman T. C. The complete amino acid sequence of the Ca2+-dependent modulator protein (calmodulin) of bovine brain. J Biol Chem. 1980 Feb 10;255(3):962–975. [PubMed] [Google Scholar]
- Watterson D. M., Vanaman T. C. Affinity chromatography purification of a cyclic nucleotide phosphodiesterase using immobilized modulator protein, a troponin C-like protein from brain. Biochem Biophys Res Commun. 1976 Nov 8;73(1):40–46. doi: 10.1016/0006-291x(76)90494-0. [DOI] [PubMed] [Google Scholar]
- Wray W., Boulikas T., Wray V. P., Hancock R. Silver staining of proteins in polyacrylamide gels. Anal Biochem. 1981 Nov 15;118(1):197–203. doi: 10.1016/0003-2697(81)90179-2. [DOI] [PubMed] [Google Scholar]
- Zurini M., Krebs J., Penniston J. T., Carafoli E. Controlled proteolysis of the purified Ca2+-ATPase of the erythrocyte membrane. A correlation between the structure and the function of the enzyme. J Biol Chem. 1984 Jan 10;259(1):618–627. [PubMed] [Google Scholar]